Abstract
We previously identified rat8 in the pathway involved in epithelial cell differentiation that occurs in the rat mammary gland at pregnancy when tubules and alveoli are formed. rat8, which encodes an IFN-inducible membrane protein, is the rat homologue of the mouse gene fragilis. By differential detergent extraction and isopycnic sucrose density gradients, we show that rat8 protein is associated to lipid membrane domains together with Lyn and Fyn, members of the Src tyrosine kinase family. We also show that recruitment of rat8 to lipid membrane domains is a necessary step in mammary epithelial cell differentiation. Immunoprecipitation analysis, performed with an anti-Fyn protein antibody, shows that rat8 was present in the Fyn immunoprecipitate. Antisense oligonucleotides, used to inhibit Fyn protein expression, block mammary cell differentiation. Taken together, these results suggest that the functional interaction, via lipid membrane domains, of rat8 and Fyn proteins is required for mammary cell differentiation. Therefore, rat8, like fragilis, may be involved in developmental decisions and the demarcation of a subset of cells in the mammary gland that cause epithelial cells to develop into a network of tubuloalveolar structures involved in secretion.
Mammary gland development is a process in which cell–cell interaction and adhesion play an important role (1, 2). The establishment of contacts among cells is a crucial prerequisite for the activation of the signaling cascade driving the subsequent differentiation that is accompanied by dramatic changes in morphology and protein expression (3). Development of the mammary gland occurs in several stages. A rudimentary system of small ducts is present in newborn mice, and, until the pubertal phase, ductal growth is moderate, and very few alveoli are formed (4). Full alveolar development occurs during pregnancy when alveoli differentiate into secretory lobules, whereas terminal differentiation of alveolar epithelial cells is completed at parturition with the production and secretion of milk (5). The end of weaning suppresses lactation, leading to the involution of the mammary gland to the nonpregnant state. The secretory alveolar cells therefore represent the final cellular state of the differentiation process within the mammary gland (6). Pregnancy and lactation steps are defined and characterized by distinct cellular and molecular changes and by activation of genes coding for the milk proteins (7). Because cellular composition varies with the functional development of the mammary gland, to study how cell adhesion and cell–cell interactions are integrated and translated into molecular signals that guide growth and differentiation is difficult in the animal in vivo. This difficulty is due to the impossibility of obtaining uncontaminated cellular samples at definite stages of differentiation.
To overcome this problem, we developed an experimental model that allowed us to study mammary gland differentiation in vitro (8). This model system is based on two cellular clones, LA7 and 106, both obtained from the parental clone Rama-25, considered to contain stem cells of the rat mammary gland (9–13). Exposure to lactogenic hormones, lipids, or differentiating agents such as cAMP or DMSO results in the formation of hemispherical polarized structures called “domes” in LA7, but not in 106, cells (14, 15). We previously showed that epithelial differentiation leading to dome formation in LA7 cells represents changes that occur in vivo in the mammary gland at pregnancy when tubules and alveoli are formed (14). The identification of genes involved in the process of dome formation has been the focus of our previous studies, carried out by using a differential expression detection approach (cDNA library subtraction) and proteomics-based technologies (8, 14, 16–18). This work has shown that the differentiation process resulting in the formation of domes requires the expression of the rat8 gene and the β-subunit of the amiloride-sensitive epithelial sodium channel (ENaC) and is negatively regulated by genes coding YMP (Y membrane protein) and maspin (8, 14, 16, 17). rat8 is the key gene regulating dome formation; rat8 is constitutively expressed in LA7, but not in 106, cells and moderately induced by DMSO treatment in both cell lines (8). Antisense oligonucleotides designed on the rat8 mRNA sequence, when added to the LA7 culture in the presence of DMSO, by blocking rat8 protein synthesis not only prevent dome formation but also cause the disappearance of existing domes and induce reversible morphological changes in the LA7 cells. These morphological changes probably reflect a redirection of the epithelial differentiation program (8).
rat8 was first identified in aortic smooth muscle cells (19). As shown by in situ hybridization and Northern analysis, this gene is expressed in many adult tissue types including ovary, liver, heart, and kidney; its expression was lowest in brain and testis (15). rat8 encodes a 14-kDa transmembrane protein and is homologous to the mouse gene fragilis, an IFN-inducible gene encoding a transmembrane protein that has been implicated recently in the acquisition of germ cell competence and specification (20, 21). It has been suggested that fragilis is involved in developmental decisions that result in the demarcation of a subset of cells during dynamic morphogenetic movements in the embryo (20, 21). Three human genes, 9-27 (Ifitm1), 1-8U (Ifitm2), and 1-8D (Ifitm3), share 58–65% homology with the rat8 gene (22). Gene 9-27 encodes the membrane protein Leu-13, which forms a multimeric complex involved in the regulation of cell aggregation and in the transduction of anti-proliferative and homotypic adhesion signals at the cell surface of human B lymphocytes (23–24).
