Abstract
Motivation: Genotype imputation has become an indispensible step in genome-wide association studies (GWAS). Imputation accuracy, directly influencing downstream analysis, has shown to be improved using re-sequencing-based reference panels; however, this comes at the cost of high computational burden due to the huge number of potentially imputable markers (tens of millions) discovered through sequencing a large number of individuals. Therefore, there is an increasing need for access to imputation quality information without actually conducting imputation. To facilitate this process, we have established a publicly available SNP and indel imputability database, aiming to provide direct access to imputation accuracy information for markers identified by the 1000 Genomes Project across four major populations and covering multiple GWAS genotyping platforms.
Results: SNP and indel imputability information can be retrieved through a user-friendly interface by providing the ID(s) of the desired variant(s) or by specifying the desired genomic region. The query results can be refined by selecting relevant GWAS genotyping platform(s). This is the first database providing variant imputability information specific to each continental group and to each genotyping platform. In Filipino individuals from the Cebu Longitudinal Health and Nutrition Survey, our database can achieve an area under the receiver-operating characteristic curve of 0.97, 0.91, 0.88 and 0.79 for markers with minor allele frequency >5%, 3–5%, 1–3% and 0.5–1%, respectively. Specifically, by filtering out 48.6% of markers (corresponding to a reduction of up to 48.6% in computational costs for actual imputation) based on the imputability information in our database, we can remove 77%, 58%, 51% and 42% of the poorly imputed markers at the cost of only 0.3%, 0.8%, 1.5% and 4.6% of the well-imputed markers with minor allele frequency >5%, 3–5%, 1–3% and 0.5–1%, respectively.
Availability: http://www.unc.edu/∼yunmli/imputability.html
Supplementary information: Supplementary data are available at Bioinformatics online.
Contact: yunli@med.unc.edu
1 INTRODUCTION
Genotype imputation has proven to be a powerful tool in genome-wide association studies (GWAS) by facilitating fine mapping and the merging of datasets from different genotyping platforms (Li et al., 2009; Marchini and Howie, 2010). It is a way to predict genotypes computationally based on linkage disequilibrium patterns instead of obtaining genotypes by laboratory-based procedure (Browning and Yu, 2009; Howie et al., 2011; Li et al., 2010). As it has been shown to directly affect downstream analysis, imputation accuracy needs to be taken into consideration when designing and performing GWAS (Zheng et al., 2011). For instance, at the study design stage, a question of interest would be which commercially available genotyping platform can provide the optimal imputation quality genome-wide or in certain genomic region(s) of interest. Such a question can be answered by assessing the imputation accuracy of relevant variants. However, there has been no resource available to provide variant imputability information without actually performing imputation.
A commonly used evaluation method is to mask a subset of markers, impute their dosages and compare those dosages with the true (masked) genotypes for those markers (Li et al., 2010). This method, however, can only be used after genotypes have already been obtained and therefore cannot help guide study design decisions. In addition, the evaluation procedure can be computationally costly because of the requirement of conducting imputation, particularly with the emergence of reference panels built through re-sequencing efforts (Sampson et al., 2012). To facilitate genetic studies in the era of genomic re-sequencing, we have built a database containing imputation accuracy information for SNPs and indels identified from the 1000 Genomes Project (The 1000 Genomes Project Consortium, 2010), a sequencing-based reference resource, which has demonstrated its potential for enhancing the power of genetic association studies in the post-GWAS era (Day-Williams et al., 2011; Holm et al., 2011; Huang et al., 2012). The assessment of marker imputability was carried out through a leave-one-out imputation procedure: a single individual serves as the imputation target, and imputation is performed using haplotypes from all the other individuals as reference. Imputation accuracy was quantified within each of the four major continental groups surveyed by the 1000 Genomes Project. We anticipate this database containing imputation accuracy information searchable by continental group and by GWAS genotyping platform will be a useful resource for geneticists in this sequencing era.
