Abstract
We report a whole-genome shotgun assembly (called WGSA) of the human genome generated at Celera in 2001. The Celera-generated shotgun data set consisted of 27 million sequencing reads organized in pairs by virtue of end-sequencing 2-kbp, 10-kbp, and 50-kbp inserts from shotgun clone libraries. The quality-trimmed reads covered the genome 5.3 times, and the inserts from which pairs of reads were obtained covered the genome 39 times. With the nearly complete human DNA sequence [National Center for Biotechnology Information (NCBI) Build 34] now available, it is possible to directly assess the quality, accuracy, and completeness of WGSA and of the first reconstructions of the human genome reported in two landmark papers in February 2001 [Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304–1351; International Human Genome Sequencing Consortium (2001) Nature 409, 860–921]. The analysis of WGSA shows 97% order and orientation agreement with NCBI Build 34, where most of the 3% of sequence out of order is due to scaffold placement problems as opposed to assembly errors within the scaffolds themselves. In addition, WGSA fills some of the remaining gaps in NCBI Build 34. The early genome sequences all covered about the same amount of the genome, but they did so in different ways. The Celera results provide more order and orientation, and the consortium sequence provides better coverage of exact and nearly exact repeats.
In 2000 Celera scientists in collaboration with the publicly funded Drosophila Genome Project published the whole-genome assembly of the Drosophila genome (1) with a description of the paired end sequencing strategy and the new algorithms (2) that enabled this historic assembly. Over the subsequent 2 years, remaining gaps in the Drosophila genome sequence were closed, and the order and orientation of the sequence were confirmed. The completed Drosophila genome sequence permitted a retrospective analysis of the quality of the initial whole-genome shotgun assembly (3). This study demonstrated that the computationally assembled genome sequence was highly accurate and served as a good substrate for finishing a eukaryotic genome (3).
In February 2001 both Celera and the International Human Genome Sequencing Consortium (IHGSC) published their first drafts of the human genome sequence (4, 5). In 2001 Celera conducted a whole-genome shotgun sequencing and assembly of the mouse genome based only on 26 million sequence reads generated at Celera (6) by using a refined version of the assembly software. The quality of the mouse assembly exceeded the quality of the reported (4) human assemblies, prompting a new assembly, called WGSA, of the human genome based on only Celera-generated data and bacterial artificial chromosome (BAC) end sequences (7, 8). In 2003 the National Center for Biotechnology Information (NCBI) released Build 34 of the human genome, hereafter referred to as NCBI-34 (9, 10). Although this new sequence is not perfect and still has gaps, it constitutes a high-quality reference against which to evaluate the other human genome constructs and assemblies. We analyzed WGSA as well as the published sequences (4, 5) to see how much of the NCBI-34 sequence they cover and how well they reconstructed the order and orientation of the sequence.
The independence of the genome assemblies reported by Celera (4) was challenged in this journal by the principal leaders of the IHGSC (11, 12). Therefore, we also show the differences in the results reported in refs. 4 and 5 by analyzing which parts of NCBI-34 are covered by each genome assembly. The assemblies cover comparable amounts of the genome but do so in clearly different patterns. As one would expect given 39 times coverage of the human genome in paired-end-sequenced plasmids, all three Celera assemblies have better order and orientation than the consortium sequence (5). The consortium's clone by clone sequencing method, using BACs (5), resulted in better coverage of exact and nearly exact sequence repeat regions. Because of the presence of both male and female donors for Celera's shotgun sequence, the coverage of the X and especially the Y chromosomes is lower than that for the other chromosomes, resulting in a lower-quality assembly for these chromosomes.
We have submitted the three Celera human genome sequences to GenBank to preserve the historical record and facilitate the ongoing analysis of the human genome.
Whole-Genome Shotgun Sequence of the Human Genome
Although whole-genome shotgun sequencing was initially considered controversial when the first genome was sequenced (13), it has now become the prevailing approach. The vast majority of genomes sequenced to date have used this method (14), including the large genomes of Drosophila (1), Anopheles (15), Mus (6, 16), Fugu (17), and Canis (18).
We present here a WGSA produced by Celera in December 2001 using only whole-genome shotgun sequence data. The Celera shotgun data set consisted of 27 million sequencing reads, of average quality-trimmed length 543 bp, organized in pairs by virtue of end-sequencing 2-kbp, 10-kbp, and 50-kbp inserts from shotgun clone libraries as described (1, 4). The trimmed sequence reads covered the genome 5.3 times, and the inserts for which pairs of reads were obtained covered the genome 39 times. In addition, 104,000 BAC-end-sequence pairs (7, 8) were used to augment the 50-kbp pairs in providing long range correlations. Assembly was performed with the Celera Assembler, originally described in ref. 2 with improvements made after publication of ref. 4.
