Abstract
The promyelocytic leukemia zinc finger (PLZF) gene, involved in rare cases of acute promyelocytic leukemia, encodes a Krüppel-type zinc finger transcription factor. It has been reported that PLZF affects myeloid cell growth, differentiation, and apoptosis. However, the function of PLZF in the lymphoid compartment, where PLZF is also expressed, remains largely unknown. To investigate a potential relationship between PLZF expression in lymphocytes and programmed cell death, an inducible model of stable clones of the lymphoid Jurkat cell line was created by using the tet-off system. Although induction of PLZF expression by itself did not produce changes in the basal levels of apoptosis, PLZF had a significant anti-apoptotic effect in Jurkat cells cultured in conditions of serum starvation, as measured by annexin V staining and terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling. In addition, retarded loss of mitochondrial transmembrane potential was observed in the PLZF-expressing clones, suggesting that PLZF protects from cell death through a mitochondrial-dependent mechanism. To identify apoptosis-related targets of PLZF, a screen for differential expression identified BID, a proapoptotic member of the Bcl2 family, as significantly down-regulated by PLZF. Furthermore, a high-affinity PLZF-binding site element was identified upstream of the BID transcriptional start site, as assessed by electrophoretic mobility-shift assays. These results suggest that BID is a target of PLZF repression and a candidate gene to mediate the PLZF-induced resistance to apoptosis.
The promyelocytic leukemia zinc finger (PLZF) gene was identified in a variant of acute promyelocytic leukemia bearing reciprocal chromosomal translocations t(11;17)(q23;q21) whose breakpoints map within the PLZF locus at chromosome 11 and the retinoic acid receptor α locus at chromosome 17 (1). As a result of these translocations, chimeric proteins PLZF–retinoic acid receptor α and retinoic acid receptor α–PLZF are expressed in the malignant cells. Transgenic animal experiments support the hypothesis that both chimeric proteins contribute to the leukemogenic phenotype of acute promyelocytic leukemia (reviewed in refs. 2 and 3).
PLZF encodes a transcription factor containing nine C-terminal Cys2–His2 zinc finger motifs and an N-terminal BTB/POZ domain. Although the zinc finger domain binds DNA in a sequence-specific manner, it has not been defined which sequences are recognized by PLZF (4, 5). The BTB/POZ domain is an evolutionarily conserved domain that appears to convey transcriptional repressor activity (6). In addition, a central domain of PLZF interacts with the nuclear corepressors (N-CoR, SMRT, and mSIN3A), which in turn recruit histone deacetylases (7–10). This interaction provides a link between PLZF and an enzymatic activity considered one of the major mechanisms of chromatin remodeling and transcriptional silencing. Thus, both the BTB/POZ domain and the N-CoR interaction domain could contribute to the transcriptional repressor function attributed to PLZF.
The PLZF protein is localized to distinct nuclear speckles and appears to interact with the promyelocytic leukemia and BCL6 proteins in large nuclear bodies of unknown function (11, 12). In the human hematopoietic system, PLZF is expressed in pluripotent progenitor cells and in peripheral blood T, B, and NK lymphocytes (13, 14). Intriguingly, a significant proportion of B cell chronic lymphocytic leukemia (B-CLL) patients exhibit down-regulation of PLZF expression compared with that in normal lymphocytes, and this pattern of expression correlates with a better prognosis (14). Apoptotic functions appear to be suppressed in B-CLL, and progression could be related to increased resistance to apoptosis. A hypothetical role of PLZF in apoptosis could link PLZF expression with B-CLL prognosis.
In this article, an inducible model of stable clones of the lymphocytic Jurkat cell line, created by using the tet-off transfection plasmids, was exploited to test a potential role of the PLZF protein in apoptosis. It was shown that in Jurkat lymphocytes PLZF expression increased resistance to apoptosis when cells were stressed by serum starvation. In addition, screening differentially expressed apoptosis-related genes based on the presence or absence of PLZF expression identified BID, a proapoptotic member of the Bcl2 family, as a target of PLZF transcriptional repression.
Materials and Methods
Generation of Stable Clones with Inducible Expression of PLZF by Using the Tet-Off System. The T lymphocytic leukemia Jurkat cell line was grown in RPMI medium 1640 (BioWhittaker) supplemented with 10% heat-inactivated FCS (BioWhittaker), 50 units of penicillin per ml, 50 units of streptomycin per ml, and 2 mM l-glutamine (“complete medium”). Stable Jurkat clones with regulatable PLZF expression were generated by using the tet-off system, following a procedure previously described (14–16), and maintained in complete medium with 250 μg of hygromycin B per ml (Roche) and 1 mg of G418 per ml (Amersham Pharmacia Biosciences) [“complete selection medium” (CSM)], and 10 ng of doxycycline (dox) per ml.
