Abstract
Transgenic expression of ceramidase suppresses retinal degeneration in Drosophila arrestin and phospholipase C mutants. Here, we show that expression of ceramidase facilitates the dissolution of incompletely formed and inappropriately located elements of rhabdomeric membranes in ninaEI17 mutants lacking the G protein receptor Rh1 in R1–R6 photoreceptor cells. Ceramidase expression facilitates the endocytic turnover of Rh1. Although ceramidase expression aids the removal of internalized rhodopsin, it does not affect the turnover of Rh1 in photoreceptors maintained in dark, where Rh1 is not activated and thus has a slower turnover and a long half-life. Therefore, the phenotypic consequence of ceramidase expression in photoreceptors is caused by facilitation of endocytosis. This study provides mechanistic insight into the sphingolipid biosynthetic pathway-mediated modulation of endocytosis and suppression of retinal degeneration.
Sphingolipids are integral components of eukaryotic cell membranes. They are essential for survival of yeast, Drosophila, and mammals. Sphingolipids have a hydrophobic backbone moiety, ceramide, linked to a variable polar head group, e.g., phosphorylcholine in sphingomyelin and a saccharide in glycosphingolipids. Ceramide itself consists of a long chain base linked to a fatty acid. Ceramide, sphingosine, and sphingosine-1-P are lipid second messengers modulating a variety of cellular functions (1–3). Ceramide is at the branch point of the sphingolipid biosynthetic pathway and serves as a substrate for several enzymes, e.g., sphingomyelin synthase, ceramidase, ceramide kinase, and glucosylceramide synthase. In Drosophila, transgenic expression of a neutral ceramidase is accompanied by a decrease in the levels of ceramide (4). We have shown that modulation of enzymes of the sphingolipid biosynthetic pathway suppressed retinal degeneration in certain phototransduction mutants (4). Transgenic expression of ceramidase or a loss of one copy of the rate-limiting enzyme in the sphingolipid biosynthetic pathway, namely, Serine Palmitoyl CoA transferase, suppressed retinal degeneration in arrestin and phospholipase C (norpA) mutants. These backgrounds also suppressed degeneration in a dynamin mutant, suggesting that ceramidase was modulating the endocytic pathway. To gain further insight into the mechanism of ceramidase action, we have investigated the effects of ceramidase expression in the rhodopsin null mutant, ninaEI17. Rhodopsin, the G protein-coupled receptor, transduces light signal in Drosophila photoreceptors R1–R6 (5, 6). Rhodopsin is also involved in the formation of a rhabdomere terminal web (RTW), an actin-based cytoskeletal scaffold implicated in rhabdomere biogenesis (7, 8). Rhabdomeres are densely organized, specialized areas of plasma membrane that house the signal transduction machinery of photoreceptors. In ninaEI17 mutants, the RTW is not organized during morphogenesis and thus photoreceptors have improperly formed rhabdomeres (8). In addition, in 3-day-old flies, rhabdomeric elements involute into the photoreceptors and are subsequently removed slowly (7, 9, 10). Here, we demonstrate that expression of ceramidase clears ninaEI17 mutant photoreceptors of these involuting rhabdomeric membranes. We show that this clearing is facilitated by a downstream process of endocytosis, which is revealed biochemically by following rhodopsin turnover in wild-type photoreceptors expressing ceramidase.
Materials and Methods
Ceramidase Construct and Expression. Ceramidase cDNA (EST SD07768) was cloned into the pUAST vector as EcoR1-XhoI fragment as described (4). Expression of ceramidase was driven by the bipartite expression system, UAS-Gal4, in transgenic flies using either eye-specific GMR-Gal4 or heat shock Gal-4 driver.
Antibodies. The 1D4 monoclonal antibody was a kind gift from Robert Molday (University of British Columbia, Vancouver). Polyclonal rabbit anti-inositol polyphosphate 1-phosphatase (IPP) antibodies were a kind gift from Charles Zuker (University of California at San Diego).
