Abstract
The tf (tufted) locus is responsible for a classic phenotype of hair loss and regrowth in mice. It is a characteristic of the BTBR strain. Here, we use a combination of positional cloning methods and complementation mapping to identify Itpr3, the inositol triphosphate receptor type 3, as the gene responsible for the tf locus.
Key words: BTBR mouse, hair, inosine triphosphate receptor type 3
When a tufted mouse is 4 weeks old, lines of hair loss begin to spread in waves from its snout to its tail. The first wave takes about a week, the second about 18 days, and subsequent waves take a month or longer (Figure 1). The tufted (tf) locus was recognized as a spontaneous mutation in the year 1956 (Lyon, 1956) and early work located it near the kinky/fused (Fu) and brachyury (T) loci (Dobrovolskaia-Zavadskaia, 1927), which are components of the “t complex” on chromosome 17. This intricate region has been studied extensively (Russell, 1985; Silver, 1985) and for many years before the advent of polymerase-chain-reaction-based genotyping, the tufted pattern of hair loss was used as a visible marker to identify carriers of the lethal TT locus (Silver, 1985). However, the identity of tf was unknown.
Figure 1.

A mature adult BTBR T+ tf/J mouse showing the characteristic waves of hair loss, which start from the dorsal surface of the head and migrate ventrally.
The BTBR mouse is a common laboratory strain carrying the tf locus. The BTBR strain was originated by Lyon, who crossed mice carrying the tf mutation with stock, maintained by Dobrovolskaia-Zavadskaia, that carried the T locus (Dobrovolskaia-Zavadskaia, 1928). Festing (1968) reported that the strain was then developed successively by Bennett and Artzt, but Artzt (personal communication) and Wahlsten et al. (2003) reported it was developed by Dunn, who originally selected it for large litter size, may have crossed it with a 129 mouse strain, and transferred it to The Jackson Laboratory around the year 1969. The strain has been maintained at The Jackson Laboratory for more than 40 generations by sibling mating (see also Clee et al., 2005).
The BTBR strain is of interest for studies of taste perception because it appears to be unable to detect tastes mediated by G-protein coupled receptors (Ellis et al., 2011). To positionally clone the underlying genes, we produced BTBR T+ tf/J × NZW/LacJ F2 hybrids, phenotyped them with a series of taste preference tests, and conducted a genome scan. The preference for saccharin and other taste compounds was strongly linked to chromosome 17 (Ellis et al., 2011). To isolate this linkage, we generated a congenic line using marker-assisted selection to repeatedly backcross BTBR·NZW mice to the BTBR strain. After 11 backcrosses, the congenic interval supporting the phenotypes involved a 0.8-Mb region on chromosome 17 containing 21 known and predicted genes (Tordoff et al., 2012).
We noticed that the tufted hair pattern was present in mice that inherited the BTBR·BTBR haplotype but not the BTBR·NZW haplotype in the congenic interval, implying that one of the 21 genes was responsible for the tf phenotype.