Induction of Apoptosis by Serum Deprivation and Quantitation by Annexin V Labeling. PLZF-expressing and nonexpressing clones were washed free of dox, and replicate aliquots were adjusted at 0.3 × 106 cells per ml in CSM with 10 ng of dox per ml or without dox. After 72 h of culture, cells were washed twice and adjusted at 0.1 × 106 cells per ml in RPMI medium 1640 with the same supplements as the CSM except that FCS was added at 0.1% [“low-serum medium” (LSM)]. dox was added again to the replicates, which had been cultured with dox, and 0.5 × 106 cells of each replicate were taken every 24 h to determine the percentage of apoptotic cells. Cells were dual-labeled with annexin V and propidium iodide (Annexin V-FITC Apoptosis Detection kit I, Pharmingen). Labeled cells were acquired in a FACSCalibur flow cytometer (Becton Dickinson), and analysis of cell populations was performed by using the cellquest package. In certain control experiments, apoptosis was induced with 1 μg of anti-CD95 Ab per ml (Immunotech, Luminy, France).
Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling. DNA fragmentation that occurs in the cell nucleus as a consequence of the apoptosis process was measured by using the in situ cell death detection kit, fluorescein (Roche). Cell populations were acquired and analyzed in the flow cytometer.
Loss of Mitochondrial Transmembrane Potential (ΔΨm). Uptake of the f luorochrome 3,3′-dihexyloxacarbocyanine iodide [DiOC6(3)] by the mitochondrial matrix decreases as a consequence of the loss of ΔΨm that occurs as an early event of apoptosis (17). Cells were labeled with DiOC6(3) (Sigma) and analyzed in the flow cytometer.
Identification of Target Gene Modulation by Semiquantitative RT-PCR. mRNA levels of 41 genes related to apoptosis were examined by a semiquantitative RT-PCR technique. Oligonucleotide primers were designed from the mRNA sequences deposited in the UniGene database (www.ncbi.nih.nlm.gov). These included members of the Bcl2 family (BCL2, BCL2L1, BCL2L2, MCL1, BCL2A1, BAG1, BAX, BAK1, BID, BAD, BIK, BCL2L11, BNIP3, and BNIP3L), caspases (CASP2, CASP3, CASP6, CASP7, CASP8, and CASP9), programmed cell death (PDCD1, PDCD2, PDCD5, PDCD6, and PDCD8), transcription factors (TP53, MYC, PML, BCL6, STAT6, and TZFP), and other factors involved in diverse apoptotic pathways (such as TNFRSF6, TNFSF6, IL3RA, CYCS, APAF1, ADPRT, HSPA1A, DIABLO, BIRC1, and CARD4). To avoid coamplification of contaminant DNA, primers were mapped to different exons. As a control for equal loading of cDNA, a fragment of β-actin mRNA was coamplified in each reaction. Sequences of the oligonucleotides are available on request. Briefly, 1 μg of total RNA, obtained by using the SV Total RNA Isolation kit (Promega), was reverse transcribed by using random hexamers and 1 unit of Moloney murine leukemia virus retrotranscriptase (Roche) at 42°C for 50 min. One-tenth of this reaction was subjected to five cycles of PCR using the primers of an apoptosis-related gene and 2.5 units of Taq DNA polymerase (Bioline, London). Each PCR cycle consisted of 30 s at 95°C, 1 min at 55°C, and 3 min at 72°C. Then, β-actin primers were added, and 23 additional cycles were performed. Amplified products were fractionated and examined on a UV transilluminator. When necessary, gels were blotted, hybridized with [γ-32P]ATP-labeled oligonucleotide probes, and autoradiographed.
Northern Blot Analysis. Total RNA (40 μg) was electrophoresed, blotted, hybridized with an [α-32P]dCTP-labeled probe, and autoradiographed as described (14). Probes included a 750-bp cDNA BID fragment, obtained from the RT-PCR product by using the TOPO TA cloning kit (Invitrogen), a 2.1-kb cDNA PLZF fragment (1), and, as a control of equivalent loading, a human glyceraldehyde-3-phosphate dehydrogenase (GAPD) probe (Clontech). An estimation of the amounts of BID mRNA was performed by optical densitometry after normalization with GAPD mRNA.
Western Blot Analysis. Immunoreactive proteins contained in 10 μg of whole-cell protein extracts were detected as described (14). A mouse anti-human PLZF mAb (Oncogene Research Products), a rabbit anti-human BID polyclonal Ab (Pharmingen), and a mouse anti-actin mAb (Santa Cruz Biotechnology) were used as primary Abs. Horseradish peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG (DAKO) was used as secondary Ab. The amounts of BID protein were estimated by optical densitometry after normalization with actin.
Quantitation of Target Gene Modulation by Flow Cytometry. Cells were labeled by using the Perm/Fix kit (Pharmingen) by using anti-PLZF or -BID primary Abs, and phycoerythrin-conjugated anti-mouse or rabbit IgG (Caltag, South San Francisco, CA) secondary Abs, respectively. Labeled cells were acquired and analyzed in the flow cytometer. Other apoptosis regulators were also analyzed by Western blot and/or flow cytometry and served as controls (anti-human BCL2, BCLX, BCLW, MCL1, BAX, BAK, BAD, BIK, BIM, BAG1, P53, CD95, or FASL, purchased from DAKO, Pharmingen, R&D Systems, or Immunotech).