Here we investigate the localization of rat8 protein in LA7 cells by immunohistochemistry and show that rat8 protein location depends on differentiation stage. rat8 protein is intracytoplasmic in undifferentiated LA7 cells and is relocated to the cell membrane on differentiation. Within the cell membrane, rat8 is associated with lipid membrane domains. Lipid membrane domains (also known as lipid rafts) are organized domains of the plasma membrane and other intracellular membranes and can be viewed as signaling platforms that serve to colocalize molecular components, facilitating their interaction and supporting signaling (25–28). Several raft-associated proteins are anchored to the cytosolic surface of the raft plasma membrane by acylation with myristate and palmitate and isoprenylation with farnesyl or geranylgeranyl moieties (29–33). Because myristic and palmitic acids induce dome formation in LA7 cells (15), we investigated the possible association of rat8 to lipid membrane domains. We found that rat8 coexists in lipid rafts with Fyn, a member of the Src family of nonreceptor tyrosine kinases. Moreover, coimmunoprecipitation experiments, performed with an anti-Fyn polyclonal antibody and probed with anti-rat8 antibody, revealed that rat8 protein was present in the Fyn immunoprecipitate and that Fyn antisense oligonucleotides, when added to the LA7 culture, block dome formation in DMSO-induced cells. Lyn, another member of the Src family, also was present in lipid domains from DMSO-induced LA7 cells, but Lyn antisense oligonucleotides had no effect on blocking dome formation in DMSO-induced LA7 cells. These results show that the incorporation of rat8 and Fyn proteins in lipid rafts plays an important role in rat mammary gland differentiation in vitro.
Materials and Methods
Chemicals. Commercial chemicals used were the purest available, common solvents were distilled before use, and water was doubly distilled in a glass apparatus. Sodium boro[3H]hydride was from Amersham Biosciences (specific radioactivity of 12.0 Ci/mmol; 1 Ci = 37 GBq).
Cells, Media, and Differentiation Inducers. The cell lines LA7 and 106, both clonal derivatives from the Rama-25 line (11), were cultured as previously described (9). After growing to confluence, 106 and LA7 cells were exposed to 1.5% DMSO, used as an inducer of cell differentiation.
Treatments of Cell Cultures with [1-3H]Sphingosine. Cells were incubated in the presence of 3 × 10–8 M [1-3H]sphingosine (5 ml per dish for 100-mm dishes) in culture media for a 2-h pulse followed by a 24-h chase. Under these conditions, free radioactive sphingosine was hardly detectable in the cells, and all sphingolipids were metabolically radiolabeled (34–36).
Sucrose Gradient Centrifugation. After metabolic radiolabeling with [1-3H]sphingosine, cells (10 × 100-mm dishes) were subjected to ultracentrifugation on discontinuous sucrose gradients as previously described (34–36). Briefly, cells were harvested, lysed in lysis buffer (1% Triton X-100/10 mM Tris buffer, pH 7.5/150 mM NaCl/5 mM EDTA/1mMNa3VO4/1 mM PMSF/2 μg/ml aprotinin; 5–8 × 107 cells per ml) and homogenized (with a Dounce homogenizer, 10 strokes, tight). Cell lysate was centrifuged (1,300 × g for 5 min) to remove nuclei and cellular debris. The postnuclear fraction was mixed with an equal volume of 85% sucrose (wt/vol) in 10 mM Tris buffer, pH 7.5/150 mM NaCl/5 mM EDTA/1 mM Na3VO4, placed at the bottom of a discontinuous sucrose concentration gradient (30–5%) in the same buffer, and centrifuged (200,000 × g for 17 h) at 4°C. After ultracentrifugation, 11 fractions were collected starting from the top of the tube. The light-scattering band located at the interface between 5% and 30% sucrose and corresponding to fraction 5 was regarded as the sphingolipid- and cholesterol-enriched membrane fraction (34). The entire procedure was performed at 0–4°C in ice immersion.
Other Analytical Methods. Radioactive lipids were detected and quantified by radioactivity imaging performed with a Beta Imager 2000 (Biospace, Paris), with an acquisition time of ≈48 h. The radioactivity associated with individual lipids was determined by using β-vision software (Biospace). The radioactivity associated with cells, cell fractions, lipids, and lipid extracts was determined by liquid scintillation counting. The protein content was determined according to the methods of Lowry et al. (37), by using BSA as the reference standard.
Immunofluorescence Microscopy. Detection of proteins was performed on cells grown for 3 days on Permanox chamber slides with cover slips (Nunc) as described (8). Cells were induced with 1.5% DMSO, fixed in 4% paraformaldehyde/PBS for 10 min, and then incubated with a commercial monoclonal antibody for α6β1-integrin (Serotec) or with an anti-rat8 polyclonal antibody (Sigma-Genosys), according to the manufacturer's protocol. Detection of Golgi bodies was performed by using a monoclonal antibody against the GM130 (Golgi membrane, 130 kDa) protein (BD Transduction/BD Pharmingen). The secondary antibodies used were rhodamine-labeled anti-mouse IgG (Vector Laboratories) for detection of the α6β1-integrin, FITC-labeled anti-rabbit IgG (Vector Laboratories) for detection of rat8 protein, and Alexa Fluor 5680-conjugated anti-mouse IgG (Molecular Probes) for detection of the GM130 protein. Cells were microscopically examined and photographed by using ×10, ×40, and ×60 objectives.