2 DATA SETUP AND RETRIEVAL
Database: The database contains imputation quality information (as measured by dosage r2, the squared Pearson correlation coefficient between the imputed dosage—ranging continuously from 0 to 2—and the observed/masked genotypes—taking values 0, 1 or 2 copies of a given allele) for every non-singleton SNP and indel discovered by and passing default quality filters in the 1000 Genomes Project (The 1000 Genomes Project Consortium, 2010). The dosage r2 of each variant reflects its potential imputation accuracy when conducting imputation using haplotypes from the 1000 Genomes Project as reference. Imputability information is available for multiple genotyping platforms, and separately for each of the four major continental groups [Europeans (EUR), Africans (AFR), Asians (ASN) and Americans (AMR)]. Details regarding sub-population constituents of the continental groups can be found at ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20110521/.
Methods: The dosage r2 of each variant was obtained using a leave-one-out imputation procedure with MaCH-Admix (Liu et al., 2012,b; http://www.unc.edu/∼yunmli/MaCH-Admix/) [high Pearson correlation (0.85–0.94) with those obtained using minimac (Howie et al., 2012) and IMPUTE2 (Howie et al., 2011), and lower correlation (0.71–0.85) with those from BEAGLE (Browning and Yu, 2009), data not shown] on samples from the latest release of the 1000 Genomes Project (version 3 March 2012 release, 2184 haplotypes). We mimicked typical GWAS imputation practice by masking genotypes at markers absent from the selected genotyping platform and treating them as untyped. These untyped markers were imputed in one individual at a time using the haplotypes of all the remaining individuals as reference (2182 haplotypes). The imputation accuracy of each marker, measured by dosage r2, was calculated separately in each of four continental groups currently available in the 1000 Genomes Project. The genotyping platforms we have evaluated include Affymetrix 5.0, Affymetrix 6.0, Affymetrix Axiom, Illumina Human1M, Illumina Omni 5 M and Illumina Omni ZhongHua. The results of the assessment are searchable through a publicly available database.
Usage: Our database can take as input either a list of marker names or the start and end position of a genomic region on a specified chromosome. Users can choose to view information corresponding to one or more specific genotyping platforms. Given the marker or region input and the choice of genotyping platform, our database returns imputability information for variants of interest ordered by their genomic location according to NCBI Build 37. Users have the option to display or to download the imputability information for each continental group or the maximum dosage r2 across the four continental groups (max-r2). Moreover, users can filter results by max-r2. Markers with no rsID follow chromosome:physical-coordinate nomenclature (Supplementary Fig. S1A). In addition, for an SNP–indel pair with the same genomic location, the SNP is always listed before the indel (Supplementary Fig. S1B).
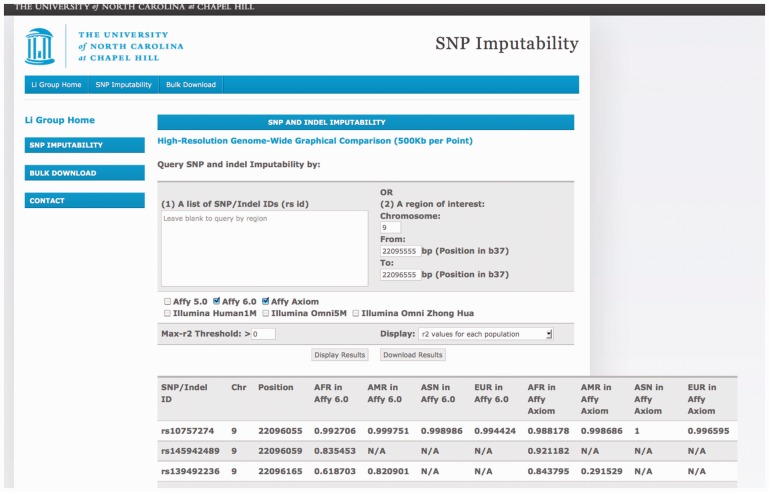
Examples: The first example shows the utility of our database at the study design stage. Specifically, suppose an investigator wants to decide between two genotyping platforms, Affymetrix 6.0 and Affymetrix Axiom, based on imputation accuracy within a 1-kb region on chromosome 9p21 (22,095,555 to 22,096,555 bp) harboring the SNP rs10757274 known to be associated with risk of coronary heart disease and multiple related phenotypes (Cunnington et al., 2010; McPherson et al., 2007). Our database interface, the example query, as well as the results of the query are shown in Figure 1. Given the regional input (start and end position 22 095 555 and 22 096 555 