Electrophoretic Mobility-Shift Assays (EMSAs). Sequences flanking the transcriptional start site (TSS) of human BID (http://elmo.ims.u-tokyo.ac.jp/dbtss/) were scanned for homologies with high-affinity PLZF-binding sites as described (4, 5). A sequence with significant homology to the PLZF-binding site contained within the yeast LEXA operator was found ≈2 kb upstream of the TSS. Nuclear extracts of the Jurkat clones containing 15 μg of protein were incubated for 20 min with a double-stranded [γ-32P]ATP-labeled probe made by using the oligonucleotide 5′-ATTTTACATACAGTAAAACTCACCATTCTTCGTGCAGAGT-3′, which maps at positions 1984–1945 nt upstream of BID TSS, and its complement. A DR5-retinoic acid response element (RARE), 5′-GATCAGGGTTCACCGAAAGTTCACTCGCATATATTAG-3′, and a p53-binding site element, 5′-CTAGGGACATGCCCGGGCATGTCCTAG-3′, were used as irrelevant competitors. For supershifts, 0.1 μg of Abs was added 10 min before the nuclear extracts. Binding reaction products were electrophoresed in 10% polyacrylamide gels for 2 h. Gels were dried and autoradiographed.
Results
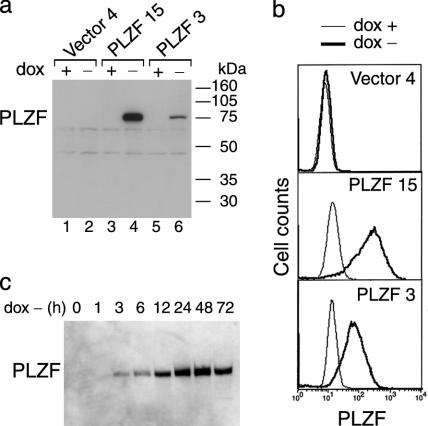
Generation of Stable Clones with Inducible Expression of PLZF. To investigate the potential role of PLZF in apoptosis, stable clones of the lymphocytic leukemia Jurkat cell line with inducible expression of PLZF were generated by using the tet-off system. Two of 21 clones selected, PLZF 3 and PLZF 15, expressed PLZF upon dox withdrawal at the expected molecular mass of 80 kDa (Fig. 1a, lanes 4 and 6). dox exerted tight repression of PLZF (Fig. 1a, lanes 3 and 5). WT Jurkat cells do not express PLZF, as measured by Western blotting or a more sensitive RT-PCR assay (data not shown). Consequently, Jurkat clones obtained by transfection with pJEF4 vector did not express PLZF in the presence or absence of dox (Fig. 1a, lanes 1 and 2). After flow cytometry analysis, comparison of the mean levels of fluorescence revealed that clone PLZF 15 exhibited ≈4-fold higher levels of PLZF expression than clone 3 (Fig. 1b). Maximum levels of PLZF expression were achieved 24 h after washing the cells free of dox and remained stable afterward (Fig. 1c).
Fig. 1.
Analysis of PLZF protein expression in the inducible clones. (a) Western blot prepared from whole-cell extracts of human Jurkat cells stably transfected with pJEF4 (clone vector 4) or pJEF4-PLZF (clones PLZF 3 and 15), obtained 72 h after washing the cells free of dox and reculturing in the presence (lanes 1, 3, and 5) or absence (lanes 2, 4, and 6) of dox, incubated with a PLZF mAb. A band at the expected molecular mass of 80 kDa was detected in clones 3 and 15 after dox withdrawal. (b) Flow cytometry analysis of the clones by using the PLZF mAb and secondary labeling with anti-mouse IgG coupled to phycoerythrin. (c) Western blot of whole-cell extracts of clone PLZF 15 showing the kinetics of induction of PLZF expression after dox withdrawal.