Antisense Oligonucleotide Methodology. For the inhibition study of Fyn protein expression, three antisense oligonucleotides, each containing 21 bases, were synthesized from the rat sequence NM012755 of Fyn mRNA: AS-FYN-1 (5′-CGTCAGTTTCGCTGCTTCTTT-3′, designed between nucleotides 275 and 255), AS-FYN-2 (5′-CGTTGTCAAGCTTGCGGATTT-3′, designed between nucleotides 861 and 841), and AS-FYN-3 (5′-TCCAGGTACCCATCCATACTT-3′, designed between nucleotides 1101 and 1081). Three sense oligomers, complementary to the three antisense oligomers, also were used.
For the inhibition study of Lyn protein expression, three antisense oligonucleotides, each containing 21 bases, were synthesized from the rat sequence L14823 of Lyn mRNA: AsLYNB-1 (5′-GCTCCTCTGGATCTTTTGCTT-3′, designed between nucleotides 151 and 131), AsLYNB-2 (5′-CTGACGGAAAGAGAGAAGCTT-3′, designed between nucleotides 470 and 450), and AsLYNB-3 (5′-ATCGCGGTGGATGTAGTTCTT-3′, designed between nucleotides 1062 and 1042). Three sense oligomers, complementary to the three antisense oligomers, also were used. The experiments were performed as previously described (8). Cells were maintained in culture for 60–72 h, inspected for dome formation, photographed, and harvested for RNA extraction or paraformaldehyde-fixed for further immunohistochemical analysis.
Analysis of Protein Patterns. Sucrose gradient fractions obtained after labeling untreated and DMSO-induced LA7 and 106 cells were analyzed by SDS/PAGE. After separation, proteins were transferred to poly(vinylidene difluoride) (PVDF) membranes (BioRad). The presence of rat8, Lyn, and Fyn proteins was assessed by Western blot analysis using an anti-rat8 polyclonal antibody (Sigma-Genosys) and anti-Fyn and anti-Lyn polyclonal antibodies (Santa Cruz Biotechnology), followed by reaction with a secondary anti-rabbit IgG horseradish peroxidase-conjugated antibody (Santa Cruz Biotechnology) and enhanced chemiluminescence detection (ECL PLUS, Amersham Pharmacia).
Immunoprecipitation Experiments. DMSO-induced LA7 and 106 cells were lysed in radioimmunoprecipitation assay (RIPA) buffer by following procedures described previously (38). Incubation of samples with anti-Fyn polyclonal antibody (20 μg/ml) was performed for 3–4 h at 4°C according to the manufacturer's recommendations. Protein A Sepharose (Sigma) was added to the samples and mixed overnight at 4°C. Beads were washed 3 times with RIPA buffer, suspended in SDS sample buffer, and heated to 95°C for 3 min. Immunoprecipitates were recovered by centrifugation and subjected to SDS/PAGE. After separation, proteins were transferred to PVDF membranes and probed with anti-Fyn and anti-rat8 antibodies.
Results
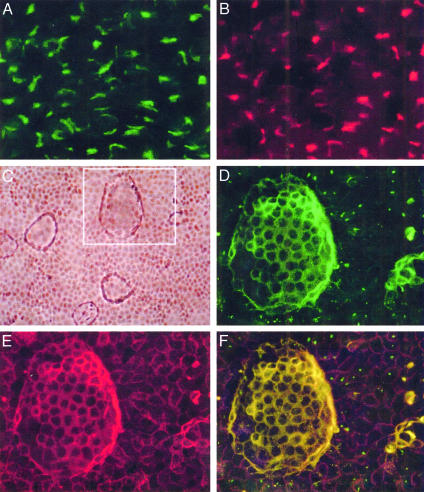
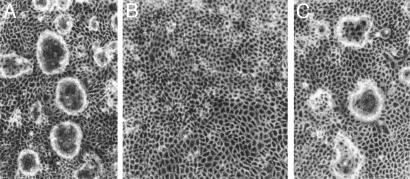
Cellular Localization of rat8 Protein. Immunoassay with a polyclonal antibody against rat8 protein showed that rat8 is expressed in both undifferentiated and differentiated LA7 cells, but at different locations. In undifferentiated LA7 cells (Fig. 1A), rat8 is present inside the cytoplasm, whereas in DMSO-induced LA7 cells, it is present in the plasma membrane of the cells involved in dome-structure formation; as shown in Fig. 1D, the strongest staining of rat8 was observed at the cell–cell contact surface areas of the dome-forming cells. The use of the monoclonal antibody against the GM130 protein, which stains the Golgi bodies, showed that in undifferentiated LA7 cells rat8 protein colocalizes with the Golgi apparatus (Fig. 1B), whereas in differentiated LA7 cells rat8 colocalizes with α6β1-integrin (Fig. 1E), a marker for the cell plasma membrane. Preimmune serum and peptide competition controls demonstrated the specificity of the staining pattern of the rat8 antibody (data not shown). In agreement with previous results, rat8 protein was not observed by immunoassaying untreated 106 cells and was weakly detected in DMSO-treated 106 cells (data not shown).