Drosophila Stocks and Crosses. ninaEI17 and the transgenic hs-Rh1–1D4 flies were kindly provided by Charles Zuker. GMR-Gal4 (stock 1104) and hs-Gal4 (stock 2077) were obtained from Drosophila Stock Center (Bloomington, IN). w; GMR-Gal4/UAS-Ceramidase; ninaEI17, w; hs-Gal4/UAS-Ceramidase; ninaEI17, w; hs-Rh1–1D4/+; +, w; hs-Rh1–1D4/GMR-Gal4, UAS-ceramidase; + flies were produced by standard genetic crosses. Unless otherwise stated, all flies were reared at 25°C on standard corn meal, agar medium. For heat shock induction of Rh1–1D4, flies were heat-shocked at 37°C for 1 h and then shifted to 25°C for different number of days. For experiments in the light, subjects were maintained in a 12-h light and 12-h dark cycle, whereas subjects of experiments in the dark were maintained in complete darkness and frozen before dissection of retina.
Histology. For all electron microscopic examinations, unless otherwise specified, 3-day-old flies grown at 25°C were decapitated under anesthesia, and their heads were dissected and fixed using 2% glutaraldehyde and 4% formaldehyde in sodium cacodylate buffer. Eyes were postfixed in 1% osmium tetroxide and dehydrated in graded ethanol and propylene oxide. Specimens were embedded in epoxy resin (Embed-812) and thin sectioned. Transverse sections (90 nm) were stained with uranyl acetate and lead citrate and examined. Four eyes were examined for each specimen. The digital images were obtained with an Hitachi H7000 electron microscope (Tokyo) equipped with a Gatan digital camera (Pleasanton, CA).
Western Analysis. At appropriate time intervals, flies were frozen and decapitated. Retina were dissected, manually homogenized in SDS-sample buffer and subjected to SDS/PAGE. Proteins were transferred to a nitrocellulose membrane, and Western blot analysis was carried out using anti-1D4 monoclonal antibody and IPP antibody. IPP served as loading controls (11).
Results and Discussion
Ceramidase Expression Facilitates the Internalization and Clearing of Inappropriately Formed Rhabdomeric Components in ninaEI17 Mutant. Rh1, a major component of rhabdomeres, is not only the receptor for transducing light signal in R1–R6, but is also required for organization of the RTW, an actin-based cytoskeletal scaffold, believed to orchestrate the biogenesis of this organelle. In Rh1 null mutant ninaEI17, the RTW is not organized during the late pupal stage, and rhabdomere biogenesis is defective (7–9, 12). Consequently, photoreceptors examined by transmission electron microscopy 3 days after eclosion show the ill formed rhabdomeres, and membranes of rhabdomeres are seen involuting into the photoreceptors. These involuting curtains of rhabdomere elements are then slowly cleared over two weeks (7, 9).
We have previously demonstrated that transgenic expression of ceramidase does not affect either the development or function of photoreceptors. However, ceramidase expression suppressed degeneration in arrestin and norpA mutants and in photoreceptors expressing a dominant-negative form of dynamin (4). This led us to propose that ceramidase was mediating its effect by modulating the endocytic pathway. We reasoned that the phenotypic consequence of ceramidase expression in Rh1 mutant ninaEI17 would be different from that observed in arrestin and dynamin mutants. If ceramidase affects endocytosis, then in a ninaEI17 mutant it should influence the process of involution and clearance of the incompletely formed and inappropriately localized rhabdomeric membranes.