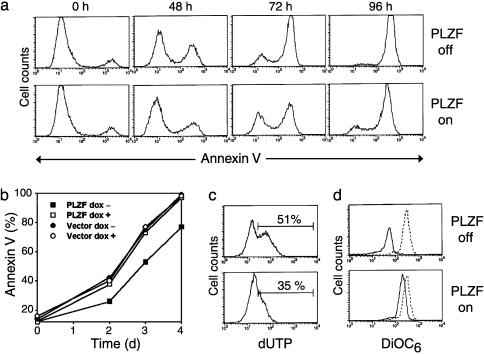
Expression of PLZF Delays the Kinetics of Apoptosis Induced by Serum Starvation. Induction of PLZF expression upon dox withdrawal did not produce changes in the basal levels of apoptosis of Jurkat cells cultured in CSM. To study whether PLZF expression could influence the response of the cells to apoptotic stimuli, replicates of the Jurkat clones grown for 72 h in the presence and absence of dox were recultured in LSM. Analysis of cell death induced by serum deprivation in Jurkat cells showed that annexin V-positive/propidium iodide-negative cells predominate in the earlier stages (24–48 h), and double-positive cells predominate in the later stages (72–96 h), suggesting a transition from single- to double-positive cells (data not shown). Therefore, to establish the kinetics of apoptosis, all annexin V-positive cells were counted as apoptotic cells. Clones PLZF 3 (Fig. 2a) and 15 (not shown) cultured in the absence of dox showed a significant delay in the kinetics of annexin V staining compared with the replicates cultured in the presence of dox. Maximal differences in the values of annexin V-positive cells were observed between 48 and 72 h of culture in LSM. These ranged between 20% and 25% less annexin V-positive cells in the replicates grown in the absence of dox. Control clones transfected with the empty vector did not show differences in the kinetics of apoptosis induced by serum starvation in the presence or absence of dox (Fig. 2b), ruling out that the differences observed in the positive clones might be due to a potential toxic effect of dox. Therefore, PLZF has an antiapoptotic effect in Jurkat cells cultured in conditions of serum starvation.
Fig. 2.
Flow cytometry analysis of apoptosis of the Jurkat clones induced by serum starvation in the presence (PLZF off) or absence (PLZF on) of dox. (a) Kinetics of annexin V staining of Jurkat clone PLZF 3. (b) Graph comparing the kinetics of annexin V staining of Jurkat clone PLZF 3 shown in a with clone vector 4. (c) Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling assay of clone PLZF 3 after 72 h of culture in LSM. (d) Loss of ΔΨm of clone PLZF 3 after 6 h of culture in LSM. The dashed line represents a control culture of clone PLZF 3 grown in CSM.
Protection Promoted by PLZF Involves Decreased Nuclear Fragmentation and Retarded Loss of ΔΨm. As a confirmation of the protective effect of PLZF, the Jurkat clones cultured in LSM were assayed by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling. PLZF-expressing clones stressed by serum deprivation in the absence of dox exhibited decreased labeling compared with replicates stressed in the presence of dox (Fig. 2c). These differences were not observed in control clones transfected with empty vector (data not shown). To test the involvement of mitochondria in PLZF down-regulation of apoptosis, loss of ΔΨm was analyzed by DiOC6(3) labeling. Replicates of the positive clones cultured with dox showed decreased ΔΨmafter6hofserumstarvation compared with those cultured without dox (Fig. 2d). Such differences were not observed in the control clones transfected with empty vector (data not shown). These data suggest that PLZF expression negatively interferes with serum deprivation-induced apoptosis upstream of the mitochondrial changes associated with this death pathway.
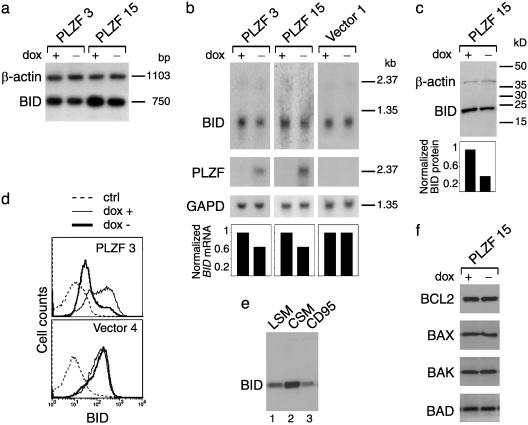
Expression of PLZF Induces Repression of BID, a Proapoptotic Member of the Bcl2 Family of Apoptosis Regulators. To identify apoptosis-related targets of the transcriptional activity of PLZF, a screen for differential expression of apoptosis-related genes was performed by semiquantitative RT-PCR. Among these genes, only BID, a proapoptotic member of the Bcl2 family of apoptosis regulators, was identified as significantly down-regulated in Jurkat clones PLZF 3 and 15 when PLZF expression was induced by culture in dox-free medium (Fig. 3a). Northern blotting confirmed BID mRNA repression in the PLZF-expressing clones upon culture in the absence of dox, whereas control clones transfected with empty vector in the presence and absence of dox showed equivalent expression (Fig. 3b). Western blotting using a polyclonal anti-human BID Ab detected a band at the expected molecular mass of BID (22 kDa), which was significantly down-regulated (2.7-fold) when PLZF expression was induced (Fig. 3c). Comparison of the mean levels of fluorescence by fluorescence-activated cell sorter analysis also revealed that expression of BID protein was ≈2.8-fold down-regulated during 96 h of culture of clone PLZF 3 in the absence of dox (Fig. 3d).
Fig. 3.