Fig. 1.
rat8 protein localization detection with immunofluorescent microscopy. Undifferentiated and DMSO-differentiated LA7 cells were stained with a polyclonal rabbit anti-rat8 antibody, a monoclonal mouse anti-α6β1-integrin antibody, and a monoclonal mouse antibody against the GM130 protein. (A) Staining for rat8 protein in undifferentiated LA7 cells showing a predominant intracytoplasmic distribution. (B) Staining of the same cells with the GM130 antibody, a Golgi body marker. (C) Phase contrast of DMSO-induced dome-forming LA7 cells. (D) A magnification of Inset in C, which shows a dome-structure stained with anti-rat8 antibody. (E) Staining of the same dome structure with a monoclonal mouse anti-α6β1-integrin antibody. (F) Merged image of D and E. Cells were microscopically photographed with a ×60 objective in A and B, a ×10 objective in C, and a ×40 objective in D–F.
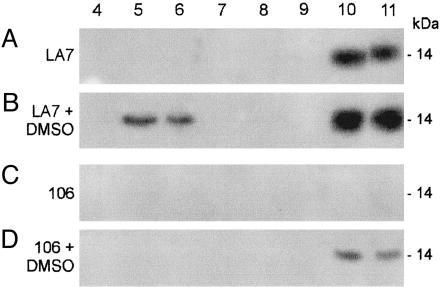
Association of rat8 Protein with Lipid Rafts. To determine whether rat8 protein was associated with lipid rafts in dome-forming cells, we used centrifugation to equilibrium on linear sucrose density gradients to isolate lipid rafts, which, as Triton X-100-insoluble complexes, float to the low-density top fractions. Western blot analyses of the sucrose density gradient fractions from untreated and DMSO-induced 106 and LA7 cells are shown in Fig. 2, wherein lane numbers correspond to the fractions collected from top to bottom of each gradient. Raft-containing fractions correspond to lanes 5 and 6, and lanes 10 and 11 correspond to detergent-soluble proteins that float near the bottom of the tube. Fig. 2B shows that in DMSO-induced dome-forming LA7 cells, a large amount of rat8 protein copurifies with lipid rafts (lanes 5 and 6). The remainder of rat8 protein is contained in the detergent-soluble fraction (lanes 10 and 11). Conversely, in untreated LA7 cells, rat8 was detected only in the detergent-soluble fractions (Fig. 2 A, lanes 10 and 11). In untreated 106 cells, rat8 protein was not detected in any of the fractions from the gradient (Fig. 2C); in DMSO-induced 106 cells, it was weakly detected in the detergent-soluble fractions (Fig. 2D, lanes 10 and 11) but not in the lipid raft-containing fractions.
Fig. 2.
Sucrose-gradient fraction analysis for rat8 protein. Western blot analysis of sucrose fractions collected from density gradients of untreated and DMSO-induced LA7 and 106 cells probed with polyclonal anti-rat8 antibody. Equal volumes from each 1-ml fraction, collected as described in Materials and Methods, were separated by SDS/PAGE (12.5% acrylamide) and probed by immunoblotting with polyclonal anti-rat8 antibody. Lane numbers correspond to fractions collected from top (fraction 1) to bottom (fraction 11) of the gradients. Fraction 5 contained a light-scattering band located at the interface between 5% and 30% sucrose. The major part of cells' sphingolipids was associated with this fraction. Fractions 9–11, containing 42.5% sucrose, represent the “loading zone” of these bottom-loaded flotation gradients and contain the bulk of cellular membrane and cytosolic proteins. Fractions 1–3 were omitted because no proteins were normally detected in these fractions of the gradients. Lipid raft-containing fractions correspond to lanes 5 and 6. Lanes 10 and 11 correspond to high-density fractions.
These data show that in DMSO-induced LA7 cells rat8 protein becomes associated with the plasma membrane only when domes are formed; this raises the question of how rat8 protein translocates from the Golgi to the cell membrane in a differentiation-dependent manner. Some proteins of the cytoplasmic leaflet become associated with lipid rafts after modification by palmitic or myristic acid acylation (31). We tested whether myristoylation affects dome formation and found that such is the case, because antisense oligonucleotides designed from the mRNA sequence of the N-myristoyl transferase-1 (NMT1) gene, when added to the LA7 cells in presence of DMSO, strongly inhibited dome formation, whereas NMT1 sense oligonucleotides had no effect on dome formation (data not shown). By structure analysis of rat8 protein, we determined that rat8 is a membrane protein with two transmembrane domains between amino acid residues 58 and 79 and 108 and 130 of the protein sequence. For the mouse homologue fragilis, it has been reported that both ends of the protein are outside the cells (20). For rat8, the location of the ends may be similar to that in fragilis. However, if the N terminus of rat8 protein is inside the cytoplasm, it is unlikely that it will be affected by myristoylation, because the two putative consensus sites for myristoylation, found in rat8 protein, are internally located, and no consensus sequence sites for proteases can be detected. Therefore, other proteins associating with rat8 must be the targets for myristoylation.