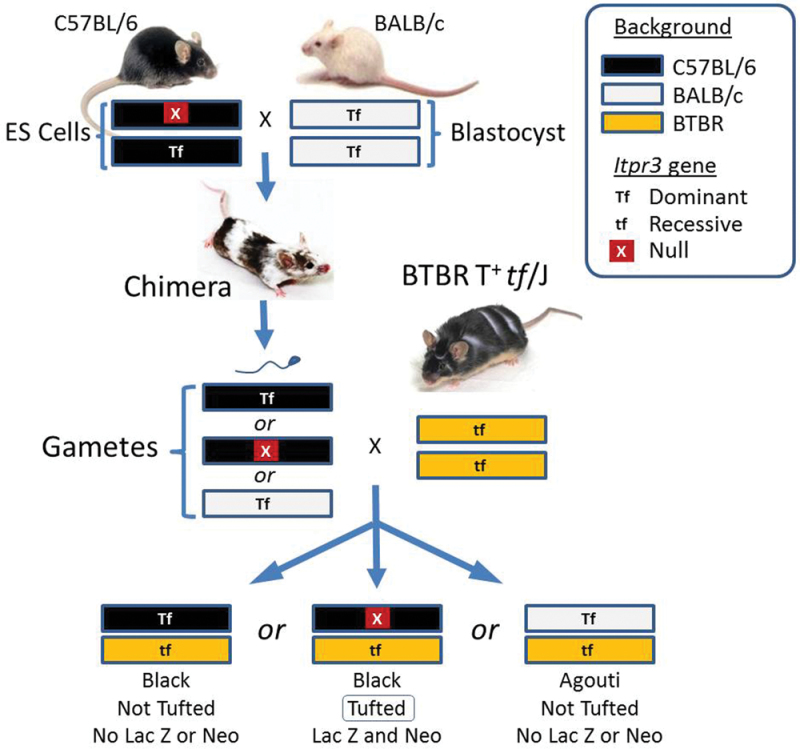
The strongest candidate gene to mediate taste deficits was Itpr3, the inositol triphosphate receptor type 3, which is a component of the taste transduction cascade (Nakashima and Ninomiya, 1999; Hisatsune et al., 2007). To evaluate the contribution of Itpr3, we produced Itpr3-knockout mice (Figure 2). We purchased C57BL/6 embryonic stem (ES) cell clones from the North American Conditional Mouse Mutagenesis project (NorCOMM; ES cell line MFGC N01293P1_W239C5). The clones involve deletion of 300bp spanning Exon 2 of Itpr3 (Chr 17: 27194249-272194548) and included both Lac and Neo as part of the construct (see http://www.knockoutmouse.org/martsearch/project/ 42456). They were verified, isolated, and then injected into BALB/c blastocysts by the Gene Targeting Service and Transgenic and Chimeric Mouse Facility at the University of Pennsylvania. The first batch of clones resulted in 12 male and 6 female offspring, which had a chimeric range between 10% and 95% (judged by coat color; white = BALB/c; black = C57BL/6).
Figure 2.
The BTBR form of Itpr3 is responsible for the tufted (tf) phenotype. Using standard technologies, a construct conferring a knockout of Itpr3 on a C57BL/6 background was inserted into embryonic stem (ES) cells, incorporated by homologous recombination. The ES cells were injected into BALB/c blastocysts, producing chimeric mice. These were mated with inbred BTBR mice. The chimeric mice could have gametes of three haplotypes and so there were three types of offspring, each heterozygous and each containing one BTBR allele, which carries tf as a recessive locus. Only the mice with a nonfunctional copy of Itpr3 displayed the tufted phenotype.
To expose the gene, we used complementation mapping (Mackay, 2004) in which the action of the recessive tf locus was visible only when unmasked by elimination of the dominant locus of the candidate gene. Mice that inherited wild-type BALB/c or C57BL/6J alleles did not display the tufted phenotype, which is to be expected because each contains a dominant Tf allele that masks the action of the tf allele inherited from the BTBR strain. However, mice with the BTBR background and a heterozygous Itpr3 null allele had the tufted phenotype (Figure 2). This unmasking of the phenotype confirms that the BTBR form of Itpr3 is responsible for tf, the tufted locus. Mice with the homozygous Itpr3 null alleles on either the BTBR or C57BL/6J backgrounds were also tufted, showing that the BTBR form of Itpr3 is nonfunctional.