For such a low level of coverage and such a large genome, the assembly is remarkably coherent, consisting of 330 large scaffolds that constitute 99% of the result, with the remaining 1% divided across 4,610 small scaffolds under 100 kbp. The comparison of WGSA against NCBI-34 allows us to measure its completeness and quality, and to gauge the effort that would be required to finish a mammalian genome from such a sequence. The scaffolds of WGSA span 96.3% of NCBI-34, and the contigs of these scaffolds reconstruct 92.7% of the NCBI-34 sequence. Gaps comprised of a missing region of sequence in an existing clone are generally trivial to close. Of 206,552 such gaps between the contigs of the scaffolds, 201,735 are spanned by at least one 2-kbp or 10-kbp end-sequenced insert (as opposed to only BACs or 50-kbp inserts). All but 651 of the spanned gaps are flanked by contigs whose order and orientation is consistent with NCBI-34. Thus, nearly half of the uncovered 7.3% of NCBI-34 (3.6%) could be obtained simply by primer directed sequencing of the gaps in WGSA's existing scaffolds. Moreover, 2,218 of the 4,610 small scaffolds under 100 kbp are subsumed by larger scaffolds and could be properly placed during insert-based gap finishing.
Both clone ordered and whole-genome shotgun sequence assemblies have had difficulties resolving the structure of large, highly identical duplications (refs. 19 and 20; Table 1). More than 83 Mbp of the 170 Mbp of NCBI-34 that are not represented in WGSA scaffolds involve such duplications. For WGSA, the largest concentration of duplicated sequence is within the unplaced scaffolds: 23% of the unplaced scaffold sequence is so annotated, accounting for 12% of the duplicated sequence that is present in WGSA. Random BAC sampling, or selected BAC sampling based on sequence-anchored probes, could be used to find clones spanning these regions in WGSA. In addition, the shotgun sequence has proven essential for evaluating the nature and extent of these duplications (20).
Table 1. Comparison of selected assemblies.
| Statistic | Notes | Assembly | |||||
|---|---|---|---|---|---|---|---|
| Assembly | a | WGA | CSA | HG06 | WGSA | NCBI-28 | NCBI-34 |
| Producer | b | Celera | Celera | UCSC | Celera | NCBI | NCBI |
| Method | c | WG | C | H | WG | H | H |
| Data source | d | Combined | Combined | IHGSC | Celera | IHGSC | IHGSC |
| Associated date | e | Nov. 2000 | Jan. 2001 | Dec. 2000 | Dec. 2001 | Dec. 2001 | Oct. 2003 |
| Intrinsic measures | |||||||
| acgt in assembly, Mbp | f | 2,587 | 2,656 | 2,742 | 2,696 | 2,853 | 2,865 |
| acgt unmapped, Mbp | g | 280 | 60 | 37 | 36 | 58 | 22 |
| No. of contigs | h | 221,036 | 169,157 | 133,667 | 211,493 | 47,117 | 512 |
| No. of scaffolds | i | 118,968 | 54,061 | 76,058 | 4,940 | 42,754 | 447 |
| N50 contig length, kbp | j | 53 | 98 | 110 | 23 | 575 | 29,105 |
| N50 scaffold length, kbp | j | 3,563 | 2,954 | 331 | 29,133 | 613 | 36,791 |
| Scaffold span, Mbp | k | 2,848 | 2,909 | 2,833 | 2,819 | 2,855 | 2,869 |
| RefSeq (50% cov, 95% id) | l | 17,348 | 18,305 | 18,122 | 19,149 | 18,810 | 19,613 |
| Segmental duplication, Mbp | m | 27.3 | 54.5 | 108.0 | 69.5 | 120.0 | 152.3 |
| Seg. dup. in unmapped, Mbp | n | 13.9 | 5.1 | 2.9 | 8.3 | 2.7 | 5.1 |
| Confirmed conflicted mates | o | 0.38% | 0.91% | 5.61% | 0.31% | 2.44% | 0.28% |
| Mates linking mapped + unmapped | p | 1.52% | 0.16% | 0.03% | 0.13% | 0.02% | 0.01% |
| Comparison to NCBI-34 | |||||||
| No. of matches | q | 256,021 | 208,148 | 150,624 | 308,371 | 60,544 | |
| No. of runs | q | 12,560 | 47,540 | 71,291 | 7,315 | 23,024 | |
| No. of clumps | q | 1,595 | 1,187 | 3,189 | 339 | 2,951 | |
| acgt in matches, Mbp | r | 2,498 | 2,520 | 2,495 | 2,657 | 2,653 | |
| Extra sequence, Mbp | s | 89 | 136 | 247 | 38 | 200 | |
| Missing sequence, Mbp | t | 367 | 345 | 370 | 208 | 212 | |
| acgt in runs, Mbp | u | 2,557 | 2,650 | 2,553 | 2,759 | 2,682 | |
| N50 match length, kbp | v | 27 | 33 | 47 | 15 | 306 | |
| N50 run length, kbp | v | 1,204 | 441 | 203 | 1,959 | 954 | |
| N50 clump length, kbp | v | 5,404 | 5,931 | 1,809 | 33,501 | 2,765 | |
| Percent of acgt in matches to NCBI-34 in: | |||||||
| Global HCS | w | 79.86% | 78.65% | 72.73% | 95.96% | 77.35% | |
| Unmapped scaffolds | x | 8.76% | 1.50% | 0.78% | 0.78% | 0.41% | |
| Mismapped scaffolds | y | 10.69% | 18.41% | 17.14% | 2.45% | 21.11% | |
| Scaffold-incompat. matches | z | 0.68% | 1.44% | 9.35% | 0.81% | 1.12% | |
| Potentially chimeric scaffolds | aa | 9 | 33 | 666 | 25 | 97 | |
| Chimeric acgt, Mbp | bb | 10 | 27 | 112 | 13 | 21 | |
| No. of small conflicts | cc | 3,474 | 6,165 | 14,582 | 3,912 | 1,586 | |
| acgt in small conflicts, Mbp | dd | 7 | 9 | 121 | 8 | 9 | |
More extensive results are contained in Tables 3 and 8 and Data Set 8 on the PNAS web site.