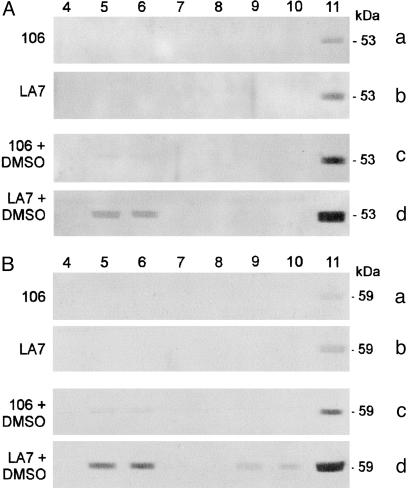
rat8-Associating Proteins. To identify possible candidate rat8-associating proteins, we screened two protein databases (www.ebi.ac.uk/swissprot/access.html and http://mendel.imp.univie.ac.at/myristate/myrbase) for proteins that can be myristoylated. We found that several proteins are modified by myristoylation, including Fyn, Lyn, and other members of the Src family of nonreceptor tyrosine kinases. Because it has been reported that members of the Src family localize to lipid rafts in many cell types (39), molecular antibodies for Fyn and Lyn were chosen for probing Western blots with LA7 and 106 cell extracts. Lyn and Fyn proteins were expressed in both untreated and DMSO-treated LA7 and 106 cells (data not shown); anti-Lyn and anti-Fyn antibodies then were used for sucrose gradient fraction analysis. In experiments similar to that presented in Fig. 2, antibodies against Lyn and Fyn tyrosine kinases revealed that these members of the Src kinase family cofractionate with lipid raft fractions of DMSO-induced LA7 and 106 cells, whereas in untreated LA7 and 106 cells Lyn and Fyn proteins are present only in the non-lipid raft fractions. (Fig. 3 A and B show gradient fraction analysis for Lyn and Fyn proteins, respectively.)
Fig. 3.
Sucrose gradient fraction analysis for Lyn and Fyn proteins. Western blot analysis of sucrose fractions collected from density gradients of untreated and DMSO-induced LA7 and 106 cells and probed with polyclonal anti-Lyn (A) and polyclonal anti-Fyn (B) antibody. Lipid raft-containing fractions correspond to lanes 5 and 6.
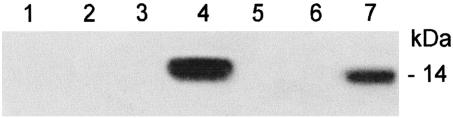
Immunoprecipitation Analysis. To determine whether rat8 interacts with Fyn, immunoprecipitation experiments were performed. Clarified immunoprecipitates were analyzed by SDS/PAGE and probed with anti-rat8 antibody. In DMSO-induced LA7 cells, rat8 protein was pulled down by using an antibody for Fyn (Fig. 4, lane 7). In Fig. 4, lanes 1–3 correspond to negative controls, lane 4 corresponds to DMSO-induced LA7 cell lysate (total cell extract before immunoprecipitation), lane 5 corresponds to unbound material from the last wash of DMSO-induced LA7 cell lysate, and lane 6 corresponds to 106 cells immunoprecipitated with Fyn antibody. These results show the coexistence of Fyn and rat8 in the same microenvironment within lipid rafts only in DMSO-induced LA7 cells.
Fig. 4.
Immunoprecipitation analysis. rat8 coimmunoprecipitates with Fyn by pull-down experiments with DMSO-induced LA7 cells using Fyn antibody. Lanes: 1 and 2, DMSO-induced LA7 cell lysate after immunoprecipitation with goat sera and preimmune sera derived from rabbit, respectively (controls); 3, DMSO-induced LA7 cell lysate after immunoprecipitation with an unrelated antibody specific for brain, Akt1/Akt2 (control); 4, DMSO-induced LA7 cell lysate (cell extract preimmunoprecipitation); 5, unbound material from last wash of DMSO-induced LA7 cell lysate after immunoprecipitation with Fyn antibody; 6, 106 cell lysate after immunoprecipitation with Fyn antibody; 7, DMSO-induced LA7 cell lysate after immunoprecipitation with Fyn antibody. All lanes were probed with anti-rat8 antibody.
Functional Role of Fyn with rat8 in Dome Formation. To determine whether Fyn, which coexists with rat8 in lipid rafts, may have a functional role in mammary gland differentiation, we determined whether dome formation was affected by blocking Fyn protein synthesis. When added to the DMSO-induced LA7 cells, antisense oligonucleotides designed from Fyn mRNA sequence resulted in the inhibition of dome formation (Fig. 5B). Similar experiments using antisense oligonucleotides designed from Lyn mRNA showed no effect on dome formation in DMSO-induced LA7 cells (Fig. 5C). The presence of the Fyn protein therefore is necessary for the formation of domes.