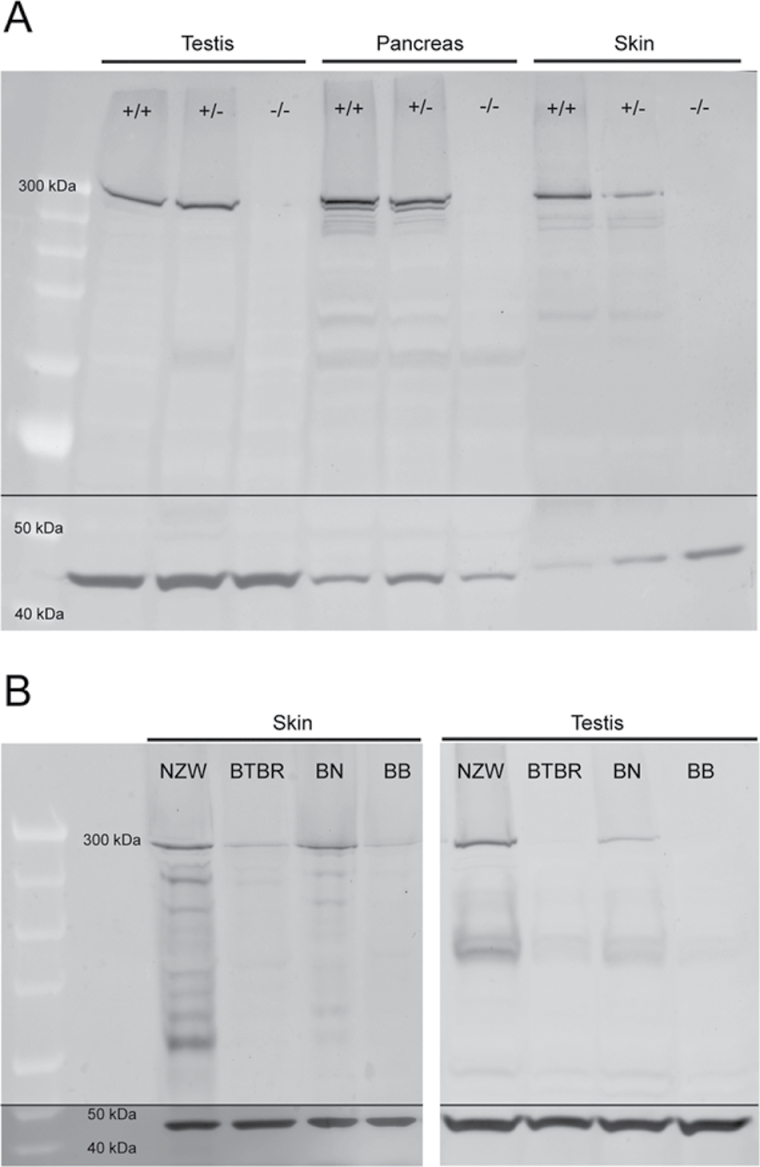
Sequencing the BTBR form of this gene revealed a 12-bp deletion in Exon 23 (Chr 17: 27238069; National Center for Biotechnology Information Build 38.1), which codes for amino acids 983–986. This is probably unique to the BTBR strain; it is not present in the C57BL/6NJ (reference sequence), C57BL6/J (our work), NZW/LacJ (our work), or any of the 17 strains sequenced by the Sanger Mouse Genomes Project (Wellcome Trust Sanger Institute, 2012). The deletion apparently causes almost complete loss of the ITPR3 protein in skin and other tissues (Figure 3; Supplementary Material online). How the loss of Itpr3 interferes with hair growth is less clear. Recent work suggests that a defect in the nuclear translocation of NFATc1 in the K6-positive bulge cells of telogen follicles may cause loose anchorage of club hairs in the early telogen–anagen transition of the hair cycle (Sato-Miyaoka et al., 2012). Whatever the mechanism, our work to investigate taste perception solves a 56-year-old mystery. The BTBR mouse has a defective Itpr3 gene to blame for its tufted hair—as well as its dysfunctional taste perception.
Figure 3.
Western blots showing expression of ITPR3 protein in mice of various genotypes: (A). ITPR3 protein is abundant in C57BL6 mice that are wild-type (+/+) or heterozygous (+/-) knockouts at Itpr3 but the protein is absent in homozygous (-/-) Itpr3 knockouts. (B). ITPR3 protein is abundant in inbred NZW mice and mice with the BTBR/NZW (BN) haplotype in the congenic interval of chromosome 17, which includes Itpr3, but the protein is absent or minimal in inbred BTBR mice and mice with the BTBR/BTBR (BB) haplotype in the congenic interval of chromosome 17. Mice with abundant ITPR3 have normal hair; those with absent or minimal ITPR3 have tufted hair. Bottom of each panel shows that all lanes were positive for the control protein, β-actin.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/
Funding
National Institute and Diabetes and Digestive and Kidney Diseases at the National Institutes of Health (grant number DK46791). Resources for genotyping: Monell Genotyping Core, which is supported by the National Institute on Deafness and Other Communication Disorders (Core Grant P30 DC011735). Production of transgenic mice: University of Pennsylvania Transgenic & Chimeric Mouse Facility and the Institute for Diabetes, Obesity, and Cardiovascular Metabolism (Grant DK019525); the Center for Molecular Studies in Digestive and Liver Diseases (Grant DK050306); and the Abramson Cancer Center (Grant CA016520). National Institute on Deafness and Other Communication Disorders (Grants T32 DC000014 and RO1 DC003055) to M.R.P.