Assembly gives the acronym used in the text.
Producers are Celera (www.celeradiscoverysystem.com), University of California, Santa Cruz (UCSC, www.genome.ucsc.edu), and NCBI (www.ncbi.nlm.nih.gov).
Method identifies the computational approach used to produce each assembly: WG, whole-genome; C, compartmental; H, hierarchical.
Data sources are Celera (shotgun reads plus public BAC ends), IHGSC (HGP data), or a combination (Celera data plus a subset of human genomic data from GenBank).
Dates shown are assembly completion date (Celera), data freeze date (UCSC), or release date (NCBI).
Unambiguous bases in the assembly consensus sequence (including “acgt unmapped”).
Unambiguous bases not assigned to specific chromosome locations.
Contiguous sequence built of overlapping sequencing reads.
Chains of linked contigs.
A base has a 50% chance of being in a contig or scaffold at least this long.
Sum of the lengths of the scaffolds, including internal Ns.
RefSeqs alignable at 50% coverage and 95% identity thresholds.
Bases in matches to segmental duplications in NCBI-34.
Subset of segmental duplication unmapped in this assembly.
Percent of mate pairs indicating a possible misassembly. Mate pair data indicate relative orientation and distance between pairs of sequencing reads. Celera fragments were aligned to each assembly. Where two or more pairs of fragments imply the same rearrangement, they are counted as a possible misassembly.
Percent of aligned mate pairs with one fragment aligned to an unmapped scaffold (see “acgt unmapped”) while the other is aligned to a mapped scaffold.
Analysis of A2Amapper's one-to-one mapping between each assembly and NCBI-34. Matches, runs, and clumps are successively less restrictive local alignments, derived from the one-to-one mapping, as described in the text. Informally, matches never include gaps > 10 bp, runs never span conflicting matches, and clumps never span a conflict >50 kbp.
Unambiguous bases within matches.
Unambiguous bases of each assembly outside all matches.
Unambiguous bases of NCBI-34 outside matches to this assembly.
Unambiguous NCBI-34 bases within runs.
A matched base has a 50% chance of being in a match/run/clump at least this long.
Percent of matched bases in the maximal set of consistent matches, defined by heaviest common subsequence (HCS).
Percent of matched bases in unmapped scaffolds.
Percent bases in matches in scaffolds disagreeing with NCBI-34 in chromosome assignment, order, or orientation.
Percent of matched bases in scaffold-incompatible matches, where a match is incompatible with its scaffold if it conflicts with the (length-weighted) majority of matches in the scaffold.
Scaffolds with a consistent subset of incompatible matches >50 kb.
Unambigous bases in minority subset(s) for potentially chimeric scaffolds.
Runs of incompatible matches not counted in “Potentially chimeric scaffolds.”
Unambigous bases in matches in small conflicts.
We saw in 1999 that for Drosophila (1), increasing the genome coverage from 6.5 times to 11.2 times increased the sequence spanned by large scaffolds by 1.7% and the sequence contained by 5.0%, and reduced the number of gaps by 73.5%. We would expect to see similar improvements if the whole-genome shotgun human data were increased from 5.3 times to 10 times. Given the increasing ratio of the cost of finishing work to shotgun sequencing, we are comfortable stipulating that this is an economical proposition. Finally, we expect WGSA algorithms to continue to improve as they have over the past 3 years. To aid in such improvements, we are making available the Celera Assembler and its source code (myscience.appliedbiosystems.com/publications/compass/index.jsp).