Fig. 5.
Fyn antisense oligonucleotides effect dome formation in DMSO-induced LA7 cells. (A) 1.5% DMSO-induced LA7 cells exhibiting domes. (B) Inhibition of dome formation seen after addition of Fyn antisense oligonucleotides to the LA7 culture in presence of DMSO. (C) Inhibition of dome formation not seen after addition of anti-Lyn oligonucleotides to the LA7 culture in the presence of DMSO. Cells were microscopically photographed with a ×10 objective.
Discussion
To study the differentiation of the mammary gland, we developed an in vitro model based on two cellular clones, LA7 and 106 (8–11). We previously showed that dome formation in LA7 cells recapitulates differentiation steps occurring in vivo, when tubules and alveoli develop in the mammary gland at pregnancy and lactation (14, 18).
In our previous work, by using a subtractive cDNA library approach to LA7 and 106 cells, we showed that rat8 is the key gene for dome formation (8). Antisense oligonucleotides designed from the rat8 mRNA sequence, when added to LA7 cell cultures in the presence of DMSO, prevent dome formation and also induce other reversible morphological changes in LA7 cells, probably reflecting a redirection of the differentiation program (8).
rat8 specifies a membrane protein with two transmembrane sites at amino acids 58–79 and 108–130 of the protein sequence; human homologues of rat8 have been implicated in mediating interaction between cells, homotypic cell adhesion, and inhibition of cell proliferation (40, 41). The immunostaining results demonstrate that in undifferentiated LA7 cells the protein is located in the Golgi apparatus (Fig. 1 A and B), whereas in DMSO-differentiated LA7 cells it is localized in the plasma membrane of the cells involved in dome-structure formation (Fig. 1D). The translocation of rat8 protein from the Golgi apparatus to the plasma membrane occurs only in DMSO-induced LA7 cells and coincides with the cell differentiation program resulting in dome formation. In agreement with previous Western and Northern analyses, the immunoassay results also show that rat8 expression is absent in untreated 106 cells and weakly detectable in these cells after DMSO treatment, highlighting the fact that 106 cells, unlike LA7 cells, cannot form domes spontaneously or by chemical induction (for instance, by DMSO).
The data show that in DMSO-induced LA7 cells, rat8 protein is associated with detergent-resistant lipid membrane domains. The localization of rat8 protein, both in the Golgi apparatus and plasma membrane (Fig. 1 A and D), is explained by its association with lipid membrane domains in the DMSO-induced LA7 cells. In fact, sucrose gradient centrifugation showed that rat8 protein cofractionates with lipid rafts only in these cells (Fig. 2B, lanes 5 and 6). Fractions corresponding to lanes 10 and 11 in Fig. 2B probably represent the portion of rat8 protein that, in DMSO-induced LA7 cells, is non-lipid raft-associated and is mostly intracytoplasmic in location. rat8 protein was found in the non-lipid raft-associated fractions of untreated LA7 cells (Fig. 2 A, lanes 10 and 11). rat8 was not detected in any fraction of untreated 106 cells (Fig. 2C), whereas in DMSO-treated 106 cells it was weakly detected in only the non-lipid raft-associated fractions (Fig. 2D, lanes 10 and 11). In these cells rat8 is induced by DMSO, but it is not recruited to lipid domains at the plasma membrane. As shown in Fig. 2 B and D, it seems that in both 106 and LA7 cells DMSO increases rat8 protein expression level. In addition, in untreated LA7 cells where rat8 is constitutively expressed (Fig. 2 A), rat8 is not seen being recruited to lipid membrane domains. We can conclude that in both cases (Fig. 2 A and C), when rat8 is not associated with lipid rafts, dome formation does not occur.
These results indicate that differentiation resulting in dome formation in LA7 cells depends on rat8's recruitment to lipid membrane domains at the cell plasma membrane; this is an essential step because the expression of rat8 alone is not sufficient to initiate this differentiation program. This conclusion is supported by previous results showing that high expression of ectopic rat8 protein in untreated and DMSO-treated 106 cells is not sufficient for dome formation (8) and that, in 106 cells transfected with rat8, DMSO does not cause the recruitment of rat8 protein to lipid membrane domains. Perhaps DMSO modifies the activities of a specific set of genes in LA7, but not in 106, cells, which allows rat8 protein to be recruited to lipid membrane domains.
