Abstract
Motivation: Data collection in spreadsheets is ubiquitous, but current solutions lack support for collaborative semantic annotation that would promote shared and interdisciplinary annotation practices, supporting geographically distributed players.
Results: OntoMaton is an open source solution that brings ontology lookup and tagging capabilities into a cloud-based collaborative editing environment, harnessing Google Spreadsheets and the NCBO Web services. It is a general purpose, format-agnostic tool that may serve as a component of the ISA software suite. OntoMaton can also be used to assist the ontology development process.
Availability: OntoMaton is freely available from Google widgets under the CPAL open source license; documentation and examples at: https://github.com/ISA-tools/OntoMaton.
Contact: isatools@googlegroups.com
1 INTRODUCTION
Well-annotated and shared bioscience research data offer new discovery opportunities and drive science of the future. Several data management plans and sharing policies have emerged, along with a growing number of community-developed guidelines and ontologies to harmonize the reporting of experiments from different domains so that these can be comprehensible and in turn, reproducible and reusable. In many research projects however, the generation and collection of experimental data occur in a multicentric, distributed fashion; and a variety of data types are generated often in a single experiment. Use of spreadsheets and related editors, such as Microsoft Excel for collecting experimental description is widespread among researchers due to their flexibility, low learning curve and above all ubiquity of tooling. However, misalignment, conflicting versions, the heterogeneity of free text, and also silent and unwanted ‘auto-corrections’ are major shortcomings to be addressed (Zeeberg et al., 2004). This scenario and the current budgetary restrictions require ‘invest to save’ solutions to promote consistent annotation and collaborative editing of bioscience experiments, assisting researchers in complying with reporting policies and community standards. OntoMaton is an open source tool that leverages the collaborative environment and editing functionalities brought by Google Spreadsheets, and provides access to ontology look-up and tagging functionalities served by the NCBO BioPortal and Annotator web services (Jonquet et al., 2010; Whetzel et al., 2011).
2 OntoMaton DESIGN AND USE CASES
Four main use cases drove the development of OntoMaton: (i) to allow collaborative, distributed and coordinated annotation while enabling configurations and restrictions to be defined; (ii) to reduce free text description in metadata tracking of experimental data; (iii) to assist design patterns-based ontology development by facilitating interaction with domain experts; and (iv) to ease mapping between models and semantic representations. Two use cases are discussed more specifically in the next sections: one insisting on the free form of the widget and its ability to integrate in any layout, agnostic of any framework; the other aligning with a standardization effort, the ISA syntax (Sansone et al., 2012). While ontology-enabled standalone tools exist (Rocca-Serra et al., 2011; Wolstencroft et al., 2011), they lack collaborative features. OntoMaton, with the aid of the Google Spreadsheet environment delivers this. OntoMaton is implemented in JavaScript upon the Google App Script API and accesses the NCBO RESTful web services. A webcast tutorial of how to use it is available at http://goo.gl/FjghA.
3 COLLABORATIVE SEMANTIC ANNOTATION
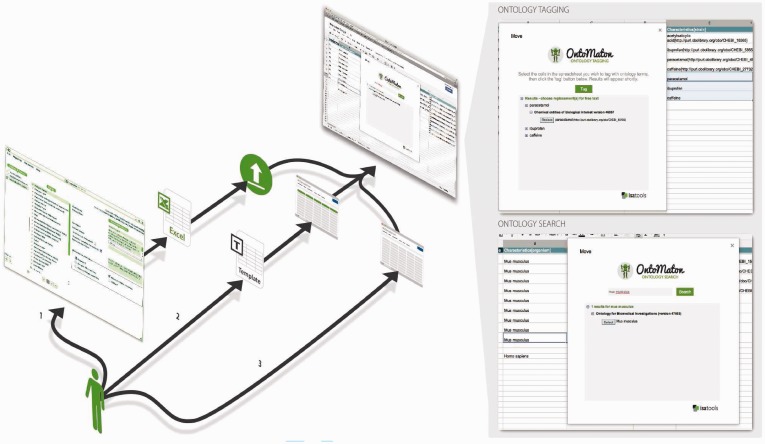
The OntoMaton Google widget can be installed and invoked from any Google Spreadsheet document or embedded in Google Templates. It provides a facility for searching ontologies hosted at NCBO BioPortal, or for calling a tagging functionality, by relying on NCBO’s Annotator services (Jonquet et al., 2010). An OntoMaton-enabled Google spreadsheet can also be configured to restrict the ontological search space to specific resources. While OntoMaton is syntax neutral, its usefulness is demonstrated when exploited by a data management infrastructure, for instance to support the creation of ISA-Tab compatible templates. The ultimate goal is to foster adoption of reporting standard conformant spreadsheets for managing biological experimental data description. Figure 1 provides detailed alternative uses of OntoMaton and a snippet of an experiment being marked-up. Several data management projects—with an existing large user base are currently using OntoMaton-based templates to assist with their data collection and management needs. These include: the Earth Microbiome Project (http://goo.gl/JLG5d); Bioplatforms Australia (http://goo.gl/uXLve)—with a focus on soil metagenomics sample collection; and Metabolights (Steinbeck et al., 2012)—a repository of metabolite profiling data at the European Bioinformatics Institute.