A comparison of WGSA to the recently published chromosome 6 sequence (21) that is part of NCBI-34 illustrates that WGSA can also contribute to the continuing effort to produce a complete human genome sequence. Along chromosome 6, the authors report 10 remaining gaps, one missing sequence tagged site (STS) marker (D6S1694), and three RefSeq genes (NM_004690, NM_018452, and NM_014034) that are only partially represented (21). The missing STS marker is present at its correct location and all three RefSeq genes are complete in WGSA. We corroborate the conjecture in ref. 21 that NM_014034 was only partially found in NCBI-34 because of a deletion/polymorphism event in the P1-derived artificial chromosome (PAC) RP3–329L24 (AL132874.30). The first exon of NM_014034 is contained in a 56,180-bp region of WGSA not present in NCBI-34, which maps between base pair 119,198,642 and its 3′ neighbor of NCBI-34 chromosome 6. Scanning the whole genome, we found evidence for more such polymorphisms/deletions. There are 573 locations where WGSA reports 1,000 or more bases in a spot where NCBI-34 reports less than 100 (see Data Set 4; Data Sets 1–8, Figs. 3–8, Tables 3–13, and supporting text files are published as supporting information on the PNAS web site). There are also 80 RefSeq genes where one finds at least 5% more of the gene in WGSA than in NCBI-34 (Tables 6 and 7).
Of the 10 gaps in NCBI-34's chromosome 6, three are due to the centromere and telomeres. The WGSA sequence in the vicinity of the two other gaps near the centromere is rearranged with respect to NCBI-34, suggesting a possible region of large-scale polymorphism. The WGSA scaffold spanning the second gap near the centromere suggests that the NCBI-34 contig just after the centromere should be inverted, leaving a 10-kbp gap (Fig. 7). One gap does not exist in WGSA, suggesting that it is an error in NCBI-34 or is due to a large, near-perfect tandem duplication. The four remaining gaps are largely closed by a total of 691 kbp of WGSA (Fig. 8), and NCBI-34 has 180 kbp that belong in these gaps but were not placed there. In addition, the missing STS marker, D6S1694, is found in the correct position within one of these gaps.
Over the entire genome, there are 196 gaps in NCBI-34 that are spanned by WGSA. Of the gaps, 38 are completely filled by 85,839 bp, and 136 are partially filled by 3.341 Mbp (Data Set 5). Furthermore, for 56 of these gaps WGSA reveals that at least 2.438 Mbp of unassigned sequence from NCBI-34 belong in those gaps (Data Set 6). Fig. 1a illustrates the ability of WGSA to resolve probable remaining errors of order and orientation of NCBI-34 contigs. Fig. 1b illustrates the potential for filling gaps between contigs. WGSA also contributes additional sequence beyond filling gaps in NCBI-34 (Table 1); as with the Drosophila genome sequencing (22), this sequence may be from heterochromatic regions not covered by the clone-by-clone approach.
Fig. 1.
Dot-plot representation of sample assembly comparison results. Horizontal axes correspond to intervals along NCBI-34, and vertical axes correspond to intervals along various assemblies, with the sequences starting from the bottom left corner. Diagonal lines show the relative positions and orientations of matches. Identical sequences would yield one diagonal line. Vertical bars represent gaps between NCBI-34 contigs. Selected regions were chosen to represent general observations regarding the assemblies; related figures of entire chromosomes are provided for all chromosomes in Data Set 7. (a) Illustration of a region in which WGSA can augment NCBI-34. Shown are the first 6 Mbp of NCBI-34 human chromosome 1 versus part of a single scaffold of WGSA. The second NCBI-34 contig is inverted, and the third and fourth contigs are interchanged, compared with WGSA. We postulate that this is an NCBI-34 contig mapping problem. Alternative explanations, such as misassembly or polymorphisms within the WGSA scaffold that coincidentally occur at the boundaries of NCBI-34 contigs, are improbable. (b–f) Comparison of the NCBI-34 human chromosome 1 region from 34–40 Mbp against the primary matching regions of WGSA (b), WGA (c), CSA (d), HG06 (e), and NCBI-28 (f). (See main text for description of assemblies.) WGSA agrees closely with NCBI-34 and spans and largely fills two gaps between NCBI-34 contigs. All other assemblies have multiple order and orientation errors. For all but HG06, the misplaced segments correspond to entire scaffolds (data not shown). For HG06, errors are a mix of within-scaffold rearrangements and scaffold order and orientation. WGA and HG06 both have a relatively large number of small, misplaced scaffolds, whereas CSA and NCBI-28 have a few, larger scaffolds that are misplaced.
The First Human Genome Reconstructions
The first human genome sequences were reported in February 2001 (4, 5). While Celera produced a whole-genome shotgun data set (as described in ref. 4 and above), the IHGSC produced and deposited into GenBank 33,000 BAC-based data entries in a variety of finished states. Twenty percent of the BACs represented a finished sequence, whereas 75% of the BACs consisted of contigs produced by a phrap (www.phrap.org) assembly of a 3–5 times shotgun sequencing of the BAC, which produced an average of 20 contigs with an average length of 8 kbp. The remaining 5% of BACs consisted of only a 1 times sampling of unassembled sequence reads.