Sucrose gradient fractionation showed that Lyn and Fyn, two members of the Src family of nonreceptor tyrosine kinases, cofractionate with lipid rafts in DMSO-induced LA7 and 106 cells. Unlike rat8, Lyn and Fyn are constitutively expressed in 106 and LA7 cells, and DMSO moderately induces their expression. DMSO is involved in Lyn and Fyn protein recruitment to lipid rafts in LA7 and 106 cells, but their recruitment alone is not sufficient for dome formation. In fact, through immunoprecipitation experiments using a Fyn tyrosine kinase antibody to pull down proteins interacting with Fyn, rat8 was detected in the immunoprecipitate of DMSO-treated LA7 cell extracts (Fig. 4, lane 3), and both proteins, rat8 and Fyn tyrosine kinase, were present in the gradient fractions of DMSO-induced LA7 cells containing lipid rafts (Figs. 2B and 3Bd). These results suggest that rat8 may form a complex with Fyn in lipid rafts.
Antisense oligonucleotide experiments showed that the loss of Fyn protein expression resulted in the specific inhibition of dome formation in LA7 cells cultured in the presence of DMSO (Fig. 5B). Conversely, antisense oligonucleotides for Lyn mRNA added to LA7 cell cultures in the presence of DMSO were ineffective in blocking dome formation (Fig. 5C).
These results suggest that the presence of Fyn and rat8 in the same microenvironment within lipid rafts is required for the differentiation program leading to dome formation in LA7 cells after DMSO treatment. Moreover, expression of either rat8 or Fyn protein in LA7 cells is not sufficient to trigger dome formation, which in fact depends on the recruitment of both Fyn and rat8 to lipid membrane domains of the cellular plasma membrane. This observation suggests that these two proteins may be part of the same functional complex. Fyn phosphorylates several target proteins associated with lipid rafts; whether and how Fyn kinase activity affects the cellular function of rat8 protein remains to be determined. As in the case of the human homologue leu-13, the cellular effects observed in our experiments may occur in the context of a protein complex involving multiple protein interactions, and Fyn may be one protein in the complex.
The data presented in this study demonstrate that in the cells involved in the dome structure formation, rat8 protein transits from the Golgi bodies to the plasma membrane, where it associates to lipid membrane domains. They also show that the recruitment of rat8 and associated proteins to lipid membrane domains in DMSO-induced LA7 cells is an essential step for the cell differentiation program resulting in dome formation. Together with previous results (8), these findings suggest that rat8, like fragilis, is involved in developmental decisions. In the case of rat8, the decision is the demarcation of a subset of cells in the mammary gland that cause epithelial cells to develop into a network of tubuloalveolar structures responsible for secretion.
Acknowledgments
We thank Drs. M. Resh and S. Maurer-Stroh for advice and Dr. M. Zoppé for sharing reagents and suggestions. This work was partially funded by Associazione Italiana per la Ricerca sul Cancro Grant 115/2003 (to I.Z.); Progetto Strategico: Genomica Funzionale, Consiglio Nazionale delle Ricerche Grant 449/97 (to P.V.); Ministero dell' Istruzione, dell'Universitá, e della Ricerca/Fondo per gli Investimenti della Ricerca di Base Grant RBME019J9W (to P.V. and I.Z.); Programmi Cofinanziati (COFIN-PRIN) Grants 2001 and 2002 (to S.S.); and a Compagnia di San Paolo grant (to P.V.) This is manuscript no. 70 of the Genoma 2000/Istituto Tecnologie Biomediche Avanzate Project funded by Cariplo.
References
- 1.Nanba, D., Nakanishi, Y. & Hieda, Y. (2001) Dev. Growth Differ. 43, 535–544. [DOI] [PubMed] [Google Scholar]
- 2.Alford, D. & Taylor-Papadimitriou, J. (1996) J. Mammary Gland Biol. Neoplasia 1, 207–218. [DOI] [PubMed] [Google Scholar]
- 3.Gumbiner, B. M. (1996) Cell 84, 345–357. [DOI] [PubMed] [Google Scholar]
- 4.Topper, Y. J. & Freeman, C. S. (1980) Physiol. Rev. 60, 1049–1106. [DOI] [PubMed] [Google Scholar]
- 5.Hennighausen, L., Robinson, G. W., Wagner, K. U. & Liu, W. (1997) J. Biol. Chem. 272, 7567–7569. [DOI] [PubMed] [Google Scholar]
- 6.Hennighausen, L. & Robinson, G. W. (1998) Genes Dev. 12, 449–455. [DOI] [PubMed] [Google Scholar]
- 7.Robinson, G. W., McKnight, R. A., Smith, G. H. & Hennighausen, L. (1995) Development (Cambridge, U.K.) 121, 2079–2090. [DOI] [PubMed] [Google Scholar]