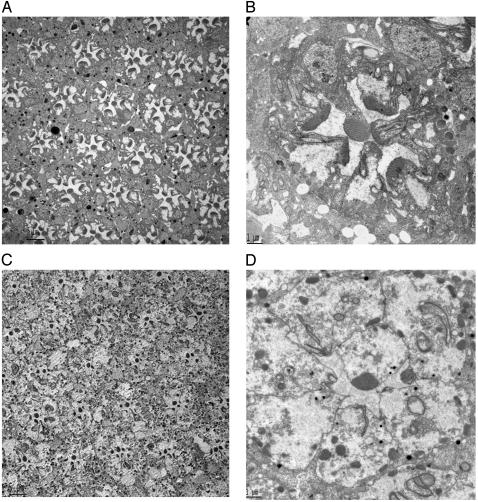
Indeed, expression of ceramidase facilitates the removal of these inappropriately positioned rhabdomeric elements as seen in Fig. 1. Three-day-old ninaEI17 flies have ill-formed rhabdomeres, and remnant rhabdomeres are seen as long contiguous elements of plasma membrane involuting into the cell body in R1–R6 photoreceptors (7–9) (Fig. 1 A and B). In contrast, 3-day-old ninaEI17 flies expressing ceramidase have very little rhabdomeric elements at the apical surface of the photoreceptors (Fig. 1 C and D). Most of the rhabdomeric membranes have been internalized and are in the process of being cleared. Although R1–R6 cells of almost all ommatidial sections of a ninaEI17 compound eye shown in Fig. 1 A have a significant density of remnant rhabdomeric membranes in the apical portion of the cells, more than 95% of the R1–R6 photoreceptors expressing ceramidase have no significant rhabdomeric membranes in the apical region (Fig. 1C). Instead, the apical regions have plasma membranes that are contiguous with the rest of the photoreceptor cells. The cell–cell adherens junction is intact, indicating the structural integrity of these photoreceptor cells.
Fig. 1.
Expression of ceramidase clears rhabdomeric remnants in ninaEI17 mutant photoreceptors. (A) ninaEI17 mutants lacking Rh1 in R1–R6 photoreceptors have defects in rhabdomere biogenesis. A cross section of a compound eye is shown. In transmission electron microscopy sections of 3-day-old fly eyes, all ommatidial units display degenerative rhabdomeres in R1–R6 photoreceptors, and rhabdomeric membranes are visible in the photoreceptor cell body. (Scale bar, 5 μm.) (B) A higher-magnification view across a single ommatidium of 3-day-old ninaEI17 shows that rhabdomeres are incompletely formed, and rhabdomeric elements are seen involuting into the photoreceptors in R1–R6 cells. (Scale bar, 1 μm.) (C) A low-magnification view of 3-day-old ninaEI17 flies expressing ceramidase. In this section, R1–R6 photoreceptors in most ommatidial units have reduced or no rhabdomere, and discrete tubulovesicular structures appear intracellularly. (Scale bar, 5 μm.) (D) A high-magnification view of one of the ommatidial units from C shows that most of the involuting rhabdomeric membranes are missing in R1–R6 cells. Some membranous structures in the cell are seen connected to the apical plasma membrane. Distinct tubulovesicular structures are also seen in the cell body. (Scale bar, 1 μm.)
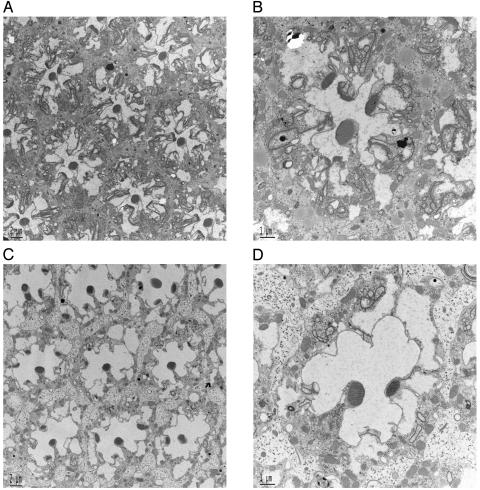
Ceramidase Facilitates the Dissolution of Rhabdomeric Elements in ninaEI17 Mutant When Expressed in Adult Photoreceptors. Because the development of rhabdomeres is initiated in the pupae and because GMR-Gal4 initiates ceramidase expression during its formation (13, 14), it can be argued that ceramidase expression could affect the biogenesis of rhabdomeric elements in ninaEI17, thus resulting in the observed phenotype. To resolve this issue, we expressed ceramidase, after eclosion, in an adult ninaEI17 mutant. In these experiments, UAS-ceramidase expression was driven by a heat shock Gal4 driver. Newly eclosed ninaEI17 flies and ninaEI17 flies with ceramidase transgene were incubated at 37°C for 1 h/day for 3 days. Control ninaEI17 flies heat shocked for 3 days showed features similar to non-heat-shocked