Celera produced two assemblies based on different approaches (4). Both used, in addition to the 5.3 times shotgun data, the GenBank data set above, shredded into 550-bp reads forming a 2 times tiling of the BAC sequence contigs. The combined whole-genome assembly (called WGA) was obtained by applying the Celera assembler to the 27 million Celera reads and 16 million shredded reads from the GenBank data. The Celera assembler used only Celera's paired reads and the BAC end reads to order and orient configs. The second assembly reported (4, 23, 24), the compartmental shotgun assembly (CSA), first used the BAC organization of the data to determine 3,800 “compartments” consisting of BACs and associated Celera data that were determined to cover a given region of a chromosome using Celera's read pairs and inferred sequence overlaps between the BACs. The GenBank data for each compartment was then shredded, combined with Celera's data for the region, and assembled with the Celera assembler, again using only the end-sequence pairs to order and orient contigs within scaffolds (4). The final step for all of the assemblies was to place the scaffolds (Celera) or the fingerprint clone contigs (IHGSC) onto chromosomal locations, based primarily on STS maps. Additional information was used for Celera's CSA (4) and consortium assemblies (5), but for Celera's WGA and WGSA assemblies only the STS maps were used.
An IHGSC result available shortly after ref. 5, herein referred to as HG06, was built from the GenBank sequence data as of December 2000, and a physical map of its 33,000 BACs (25). The physical map was assembled by using HindIII restriction digest fingerprints of 354,000 BACs, including the 33,000 selected for sequencing. As described in ref. 5, contigs from adjoining BAC assemblies were partially ordered and merged based on the BAC overlaps in the physical map. Contigs were further ordered by mapping exons of RefSeq sequences (26) and ESTs, and 1.8 million read pairs from inserts ranging between 2 kbp and 6 kbp that were stored in the SNP consortium database (27).
In addition to these reported assemblies (4, 5), we also evaluate two assemblies contemporaneous with WGSA. The first is NCBI Build 28 (NCBI-28), based on the consortium data available in December 2001 (when WGSA was produced). The second assembly is another combined whole-genome assembly (WGA2) which was produced at the same time as WGSA to take full advantage of all of the data available. The set of GenBank sequence from September 2001 used for WGA2 had 1.7% more basepairs than the December 2000 set used for WGA. Comparing WGSA, which used only whole-genome shotgun data, and WGA2 shows how much additional sequence of the genome is recovered by adding GenBank data to Celera's shotgun data, because both were assembled with the same version of the software.
Evaluation of the Assemblies Against NCBI-34
Methods and Summary Statistics. We have developed a suite of tools, A2Amapper, for constructing a one-to-one correspondence between pairs of assemblies. Like other whole-genome comparison methods (28–31), A2Amapper is based on the identification of seed alignments, in this case unique exact matches, followed by a more aggressive local alignment phase between seeds within nonoverlapping chains of seeds. Cutoffs were carefully tuned to balance sensitivity (finding all correlations), specificity (finding only the true ones), and computational requirements (see Data Set 1). Details about A2Amapper will be presented elsewhere (H.S., J.R.M., C.M.M., M.J.F., S.Y., and G.G.S., unpublished work; R.L., X. Zhao, L.F., C.M.M., and S.I., unpublished work). A2Amapper produces a set of one-to-one matches that are alignments of nearly identical pairs of segments imputed to be analogous up to polymorphisms. Each match aligns a segment of the target genome against a segment of NCBI-34. The segments are nonoverlapping by construction, and we consider the coverage of NCBI-34 to be the sum of the lengths of these segments. This set of matches is the basis for further analysis regarding correctness of order and orientation for which we develop three concepts: runs, heaviest common subsequence, and clumps. One match is consistent with another if in each assembly the segments of the matches are in the same relative order and orientation with no intervening matches between them. A run is a maximal chain of consistent matches. The heaviest common subsequence between two genomes is a subset of the matches for which the sum of the lengths of the matches is maximal and removing all other matches from consideration leaves a single run. Intuitively, the heaviest common subsequence is a global measure of the largest subset of the two assemblies that agree with each other. A clump is a run of 50 kbp or more that can be obtained by eliminating out-of-order matches, giving a local equivalent of the heaviest common subsequence (Supporting Text 1).