Analysis of BID mRNA and protein expression in the Jurkat clones after 72 h of culture in the presence (+) or absence (–) of dox. (a) RT-PCR showing BID mRNA down-regulation in clones PLZF 3 and 15 after culture in the absence of dox. (b) Northern blot showing BID mRNA down-regulation in clones PLZF 3 and 15 but not in control clone vector 1 after culture in the absence of dox. Bar graphs depict estimations of BID mRNA amount after normalization with GAPD, expressed in relative units, 1 being the value given to the culture in the presence of dox of each clone. (c) Western blot showing BID protein down-regulation in whole-cell extracts of clone PLZF 15 after culture in the absence of dox. The bar graph depicts an estimation of BID protein amount after normalization with actin, expressed in relative units as in b. (d) Flow cytometry analysis showing BID protein down-regulation in clone PLZF 3 after culture in the absence of dox. (e) Western blot showing BID protein cleavage in whole-cell extracts of clone PLZF 15 in the presence of after culture in LSM for 48 h (lane 1) or in CSM with anti-CD95 for 4 h (lane 3) compared with control grown in CSM (lane 2). (f) Western blot showing no changes in BCL2, BAX, BAK, and BAD protein expression in whole-cell extracts of clone PLZF 15 after 72 h of culture in the presence (+) or absence (–) of dox.
BID plays an important role in apoptosis induced by diverse stimuli in different cell types, where it is cleaved to produce an active truncated form that translocates from the cytosol to the mitochondria (18–21). Among these stimuli, CD95 (FASR/TNFRSF6) stimulation induces a rapid apoptotic process in Jurkat cells, and BID cleavage contributes to this pathway (22). To establish whether BID may be involved in serum deprivation-induced apoptosis of Jurkat cells, whole-cell extracts of serum-deprived Jurkat cells were compared with nondeprived or anti-CD95 Ab-stimulated Jurkat cells by Western blot analysis. Dramatic decreases of full-length BID were observed after culture in LSM for 48 h or with anti-CD95 for 4 h (Fig. 3e, lanes 1 and 3, respectively) compared with culture in CSM (Fig. 3e, lane 2). These results suggest that, as in CD95-induced apoptosis, BID is cleaved in serum deprivation-induced apoptosis. Furthermore, in concordance with RT-PCR data, no regulation of other apoptosis regulators was observed in the PLZF-expressing or nonexpressing Jurkat clones cultured in the presence or absence of dox by Western blotting (Fig. 3f) and/or flow cytometry (data not shown). These results indicate that BID is a specific target of PLZF transcriptional repression and a candidate protein to mediate the increased resistance to serum deprivation-induced apoptosis conferred to Jurkat cells by the expression of PLZF.
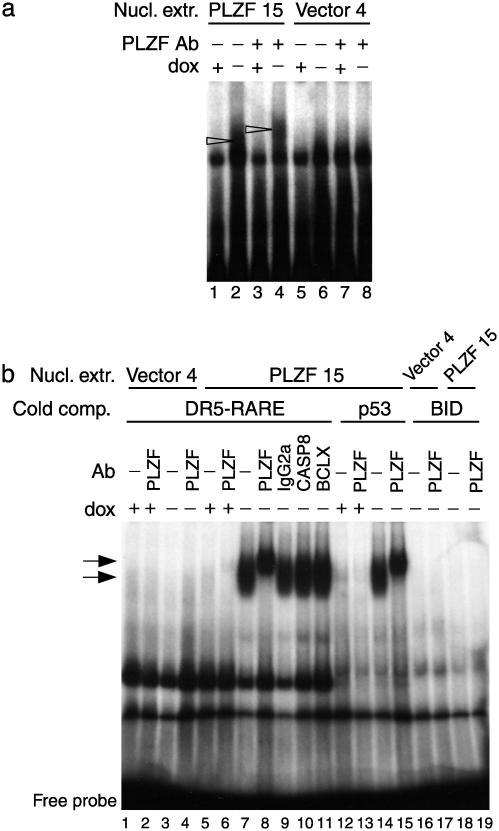
PLZF Is Found in a Complex That Binds a Specific Sequence Within the BID Promoter. To investigate the mechanism of repression, the BID promoter was examined. A sequence that maps ≈2 kb upstream of the human BID TSS, significantly homologous to the PLZF-binding site contained in the yeast LEXA operator, was tested by EMSA. A retarded complex was detected when nuclear extracts of clone PLZF 15 cultured in the absence of dox were incubated with the labeled probe (Fig. 4a, lane 2). The complex was shifted with a PLZF mAb (Fig. 4a, lane 4) and was not observed in the presence of dox (Fig. 4a, lanes 1 and 3) or when extracts of clone vector 4 were used (Fig. 4a, lanes 5–8). The specificity of PLZF binding to this sequence was shown in competition EMSA (Fig. 4b). The retarded complex was detected in the presence of a 1,000-fold molar excess of inlabeled competitors: a DR5-RARE (Fig. 4b, lanes 7 and 9–11) or a p53-binding site probe (Fig. 4b, lane 14), whereas it disappeared in the presence of the specific competitor (Fig. 4b, lanes 18 and 19). This complex was not observed when the clone was cultured in the presence of dox (Fig. 4b, lanes 5, 6, 12, and 13) or when nuclear extracts of clone vector 4 were used (Fig. 4b, lanes 1–4). The complex was shifted with a PLZF mAb (Fig. 4b, lanes 8 and 15) but not with irrelevant isotype Abs (Fig. 4b, lanes 9–11). These findings indicate that the PLZF protein could target BID for transcriptional repression through binding to specific sequences of its promoter.