- 8.Zucchi, I., Montagna, C., Susani, L., Vezzoni, P. & Dulbecco, R. (1998) Proc. Natl. Acad. Sci. USA 95, 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dulbecco, R., Bologna, M. & Unger, M. (1979) Proc. Natl. Acad. Sci. USA 76, 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulbecco, R. & Okada, S. (1980) Proc. R. Soc. London Ser. B 208, 399–408. [DOI] [PubMed] [Google Scholar]
- 11.Bennett, D. C., Peachey, L. A., Durbin, H. & Rudland, P. S. (1978) Cell 15, 283–298. [DOI] [PubMed] [Google Scholar]
- 12.Paterson, F. C., Warburton, M. J. & Rudland, P. S. (1985) Dev. Biol. 107, 301–313. [DOI] [PubMed] [Google Scholar]
- 13.Rudland, P. S. (1992) J. Cell. Physiol. 153, 157–168. [DOI] [PubMed] [Google Scholar]
- 14.Zucchi, I., Bini, L., Albani, D., Valaperta, R., Liberatori, S., Raggiaschi, R., Montagna, C., Susani, L., Barbieri, O., Pallini, V., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 8660–8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dulbecco, R., Bologna, M. & Unger, M. (1980) Proc. Natl. Acad. Sci. USA 77, 1551–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zucchi, I., Montagna, C., Susani, L., Montesano, R., Affer, M., Zanotti, S., Redolfi, E., Vezzoni, P. & Dulbecco, R. (1999) Proc. Natl. Acad. Sci. USA 96, 13766–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zucchi, I., Bini, L., Valaperta, R., Ginestra, A., Albani, D., Susani, L., Sanchez, J. C., Liberatori, S., Magi, B., Raggiaschi, R., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 5608–5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zucchi, I. & Dulbecco, R. (2002) J. Mammary Gland Biol. Neoplasia 7, 373–384. [DOI] [PubMed] [Google Scholar]
- 19.Hayzer, D. J., Brinson, E. & Runge, M. S. (1992) Gene 117, 277–278. [DOI] [PubMed] [Google Scholar]
- 20.Saitou, M., Barton, S. C. & Surani, M. A. (2002) Nature 418, 293–300. [DOI] [PubMed] [Google Scholar]
- 21.Lange, U., Saitou, M., Western, P., Barton, S. & Surani, M. (2003) BMC Dev. Biol. 3, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewin, A. R., Reid, L. E., McMahon, M., Stark, G. R. & Kerr, I. M. (1991) Eur. J. Biochem. 199, 417–423. [DOI] [PubMed] [Google Scholar]
- 23.Deblandre, G. A., Marinx, O. P., Evans, S. S., Majjaj, S., Leo, O., Caput, D., Huez, G. A. & Wathelet, M. G. (1995) J. Biol. Chem. 270, 23860–23866. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi, S., Doss, C., Levy, S. & Levy, R. (1990) J. Immunol. 145, 2207–2213. [PubMed] [Google Scholar]
- 25.Simons, K. & Ikonen, E. (1997) Nature 387, 569–572. [DOI] [PubMed] [Google Scholar]
- 26.Simons, K. & Toomre, D. (2000) Nat. Rev. Mol. Cell Biol. 1, 31–39. [DOI] [PubMed] [Google Scholar]
- 27.Brown, D. A. & Rose, J. K. (1992) Cell 68, 533–544. [DOI] [PubMed] [Google Scholar]
- 28.Brown, D. A., Crise, B. & Rose, J. K. (1989) Science 245, 1499–1501. [DOI] [PubMed] [Google Scholar]
- 29.Casey, P. J. (1995) Science 268, 221–225. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, F. L. & Casey, P. J. (1996) Annu. Rev. Biochem. 65, 241–269. [DOI] [PubMed] [Google Scholar]
- 31.Resh, M. D. (1994) Cell 76, 411–413. [DOI] [PubMed] [Google Scholar]
- 32.van't Hof, W. & Resh, M. D. (1997) J. Cell Biol. 136, 1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilcox, C., Hu, J. S. & Olson, E. N. (1987) Science 238, 1275–1278. [DOI] [PubMed] [Google Scholar]
- 34.Prinetti, A., Chigorno, V., Tettamanti, G. & Sonnino, S. (2000) J. Biol. Chem. 275, 11658–11665. [DOI] [PubMed] [Google Scholar]
- 35.Panigone, S., Bergomas, R., Fontanella, E., Prinetti, A., Sandhoff, K., Grabowski, G. A. & Delia, D. (April 27, 2001) FASEB J., 10.1096/fj.00-0531fje. [DOI] [PubMed]
- 36.Prinetti, A., Chigorno, V., Prioni, S., Loberto, N., Marano, N., Tettamanti, G. & Sonnino, S. (2001) J. Biol. Chem. 276, 21136–21145. [DOI] [PubMed] [Google Scholar]
- 37.Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. (1951) J. Biol. Chem. 193, 265–275. [PubMed] [Google Scholar]
- 38.van't Hof, W. & Resh, M. D. (1999) J. Cell Biol. 145, 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster, L. J., De Hoog, C. L. & Mann, M. (2003) Proc. Natl. Acad. Sci. USA 100, 5813–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans, S. S., Collea, R. P., Leasure, J. A. & Lee, D. B. (1993) J. Immunol. 150, 736–774. [PubMed] [Google Scholar]
- 41.Evans, S. S., Lee, D. B., Han, T., Tomasi, T. B. & Evans, R. L. (1990) Blood 76, 2583–2593. [PubMed] [Google Scholar]