Coverage and Order and Orientation. Although the more recent assemblies (WGSA and WGA2) have distanced themselves significantly from the earlier ones (CSA, WGA, and HG06) in terms of quality, the earlier assemblies covered 86–88% of NCBI-34 (Table 1). CSA and WGA placed 79–80% of the covered sequence in the correct order and orientation, whereas HG06 positioned 74% correctly. This improved order and orientation is also demonstrated by longer runs and clumps and higher mate pair satisfaction rates (see Supporting Text 2 and Tables 9 and 10) for CSA and WGA relative to HG06 (Table 1). HG06 displayed a greater match length, mostly reflecting the larger numbers of gaps between contigs in the Celera drafts. In the case of WGA, nearly 9% of matches to NCBI-34 are found on unmapped scaffolds (Table 1), with an additional 10% being in mismapped scaffolds, whereas less than 1% of sequence involved an intra-scaffold conflict. This implies that most of the order and orientation conflicts were due to incorrect mapping of scaffolds and not the order of contigs within a scaffold. HG06 shows a large amount, more than 16%, of sequence in scaffolds that are in the wrong location or orientation, and has more than 9% of the total sequence in conflict with the majority of the containing scaffold. In addition to mismapped scaffolds, all assemblies had subscaffold segments that were misplaced (Table 1). Many such small discrepancies were assembly errors that could be corrected by routine gap closure, as discussed above.
WGSA and WGA2 provide 93% and 96% coverage of NCBI-34, of which ≈97% is in globally consistent order and orientation. The quality of WGSA is remarkable in light of it having less input data than any other assembly, although it does have three clear limitations: relatively short contigs largely reflecting low coverage, unresolved ubiquitous repeats, and missing segmental duplications. The latter is reflected by low run coverage near NCBI-34's centromeres (Data Set 3). Manual curation of WGSA identified 16 clearly chimeric scaffolds and 3,912 smaller segments totaling 8.1 Mbp that were also out of order, reflecting some combination of misassembled contigs, transpositions within a scaffold, structural polymorphism between donors, and errors in the one-to-one mapping produced by A2Amapper. (Fig. 1a illustrates why a manual curation of order and orientation discrepancies is necessary.) Probably because of low shotgun sequence coverage, a disproportionate number of the discrepancies are on the X and Y chromosomes. NCBI-28, a contemporary of WGSA, had similarly high coverage, but despite generally longer contigs and scaffolds, its order and orientation results were closer to those of the earlier assemblies, reflecting problems with mapping scaffolds onto chromosomes. The general patterns described above regarding order and orientation are illustrated in Fig. 1.
CSA, WGA, and HG06 all cover 86–88% of NCBI-34 (Table 1), yet their union covers 96.7%. Since the input to the CSA and WGA assemblies included a representation of almost all of the data input to the HG06 assembly, one must conclude that the differing methods of construction reproduced different parts of the genome. If the CSA and WGA assemblies were merely reconstituting the shredded BAC data and adding a little additional data, then both CSA and WGA should be slight supersets of HG06 and they clearly are not. Table 2 shows a statistical measure of similarity for various pairs of assemblies. Despite the fact that WGA and CSA involved the GenBank data, WGA, CSA, and WGSA are all quite different from HG06 and quite similar to each other. One can get a picture of the impact of adding shredded GenBank data to Celera's 5.3 times shotgun data set by examining a pair of assemblies performed with the same version of the assembly software where one uses just Celera data, and the other uses the combined data set. The only pair of assemblies satisfying this property is WGSA and WGA2. Close examination reveals that WGA2 is largely a superset of WGSA. There are 12.7 Mbp in WGSA lost in WGA2 because of the addition of the shredded data, but 114.4 Mbp are gained in WGA2, 90.0 Mbp of which are filling gaps in WGSA scaffolds. Thus adding the shredded reads resulted in 3.4% more of the genome being reconstructed with contig statistics improving as expected for the given increase in coverage.
Table 2. Similarity of genome content I(A, B) between pairs of assemblies A and B.
| I(A, B) | HG06 | WGA | CSA | WGSA |
|---|---|---|---|---|
| WGA | 0.20 | |||
| CSA | 0.37 | 0.49 | ||
| WGSA | 0.21 | 0.58 | 0.50 | |
| WGA2 | 0.46 | 0.91 | 0.81 | 0.87 |
Cells are coded for low, medium, and high similarity by plain, bold, and underlined bold text, respectively. Let cA be the fraction of NCBI-34 covered by assembly A, and let cA∩B be the fraction shared by both assemblies A and B. If both assemblies A and B cover large amounts of the genome (cA and cB), then they must also share a large portion of the genome (cA∩B). If A and B are unrelated samplings, then one would expect cA and cB to be randomly chosen fractions, and cA∩B would equal cA·cB. If A and B are maximally similar, then one would be completely subsumed by the other and cA∩B would equal min(cA, cB). So let I(A, B) = (cA∩B - cA·cB)/(min(cA, cB) - cA·cB) ∈ [0, 1] be a normalized measure between these two extremes, where I(A, B) is 0 if A and B are unrelated, and 1 if they are maximally similar.