Fig. 1.
Uses of OntoMaton: (1) open the OntoMaton-enabled Google Spreadsheet template from the online gallery; (2) create a standard Google Spreadsheet and install OntoMaton within that; or (3) as part of the ISA suite, export an Excel template from ISAconfigurator and upload it into Google Spreadsheets
4 COLLABORATIVE ONTOLOGY ENGINEERING
Developing ontologies and knowledge representation artefacts requires the interaction of domain experts and computer scientists. The core interaction consists of converting domain expert vetted representations (a.k.a a design pattern) to OWL representations through the intervention of knowledge engineers. Tools such as Populous (Jupp et al., 2012) and Protege Mapping Master (http://protege.stanford.edu) have been developed to support these activities. The developers of the Ontology of Biomedical Investigations (OBI) (Brinkman et al., 2010), currently rely on the Quick Term Template (Rocca-Serra et al., 2011) approach to quickly add defined classes based on a template and the Manchester OWL Syntax for the mapping. However, owing to the collaborative nature of OBI development, the approach has been hindered by the lack of tools. OntoMaton closes this gap and several templates have now been documented to support different design patterns. Those templates unfold the restrictions of a class model in a table: fields correspond to facet fillers and cell values should be class names or URIs. OntoMaton, by enabling in situ resource lookups, simplifies development, review and curation by the pool of OBI editors.
5 DISCUSSION
Developed by harnessing the Google Spreadsheet environment and the term lookup and annotation power of the NCBO Web services, OntoMaton is an effective tool assisting both collaborative semantic annotation of experiments and the ontology development process. Several annotation tools exist (Nelson et al., 2011) but not all have support for community-driven guidelines and ontologies, and none of them allow collaborative annotation. Moreover, Excel-based tools (Jones and Côté, 2008) tend to be platform and version dependent. Google Spreadsheets on the other hand work across all platforms. A comparison of tools attempting to mix spreadsheets with access to vocabulary servers is available at http://goo.gl/NV3lZ. Ongoing development of OntoMaton focuses on: (i) transformation of data into the Resource Description Framework and Linked Data; (ii) support for cell level, vocabulary drop-down list as soon as the Google API supports it; and (iii) further integration with the ISA software suite as requested by users.
Funding: This work was supported by the Biotechnology and Biological Sciences Research Council [grant BB/I025840/1, BB/I000771/1 and BB/I000917/1 to S.A.S.] and the National Institutes of Health [grant U54 HG004028 supporting TW].
Conflict of Interest: none declared.
REFERENCES
- Brinkman RR, et al. Modeling biomedical experimental processes with OBI. J. Biomed. Semantics. 2010;1(Suppl. 1):S7. doi: 10.1186/2041-1480-1-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Côté R. The PRIDE proteomics identifications database: data submission, query, and dataset comparison. Methods Mol. Biol. 2008;484:287–303. doi: 10.1007/978-1-59745-398-1_19. [DOI] [PubMed] [Google Scholar]
- Jonquet C, et al. Building a biomedical ontology recommender web service. J. Biomed. Semantics. 2010;1(Suppl 1):S1. doi: 10.1186/2041-1480-1-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupp S, et al. Populous: a tool for building OWL ontologies from templates. BMC Bioinformatics. 2012;13(Suppl. 1):S5. doi: 10.1186/1471-2105-13-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EK, et al. Labkey server: an open source platform for scientific data integration, analysis and collaboration. BMC Bioinformatics. 2011;12:71. doi: 10.1186/1471-2105-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca-Serra P, et al. ISA software suite: supporting standards-compliant experimental annotation and enabling curation at the community level. Bioinformatics. 2010;26:2354–2356. doi: 10.1093/bioinformatics/btq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca-Serra P, et al. Overcoming the ontology enrichment bottleneck with quick term templates. J. Appl. Ontol. 2011;6:13–22. [Google Scholar]
- Sansone SA, et al. Toward interoperable bioscience data. Nat. Genet. 2012;44:121–126. doi: 10.1038/ng.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbeck C, et al. MetaboLights: towards a new COSMOS of metabolomics data management. Metabolomics. 2012;8:757–760. doi: 10.1007/s11306-012-0462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetzel PL, et al. BioPortal: enhanced functionality via new Web services from the NCBO to access and use ontologies in software applications. Nucleic Acids Res. 2011;39:541–545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstencroft K, et al. Rightfield: embedding ontology annotation in spreadsheets. Bioinformatics. 2011;27:2021–2022. doi: 10.1093/bioinformatics/btr312. [DOI] [PubMed] [Google Scholar]
- Zeeberg BR, et al. Mistaken identifiers: gene name errors can be introduced inadvertently when using excel in bioinformatics. BMC Bioinformatics. 2004;5:80. doi: 10.1186/1471-2105-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]