on chromosome 9), our database returns a list of markers within the region (only the top three are shown). For each marker, the database shows its marker name, genomic location and dosage r2 for the two selected genotyping platforms across four continental groups. To ease comparison, users can choose to display max-r2 instead of r2 values for each population separately and/or filter by setting non-zero max-r2 threshold. Based on what is shown in Figure 1, we would recommend the Axiom over the 6.0 panel, unless the samples under study are Americans (e.g. Hispanic or African Americans) and the SNP of primary interest is rs139492236. Note that this is a toy example mainly meant to introduce the interface of our database where we show only the top three SNPs. For more realistic settings where the region of interest typically includes many more markers, we recommend prioritization of markers in the region (e.g. according to functional annotation and/or evidence from existing association or functional studies, if available), followed by the examination and comparison of the max-r2 distribution through ‘Download Results’ or ‘Genome-wide Graphical Comparison’. Such comparison of imputation accuracy across platforms will facilitate decision making regarding the choice of genotyping assays.
Fig. 1.
The SNP and indel imputability database interface
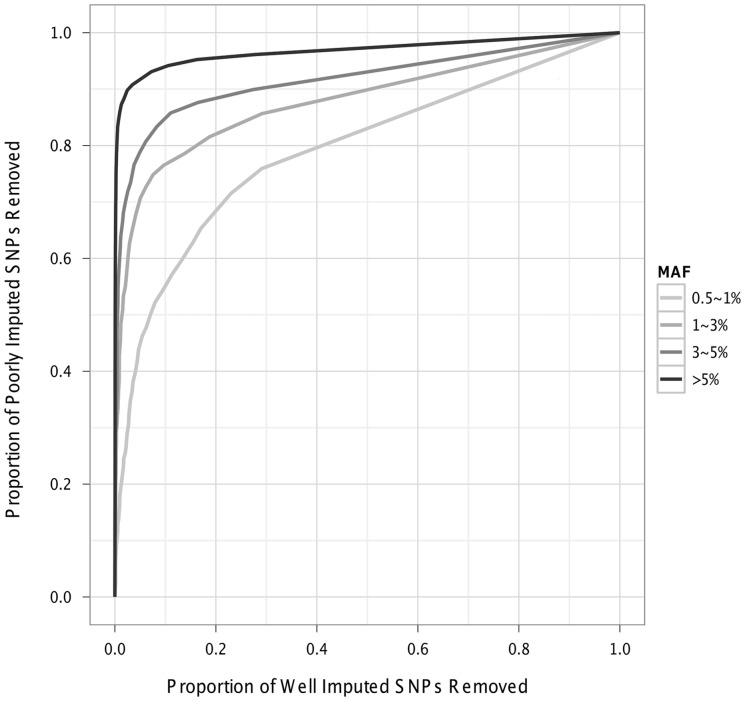
Once the investigator has decided on the genotyping platform, a typical question is whether specific markers or markers in specific regions of interest can be imputed well (e.g. novel variants or associated regions identified in other cohorts). When computational resources are limited or when an investigator is interested in a considerable number of markers/regions, imputability information can help prioritize markers/regions that have the potential to be well-imputed as well as avoid wasting resources on markers/regions that have little potential for high-quality imputation. As shown in Figure 1, our database contains four dosage r2 values (one for each continental group) for each marker, given a genotyping platform. As false-negatives (markers that can be well-imputed but with bad predicted imputation accuracy such that one would not perform actual imputation) are typically more costly than false-positives (the consequence would be wasted computational resources on markers/regions that are truly not imputable), we recommend using the maximum dosage r2 across the four continental groups (max-r2) to guide decisions, particularly for samples involving admixed individuals. Figure 2 shows the receiver-operating characteristic curve for data from the Cebu Longitudinal Health and Nutrition Survey (CLHNS) when max-r2 is used for thresholding. In this cohort of Filipinos (Adair et al., 2011; Marvelle et al., 2007), we have 81 individuals who have both Affymetrix 5.0 (Lange et al., 2010) and Metabochip (Croteau-Chonka et al., 2012) genotypes. We imputed the Metabochip SNPs from the Affymetrix 5.0 data, using haplotypes from the 1000 Genomes Project as reference. We computed the imputation accuracy in this sample (CLHNS-specific dosage r2) by comparing the imputed dosages with the genotypes obtained through genotyping using Metabochip. The y-axis shows the proportion of poorly imputed SNPs (CLHNS-specific dosage r2 < 0.2) removed and the x-axis shows the proportion of well-imputed SNPs (CLHNS-specific dosage r2 > 0.8) sacrificed for SNPs in different minor allele frequency (MAF) categories (defined within CLHNS). Using a max-r2 threshold of 0.7, which removes ∼15 million of the ∼31 million markers in the latest release from the 1000 Genomes Project, we found that the database filters out 77%, 58%, 51% and 42% of the poorly imputed SNPs (again, SNPs with CLHNS-specific dosage r2 < 0.2) at the cost of 0.3%, 0.8%, 1.5% and 4.6% well-imputed markers (SNPs with CLHNS-specific dosage r2 > 0.8) in the MAF categories of >5%, 3–5%, 1–3% and 0.5–1%, respectively. Using a different threshold of 0.5 (0.9), which removes ∼12 (∼20) million of the ∼31 million markers, we can filter out 54%, 32%, 29% and 26% (92%, 80%, 75% and 66%) of the poorly imputed SNPs at the cost of 0.1%, 0.3%, 0.2% and 2.1% (4.8%, 6.1%, 7.5% and 17.2%) well-imputed SNPs. We also confirmed in samples of Caucasians and samples of African Americans (data not shown) that a max-r2 in the range of 0.5–0.8 serves as a reasonable threshold in terms of a trade-off between sensitivity and specificity. The actual threshold an investigator selects can be tailored according to MAF and available computational resources (including both CPU times and disk space). We and others have previously observed lower imputation quality for rarer variants (Li, et al., 2011a; Liu et al., 2012a; The International HapMap 3 Consortium, 2010). Our database now shows that imputation quality of rarer variants is also more challenging for prediction estimation: the total area under the receiver-operating characteristic curve is 0.97, 0.91, 0.88 and 0.79, respectively, for markers with MAF >5%, 3–5%, 1–3% and 0.5–1%.
Fig. 2.
Receiver-operating characteristic curve in the Cebu Longitudinal Health and Nutrition Survey
3 CONCLUSION
In summary, we have built a publicly available database for marker imputability to aid genetic association studies in the re-sequencing era (Fridley et al., 2010; Li, et al., 2011b; Sampson et al., 2012). Reference panels built from re-sequencing studies bring us the benefits of improved imputation accuracy and the potential to impute low-frequency variants. These benefits come, however, at the cost of heavy computational burden for imputation if we impute every marker discovered by sequencing, which is >30 million in the latest release from the 1000 Genomes Project. It is therefore desirable to have direct access to marker imputability information without actually conducting genotype imputation. Our marker imputability database provides direct access to imputation accuracy information for SNPs and indels identified from the 1000 Genomes Project across four major continental groups using multiple genotyping platforms. We anticipate that this database will serve as a useful resource for researchers in this re-sequencing era in terms of design and analysis of genetic association studies. In addition, although the database is developed mainly for guidance before actual imputation, it can be used for post-imputation quality assurance by comparing estimated r2 values in the imputed study sample with those in our database in an SNP-specific manner. Using a cohort of Filipinos, we estimate that we can, with up to 48.6% reduced computation efforts (by imputing only the top 51.4% markers according to imputation quality estimated from individuals in the 1000 Genomes Project), filter out 42–77% of poorly imputed markers at the cost of 0.3–4.6% well-imputed markers. Finally, two caveats should be kept in mind by database users. First, we record results from the MaCH-Admix software. Although more than moderate level of correlation is observed with results from other imputation software, caution needs to be taken when generalizing to other imputation methods, particularly those that are not based on the Li and Stephens model (Li and Stephens, 2003). Second, loss of some typed markers due to quality control in real studies could lead to reduced imputation quality of specific markers, which cannot be modeled generically and are thus not reflected by our database. We will update the database when new data releases of the 1000 Genomes Project or new genotyping platforms become available.