Evaluation of the Assemblies Against RefSeq
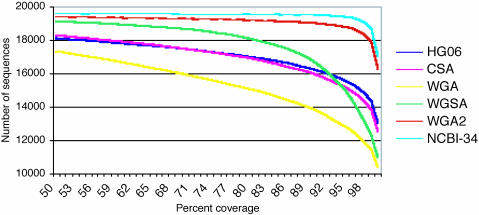
A good indicator for the annotation potential of a genome is the rate and quality of mapping of known full-length mRNA sequences, for instance those contained in the RefSeq repository (26). We developed a high-throughput mapping tool, called ESTmapper (L.F. and B.W., unpublished work), to efficiently align full-length and first-pass cDNA (mRNA, EST) sequences to a sequence assembly. Like its predecessor sim4 (32), ESTmapper generates a nucleotide-level alignment between the query sequence and the target genome. We mapped the 19,667 human mRNA sequences in the August 2003 RefSeq data set to each of the genomes at different coverage cutoffs (Fig. 2 and Data Set 2). With small exceptions, the order that this measure induces on the set of assemblies does not change with varying coverage cutoffs. The more complete assemblies (WGA2 and NCBI-34) performed better than WGSA, which in turn shows considerably higher integrity than the earlier assemblies (WGA, CSA, and HG06) at all but the highest coverage thresholds. As the performance of WGSA versus WGA2 reveals, it is completeness rather than continuity of order and orientation that is the main issue: WGSA has a part of almost every gene that WGA2 and NCBI-34 do, but because it is an assembly of only 5.3 times data, enough sequence is missing to cause a larger drop as the coverage threshold increases. Further evidence of this observation is that 470 RefSeq sequences have less than 95% of their base pairs mapped to NCBI-34, whereas only 89 sequences were inconsistent with NCBI-34's sequence order. The same pattern holds for all of the other assemblies. WGA2, based only on a slight update on the original combined data sets, is nearly as complete as NCBI-34.
Fig. 2.
The proportion of the 19,667 RefSeq mRNA sequences that can be aligned to each of the genomes at various coverage thresholds and more than 95% sequence identity.
Conclusion
The Celera Assembler, first described in 2000 with the successful assembly of the Drosophila genome (1), was used with modification for the initial assemblies of the human genome reported in (4), and with further modification was used for the successful assembly of the mouse genome (6), the dog genome (18), and the Anopheles mosquito genome (15). The same assembler was used for the whole-genome shotgun assembly of the human genome reported here. With coverage of 92.7% of the NCBI-34 sequence, and continuity close to that of NCBI-34, WGSA clearly shows that a high-quality genome sequence can be assembled from the Celera proprietary data alone, independently of the IHGSC data and methods. Indeed, WGSA provides valuable additions and corrections to the nearly complete human genome, NCBI-34. Thus, whole-genome shotgun assembly can give a high-quality draft, much higher than that originally released by either Celera or the IHGSC, of a higher eukaryote at a remarkably modest level of coverage.
Supplementary Material
Acknowledgments
We thank all of our colleagues who have contributed to our efforts to sequence, assemble, map, and analyze the human genome. In particular we wish to thank Royden A. Clark, ZhenYuan Wang, Allison Yao, Qing Zhang, Zhongwu Lai, Richard Mural, Peter Li, and Robert Sanders.
Abbreviations: WGSA, whole-genome shotgun assembly; IHGSC, the International Human Genome Sequencing Consortium; BAC, bacterial artificial chromosome; NCBI-34, National Center for Biotechnology Information (NCBI) Build 34 of the human genome; STS, sequence tagged site; CSA, compartmental shotgun assembly; WGA, whole-genome assembly.
Data deposition: The sequences of the assemblies herein referred to as WGSA, CSA, and WGA have been deposited in the GenBank database (whole-genome assembly project accession nos. AADD00000000, AADC00000000, and AADB00000000).
References
- 1.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 2.Myers, E. W., Sutton, G. G., Delcher, A. L., Dew, I. M., Fasulo, D. P., Flanigan, M. J., Kravitz, S. A., Mobarry, C. M., Reinert, K. H. J., Remington, K. A., et al. (2000) Science 287, 2196–2204. [DOI] [PubMed] [Google Scholar]
- 3.Celniker, S. E., Wheeler, D. A., Kronmiller, B., Carlson, J. W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S. P., Frise, E., et al. (2002) Genome Biol. 3, research0079.1–0079.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 5.International Human Genome Sequencing Consortium. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 6.Mural, R. J., Adams, M. D., Myers, E. W., Smith, H. O., Miklos, G. L. G., Wides, R., Halpern, A., Li, P. W., Sutton, G. G., Nadeau, J., et al. (2002) Science 296, 1661–1671. [DOI] [PubMed] [Google Scholar]
- 7.Venter, J. C., Smith, H. O. & Hood, L. (1996) Nature 381, 364–366. [DOI] [PubMed] [Google Scholar]
- 8.Zhao, S., Malek, J., Mahairas, G., Fu, L., Nierman, W., Venter, J. C. & Adams, M. D. (2000) Genomics 63, 321–332. [DOI] [PubMed] [Google Scholar]
- 9.Collins, F. S., Green, E. D., Guttmacher, A. E. & Guyer, M. S. (2003) Nature 422, 835–847. [DOI] [PubMed] [Google Scholar]
- 10.The International Human Genome Sequencing Consortium (April 14, 2003) News Release: International Consortium Completes Human Genome Project, www.genome.gov/11006929.