Fig. 4.
EMSA analysis using a sequence contained upstream of the BID TSS and nuclear extracts (Nucl. extr.) of the Jurkat clones PLZF 15 and vector 4. (a) EMSA using the BID radiolabeled probe without competitors. Arrowheads denote the presence of a retarded complex observed when nuclear extracts of clone PLZF 15 cultured in the absence of dox were used (lane 2) and the shifted complex when a PLZF mAb was used (lane 4). (b) EMSA using BID-radiolabeled probe with a 1,000-fold molar excess of unlabeled competitors (a DR5-RARE, lanes 1–11; a p53-binding site probe, lanes 12–15; or the specific unlabeled probe, lanes 16–19). Arrows denote the presence of a retarded complex observed when nuclear extracts of clone PLZF 15 cultured in the absence of dox were used (lanes 7, 9–11, and 14), and the shifted complex when a PLZF mAb was used (lanes 8 and 15). Irrelevant Abs failed to produce a shift (lanes 9–11). The complex was detected in the presence of a 1,000-fold molar excess of the irrelevant unlabeled competitors (lanes 7–11, 14, and 15), whereas it disappeared in the presence of the specific unlabeled probe (lanes 18 and 19).
Discussion
PLZF, like other members of a protein family characterized by a structure that consists of an N-terminal BTB/POZ domain and C-terminal Cys2–His2 zinc finger domains, e.g., BCL6 and TZFP (23, 24), acts as a transcriptional repressor (4, 25). It is likely that PLZF binds specific DNA sequences within the regulatory regions of its target genes through its zinc finger domain and recruits histone deacetylase through its interaction with nuclear corepressors, e.g., N-CoR, SMRT, and mSIN3A (7–10, 26). Histone deacetylase enzymatic activity would render the regulatory regions of PLZF target genes transcriptionally inactive. Other nuclear factors that interact with PLZF, such as ETO (25), might participate in these multiprotein complexes and cooperate with PLZF to repress transcription.
Shaknovich et al. (27), by means of retrovirally induced overexpression of PLZF in the murine IL-3-dependent hematopoietic progenitor 32D cell line, concluded that PLZF may be an important regulator of myeloid cell growth, differentiation, and apoptosis. However, the role of PLZF in apoptosis was reported to be paradoxical. Thus, when 32D cells were grown in the presence of IL-3, a moderate proapoptotic effect of PLZF was exhibited. In striking contrast, a strong antiapoptotic effect of PLZF was observed when 32D cells were grown in the absence of IL-3. In B-CLL, the observation that decreased levels of PLZF expression are associated with a better prognosis suggested the possibility that PLZF may affect the apoptotic phenotype of B-CLL cells (14). Although the mechanism of PLZF dysregulation in B-CLL is not known, a potential antiapoptotic function of PLZF would be concordant with a lower survival of PLZF low-expressor malignant cells and a better prognosis for these patients.
The Jurkat lymphoid cell line, which does not express PLZF, has been widely used in apoptosis studies because the cells are sensitive to diverse stress stimuli, such as serum deprivation, genotoxic agents, or death receptor ligation. In addition, the frequency of apoptotic Jurkat cells can be readily quantified by flow cytometry. The tet-off inducible system developed here allows the examination of the functional consequences of PLZF expression within the same clone, thus eliminating clonality biases. By means of such a cell system, it was observed that Jurkat clones induced to express PLZF produced an antiapoptotic response of the cells stressed by serum deprivation, measured by preservation of plasma membrane integrity, nuclear DNA fragmentation, or mitochondrial membrane depolarization.
We sought to identify potential apoptosis-related PLZF targets for transcriptional regulation in Jurkat cells. A semiquantitative RT-PCR method that uses β-actin as an internal control was developed to detect modulation in the levels of mRNA expression of a variety of apoptosis regulators or effectors, including Bcl2 family members, caspases, and transcription factors, among others. Only BID was shown to be significantly repressed in the PLZF-expressing clones upon dox removal. The repression of BID was confirmed at the mRNA level by Northern blotting and at the protein level by Western blotting and flow cytometry. Furthermore, ≈2 kb upstream of the BID TSS we detected the sequence TTTTACATACAGTA, which is significantly homologous to the sequence TTATATGTACAGTA contained within the PLZF-binding site of the yeast LEXA operator (4). EMSA using Jurkat nuclear protein extracts showed that PLZF participates in a complex that binds with high affinity to a double-stranded oligonucleotide synthesized from this region of the BID promoter. These findings suggest that PLZF targets BID, an apoptosis agonist, for transcriptional repression. BID activation during serum deprivation-induced apoptosis suggests that this mechanism may contribute to this form of apoptosis in Jurkat cells.