Supplementary Material
References
- Clee SM, Nadler ST, Attie AD. 2005. Genetic and genomic studies of the BTBR ob/ob mouse model of type 2 diabetes. Am J Ther. 12: 491–498 [DOI] [PubMed] [Google Scholar]
- Dobrovolskaia-Zavadskaia NCR. 1927. Sur la mortification spontanée de la que chez la souris nouveau-née sur l’existence d’un caractere (facteur) héréditaire, non viable. Compt Rend Seances Soc Biol. 97: 114–116 [Google Scholar]
- Dobrovolskaia-Zavadskaia N. 1928. L’irradiation des testicules et l’heredite chez la souris. Arch Biol (Liege). 38: 457–501 [Google Scholar]
- Ellis HT, Shao H, Reed DR, Tordoff MG. 2011. Individual differences in the avidity for calcium and saccharin are influenced by variation in Itpr3 or in a nearby gene on mouse chromosome 17 [abstract]. Chem Senses. 31: A22 [Google Scholar]
- Festing MFW. Inbred strains of mice: BTBRF. MGI [cited 1998. April 9] Available from: http://www.informatics.jax.org/external/festing/mouse/docs/BTBRTF.shtml
- Hisatsune C, Yasumatsu K, Takahashi-Iwanaga H, Ogawa N, Kuroda Y, Yoshida R, Ninomiya Y, Mikoshiba K. 2007. Abnormal taste perception in mice lacking the type 3 inositol 1,4,5-trisphosphate receptor. J Biol Chem. 282: 37225–37231 [DOI] [PubMed] [Google Scholar]
- Lyon MF. 1956. Hereditary hair loss in the tufted mutant of the house mouse. J Hered. 47: 101–103 [Google Scholar]
- Mackay TF. 2004. Complementing complexity. Nat Genet. 36: 1145–1147 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Ninomiya Y. 1999. Transduction for sweet taste of saccharin may involve both inositol 1,4,5-trisphosphate and cAMP pathways in the fungiform taste buds in C57BL mice. Cell Physiol Biochem. 9: 90–98 [DOI] [PubMed] [Google Scholar]
- Russell ES. 1985. A history of mouse genetics. Annu Rev Genet. 19: 1–28 [DOI] [PubMed] [Google Scholar]
- Sato-Miyaoka M, Hisatsune C, Ebisui E, Ogawa N, Takahashi-Iwanaga H, Mikoshiba K. 2012. Regulation of hair shedding by the Type 3 IP3 receptor. J Invest Dermatol. 132: 2137–2147 [DOI] [PubMed] [Google Scholar]
- Silver LM. 1985. Mouse t haplotypes. Annu Rev Genet. 19: 179–208 [DOI] [PubMed] [Google Scholar]
- Tordoff MG, Jaji SA, Marks JM, Ellis HT. 2012. Macronutrient choice of BTBR.NZW mice congenic for a 21-gene region of chromosome 17. Physiol Behav. 106: 556–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlsten D, Metten P, Crabbe JC. 2003. Survey of 21 inbred mouse strains in two laboratories reveals that BTBR T/+ tf/tf has severely reduced hippocampal commissure and absent corpus callosum. Brain Res. 971: 47–54 [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Sanger Institute Mouse genomes project. [cited 2012. September 10] Available from: http://www.sanger.ac.uk/resources/mouse/genomes/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.