- 11.Waterston, R. H., Lander, E. S. & Sulston, J. E. (2002) Proc. Natl. Acad. Sci. USA 99, 3712–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waterston, R. H., Lander, E. S. & Sulston, J. E. (2003) Proc. Natl. Acad. Sci. USA 100, 3022–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischmann, R. D., Adams, M. D., White, O., Clayton, R. A., Kirkness, E. F., Kerlavage, A. R., Bult, C. J., Tomb, J. F., Dougherty, B. A., Merrick, J. M., et al. (1995) Science 269, 496–512. [DOI] [PubMed] [Google Scholar]
- 14.Venter, J. C., Levy, S., Stockwell, T., Remington, K. & Halpern, A. (2003) Nat. Genet. 33, Suppl., 219–227. [DOI] [PubMed] [Google Scholar]
- 15.Holt, R. A., Subramanian, G. M., Halpern, A., Sutton, G. G., Charlab, R., Nusskern, D. R., Wincker, P., Clark, A. G., Ribeiro, J. M. C., Wides, R., et al. (2002) Science 298, 129–149.12364791 [Google Scholar]
- 16.Mouse Genome Sequencing Consortium (2002) Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 17.Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J. M., Dehal, P., Christoffels, A., Rash, S., Hoon S., Smit, A., et al. (2002) Science 297, 1301–1310. [DOI] [PubMed] [Google Scholar]
- 18.Kirkness, E. F., Bafna, V., Halpern, A. L., Levy, S., Remington, K., Rusch, D. B., Delcher, A. L., Pop, M., Wang, W., Fraser, C. M., et al. (2003) Science 301, 1898–1903. [DOI] [PubMed] [Google Scholar]
- 19.Eichler, E. E. (1998) Genome Res. 8, 758–762. [DOI] [PubMed] [Google Scholar]
- 20.Bailey, J. A., Gu, Z., Clark, R. A., Reinert, K., Samonte, R. V., Schwartz, S., Adams, M. D., Myers, E. W., Li, P. W., Eichler, E. E. (2002) Science 297, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 21.Mungall, A. J., Palmer, S. A., Sims, S. K., Edwards, C. A., Ashurst, J. L., Wilming, L., Jones, M. C., Horton, R., Hunt, S. E., Scott, C. E., et al. (2003) Nature 425, 805–811. [DOI] [PubMed] [Google Scholar]
- 22.Hoskins, R. A., Smith, C. D., Carlson, J. W., Carvalho, A. B., Halpern, A., Kaminker, J. S., Kennedy, C., Mungall, C. J., Sullivan, B. A., Sutton, G. G., et al. (2002) Genome Biol. 3, research0085.1–0085.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huson, D. H., Reinert, K., Kravitz, S. A., Remington, K. A., Delcher, A. L., Dew, I. M., Flanigan, M., Halpern, A. L., Lai, Z., Mobarry, C. M., et al. (2001) Bioinformatics 17, S132–S139. [DOI] [PubMed] [Google Scholar]
- 24.Huson, D. H., Reinert, K. & Myers, E. W. (2002) J. Assoc. Comput. Mach. 49, 603–615. [Google Scholar]
- 25.McPherson, J. D., Marra, M., Hillier, L., Waterston, R. H., Chinwalla, A., Wallis, J., Sekhon, M., Wylie, K., Mardis, E. R., Wilson, R. K., et al. (2001) Nature 409, 934–941. [DOI] [PubMed] [Google Scholar]
- 26.Pruitt, K. D. & Maglott, D. R. (2001) Nucleic Acids Res. 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorisson, G. A. and Stein, L. D. (2003) Nucleic Acids Res. 31, 124–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brudno, M., Do, C. B., Cooper, G. M., Kim, M. F., Davydov, E., NISC Comparative Sequencing Program, Green, E. D., Sidow, A. & Batzoglou, S. (2003) Genome Res. 13, 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brudno, M., Malde, S., Poliakov, A., Do, C. B., Couronne, O., Dubchak, I. & Batzoglou, S. (2003) Bioinformatics 19, 154–162. [DOI] [PubMed] [Google Scholar]
- 30.Bray, N., Dubchak, I. & Pachter, L. (2003) Genome Res. 13, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz, S., Kent, W. J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R. C., Haussler, D. & Miller, W. (2003) Genome Res. 1, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florea, L., Hartzell, G., Zhang, Z., Rubin, G. M. & Miller, W. (1998) Genome Res. 8, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.