BID is a cytosolic protein that induces cytochrome c release from mitochondria in response to caspase 8, the caspase activated by cell surface death receptors such as FAS receptor and tumor necrosis factor receptor. Activated caspase 8 cleaves BID, and the C-terminal fragment translocates to mitochondria, where it triggers cytochrome c release, loss of ΔΨm, cell shrinkage, and nuclear condensation (18–20). The caspase-activated form of BID activates proapoptotic BAX or BAK to produce large membrane openings that allow the translocation of large mitochondrial proteins during apoptosis (21, 28). Krajeswska et al. (29) reported that BID expression in normal tissues varies widely, being prominent in several types of shortlived cells and in apoptosis-sensitive cells. Evidence of BID dysregulation in B-CLL has also been reported (30). Little is known about regulation of BID expression. Sax et al. (31) reported that p53 up-regulates BID expression and increases cellular chemosensitivity. Taken together, these data suggest that BID regulation may be relevant in human cancer.
Our findings support the notion that PLZF can repress programmed cell death in lymphoid cells stressed by nutrient deprivation and thus are consistent with previously reported data obtained by using a different cell type stressed by growth factor deprivation (27). Whether PLZF may control the response of lymphoid cells to other apoptotic stimuli remains to be studied. It is curious that both Jurkat clones expressed different PLZF levels but none of the effects observed were significantly different. One possible explanation is that PLZF expression in both clones reaches levels higher than those required for its maximal activity in Jurkat cells under these physiological conditions. Thus, whether PLZF involvement in apoptosis may depend on its level of expression or the cellular context also remains to be elucidated.
We conclude that BID repression could be a mechanism to mediate the antiapoptotic effect of PLZF, and we hypothesize that BID repression by PLZF plays a role in lymphoid malignancies where PLZF is dysregulated, such as B-CLL. Therefore, analysis of the potential role of PLZF and BID in lymphoid malignancies may be promising fields for future research.
Acknowledgments
We thank Dr. Zhu Chen for the pSG5-PLZF plasmid, Drs. Jane Eike Floettmann and Martin Rowe for the pJEF3 and pJEF4 plasmids, and Dr. Hermann Bujard for the pUHC13-3 luciferase reporter. This work was suported by grants from the Fondo de Investigaciones Sanitarias of Spain (personal grant 99/3039 to A.P., project grant 01/0745, and equipment grant 01/3622), the Leukaemia Research Fund of Great Britain, the Welsh Bone Marrow Transplant Research Fund, the Eli Lilly International Foundation, and the Fondation de France.
Abbreviations: PLZF, promyelocytic leukemia zinc finger; DiOC6(3), 3,3′-dihexyloxacarbocyanine iodide; EMSA, electrophoretic mobility-shift assay; CSM, complete selection medium; LSM, low-serum medium; ΔΨm, mitochondrial transmembrane potential; dox, doxycycline; TSS, transcriptional start site; B-CLL, B cell chronic lymphocytic leukemia; RARE, retinoic acid response element.
References
- 1.Chen, Z., Brand, N. J., Chen, A., Chen, S.-J., Tong, J.-H., Wang, Z. Y., Waxman, S. & Zelent, A. (1993) EMBO J. 12, 1161–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He, L.-Z., Merghoub, T. & Pandolfi, P. P. (1999) Oncogene 18, 5278–5292. [DOI] [PubMed] [Google Scholar]
- 3.Parrado, A., Chomienne, C. & Padua, R. A. (2000) Leuk. Lymphoma 39, 271–282. [DOI] [PubMed] [Google Scholar]
- 4.Sitterlin, D., Tiollais, P. & Transy, C. (1997) Oncogene 14, 1067–1074. [DOI] [PubMed] [Google Scholar]
- 5.Li, J.-Y., English, M. A., Ball, H. J., Yeyati, P. L., Waxman, S. & Licht, J. D. (1997) J. Biol. Chem. 272, 22447–22455. [DOI] [PubMed] [Google Scholar]
- 6.Bardwell, V. J. & Treisman, R. (1994) Genes Dev. 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- 7.Hong, S.-H, David, G., Wong, C.-W., Dejean, A. & Privalsky, M. L. (1997) Proc. Natl. Acad. Sci. USA 94, 9028–9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin, R. J., Nagy, L., Inoue, S., Shao, W., Miller, W. H., Jr., & Evans, R. M. (1998) Nature 391, 811–814. [DOI] [PubMed] [Google Scholar]