ACKNOWLEDGEMENTS
The authors thank Drs Margaret G. Ehm, Matthew R. Nelson, Li Li, Liling Warren and Toby Johnson for providing feedback on our database. They also thank Drs Mingyao Li, Alexander P. Reiner and Guillaume Lettre for comments on the manuscript.
Funding: This research was supported by the National Institute of Health grants R01-HG006292 and R01-HG006703 (awarded to Y.L.) and R01-DK078150 (awarded to K.L.M.).
Conflict of interest: none declared.
REFERENCES
- Adair LS, et al. Cohort profile: the Cebu longitudinal health and nutrition survey. Int. J. Epidemiol. 2011;40:619–625. doi: 10.1093/ije/dyq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am. J. Hum. Genet. 2009;85:847–861. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau-Chonka DC, et al. Population-specific coding variant underlies genome-wide association with adiponectin level. Hum. Mol. Genet. 2012;21:463–471. doi: 10.1093/hmg/ddr480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnington MS, et al. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6:e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day-Williams A, et al. A variant in MCF2L is associated with osteoarthritis. Am. J. Hum. Genet. 2011;89:446–450. doi: 10.1016/j.ajhg.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley BL, et al. Utilizing genotype imputation for the augmentation of sequence data. PloS ONE. 2010;5:e11018. doi: 10.1371/journal.pone.0011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, et al. Genotype imputation with thousands of genomes. G3 (Bethesda, MD.) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, et al. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm H, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat. Genet. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, et al. 1000 Genomes-based imputation identifies novel and refined associations for the. Wellcome Trust Case Control Consortium phase 1 Data. Eur. J. Hum. Genet. 2012;20:801–805. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange LA, et al. Genome-wide association study of homocysteine levels in Filipinos provides evidence for CPS1 in women and a stronger MTHFR effect in young adults. Hum. Mol. Genet. 2010;19:2050–2058. doi: 10.1093/hmg/ddq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, et al. Performance of genotype imputation for rare variants identified in exons and flanking regions of genes. PloS ONE. 2011a;6:e24945. doi: 10.1371/journal.pone.0024945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. Genotype imputation. Ann. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011b;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, et al. Genotype imputation of metabochip SNPs using a study-specific reference panel of ∼4,000 haplotypes in African Americans from the women’s health initiative. Genet. Epidemiol. 2012a;117:107–117. doi: 10.1002/gepi.21603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, et al. MaCH-admix: genotype imputation for admixed populations. Genet. Epidemiol. 2012b;00:1–13. doi: 10.1002/gepi.21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nature reviews. Genetics. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Marvelle AF, et al. Comparison of ENCODE region SNPs between Cebu Filipino and Asian HapMap samples. J. Hum. Genet. 2007;52:729–737. doi: 10.1007/s10038-007-0175-9. [DOI] [PubMed] [Google Scholar]
- McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JN, et al. A two-platform design for next generation genome-wide association studies. Genet. Epidemiol. 2012;36:400–408. doi: 10.1002/gepi.21634. [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, et al. A comparison of approaches to account for uncertainty in analysis of imputed genotypes. Genet. Epidemiol. 2011;35:102–110. doi: 10.1002/gepi.20552. [DOI] [PMC free article] [PubMed] [Google Scholar]