- 9.Grignani, F., De Matteis, S., Nervi, C., Tomassoni, L., Gelmetti, V., Cioce, M., Fanelli, M., Ruthardt, M., Ferrara, F. F., Zamir, I., et al. (1998) Nature 391, 815–818. [DOI] [PubMed] [Google Scholar]
- 10.Guidez, F., Ivins, S., Zhu, J., Söderström, M., Waxman, S. & Zelent, A. (1998) Blood 91, 2634–2642. [PubMed] [Google Scholar]
- 11.Koken, M. H. M., Reid, A., Quignon, F., Chelbi-Alix, M. K., Davies, J. M., Kabarowski, J. H. S., Zhu, J., Dong, S., Chen, S.-J., Chen, Z., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 10255–10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhordain, P., Albagli, O., Honoré, N., Guidez, F., Lantoine, D., Schmid, M., de Thé, H., Zelent, A. & Koken, M. H. M. (2000) Oncogene 19, 6240–6250. [DOI] [PubMed] [Google Scholar]
- 13.Reid, A., Gould, A., Brand, N., Cook, M., Strutt, P., Li, J., Licht, J., Waxman, S., Krumlauf, R. & Zelent, A. (1995) Blood 86, 4544–4552. [PubMed] [Google Scholar]
- 14.Parrado, A., Noguera, M. E., Delmer, A., McKenna, S., Davies, J., Le Gall, I., Bentley, P., Whittaker, J., Sigaux, F., Chomienne, C. & Padua, R. A. (2000) Hematol. J. 1, 15–27. [DOI] [PubMed] [Google Scholar]
- 15.Floettmann, J. E., Ward, K., Rickinson, A. B. & Rowe, M. (1996) Virology 223, 29–40. [DOI] [PubMed] [Google Scholar]
- 16.Gossen, M. & Bujard, H. (1992) Proc. Natl. Acad. Sci. USA 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamzami, N., Marchetti, P., Castedo, M., Zanin, C., Vayssiere, J. L., Petit, P. X. & Kroemer, G. (1995) J. Exp. Med. 181, 1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo, X., Budihardjo, I., Zou, H., Slaughter, C. & Wang, X. (1998) Cell 94, 481–490. [DOI] [PubMed] [Google Scholar]
- 19.Li, H., Zhu, H., Xu, C. & Yuan, J. (1998) Cell 94, 491–501. [DOI] [PubMed] [Google Scholar]
- 20.Desagher, S., Osen-Sand, A., Nichols, A., Eskes, R., Montessuit, S., Lauper, S., Maundrell, K., Antonsson, B. & Martinou, J. C. (1999) J. Cell Biol. 144, 891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei, M. C., Zong, W.-X., Cheng, E. H.-Y., Lindsten, T., Panoutsakopoulou, V., Ross, A. J., Roth, K. A., MacGregor, G. R., Thompson, C. B. & Korsmeyer, S. J. (2001) Science 292, 727–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner, A. B., de Vries, E., Tait, S. W. G., Bontjer, I. & Borst, J. (2002) J. Biol. Chem. 277, 40760–40767. [DOI] [PubMed] [Google Scholar]
- 23.Baron, B. W., Anastasi, J., Thirman, M. J., Furukawa, Y., Fears, S., Kim, D. C., Simone, F., Birkenbach, M., Montag, A., Sadhu, A., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 2860–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miaw, S. C., Choi, A., Yu, E., Kishikawa, H. & Ho, I. C. (2000) Immunity 12, 323–333. [DOI] [PubMed] [Google Scholar]
- 25.Melnick, A. M., Westendorf, J. J., Polinger, A., Carlile, G. W., Arai, S., Ball, H. J., Lutterbach, B., Hiebert, S. W. & Licht, J. D. (2000) Mol. Cell. Biol. 20, 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.David, G., Alland, L., Hong, S.-H., Wong, C.-W., DePinho, R. A. & Dejean, A. (1998) Oncogene 16, 2549–2556. [DOI] [PubMed] [Google Scholar]
- 27.Shaknovich, R., Yeyati, P. L., Ivins, S., Melnick, A., Lempert, C., Waxman, S., Zelent, A. & Licht, J. D. (1998) Mol. Cell. Biol. 18, 5533–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuwana, T., Mackey, M. R., Perkins, G., Ellisman, M. H., Latterich, M., Schneiter, R., Green, D. R. & Newmeyer, D. D. (2002) Cell 111, 331–342. [DOI] [PubMed] [Google Scholar]
- 29.Krajewska, M., Zapata, J. M., Meinhold-Heerlein, I., Hedayat, H., Monks, A., Bettendorf, H., Shabaik, A., Bubendorf, L., Kallioniemi, O. P., Kim, H., et al. (2002) Neoplasia 4, 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein, U., Tu, Y., Stolovitzky, G. A., Mattioli, M., Cattoretti, G., Husson, H., Freedman, A., Inghirami, G., Cro, L., Baldini, L., et al. (2001) J. Exp. Med. 194, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sax, J. K., Fei, P., Murphy, M. E., Bernhard, E., Korsmeyer, S. J. & El-Deiry, W. S. (2002) Nat. Cell Biol. 4, 842–849. [DOI] [PubMed] [Google Scholar]