mutant flies. Rhabdomeres were incompletely formed but slightly compact, and membranes were seen involuting into the R1–R6 photoreceptors (Fig. 2 A and B, compare with Fig. 1 A and B). On the other hand, flies expressing ceramidase cleared most of the rhabdomere from the apical region (Fig. 2 C and D). Thus, expression of ceramidase specifically accelerates the intracellular dissolution of rhabdomeric membranes in ninaEI17 mutant photoreceptor cells. We also examined the internalized tubular and vesicular elements generated by expression of ceramidase in ninaEI17 for an antigen specific to rhabdomere. Chaoptin is a photoreceptor specific, leucine-repeat containing, cell adhesion plasma membrane protein that localizes to the outer leaflet and is enriched in rhabdomeres of photoreceptors (15). ninaEI17 flies expressing ceramidase were examined by immunoelectron microscopy for chaoptin. We localized chaoptin immunoprecipitates on internalized membranous tubular and vesicular structures (data not shown), providing additional evidence of their rhabdomeric origin.
Fig. 2.
Ceramidase facilitates the removal of rhabdomeric elements when expressed in adult photoreceptors. (A) Transmission electron microscopy of ninaEI17 ommatidium subjected to heat shock for 3 days at 37°C for 1 h/day. The heat shock was induced from day 1 to day 3 after eclosion, and the compound eyes were examined on day 4. Heat shock resulted in slight compacting of incompletely formed rhabdomeres. However, involuting rhabdomeric elements were still distinguished in these mutant photoreceptors. (Scale bar, 2 μm.) (B) Higher-magnification view of one ommatidium from ninaEI17 photoreceptors treated as described in A. (Scale bar, 1 μm.) (C) TEM of ninaEI17 with ceramidase photoreceptors subjected to heat shock for 3 days at 37°C for 1 h/day to induce ceramidase expression. Heat shock was induced from day 1 to day 3, and eyes were examined on day 4. Induction of ceramidase resulted in dissolution of rhabdomeric elements and clearing of the photoreceptors. (Scale bar, 2 μm.) (D) Higher-magnification of a single ommatidial unit from photoreceptors shown in C. (Scale bar, 1 μm.)
Like other G protein-coupled receptors, rhodopsins undergo a ligand-dependent endocytic turnover (16–18). Given our previous data on the consequence of ceramidase expression in endocytic mutants and our current observations with ninaEI17 mutant, we decided to evaluate the effects of ceramidase expression on ligand (light)-induced rhodopsin turnover in wild-type photoreceptors.
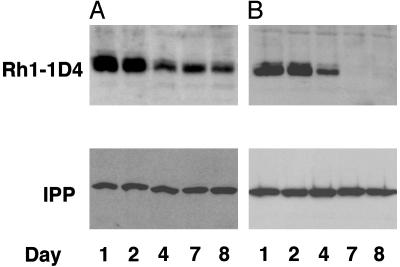
Ceramidase Facilitates Turnover of Rh1, a Major Component of the Rhabdomere. The endocytic turnover of Rh1 can be followed using an inducible and uniquely tagged Rh1. Rh1–1D4 transgenic flies express a heat-shock inducible Drosophila Rh1 carrying a specific tag, 1D4, derived from the C terminus of bovine Rh1 (19). Rh1–1D4 transgene is functional, because it rescues the phenotypic defects in ninaEI17 (20, 21). Rh1–1D4 synthesis was initiated in wild-type and ceramidase transgenic flies by a single heat shock at 37°C, and was followed by western analysis for 1D4 tag over a period while being maintained in a normal 12-h light/12-h dark cycle at 25°C (Fig. 3 A and B). Under these conditions, heat shock induction initiated the synthesis of Rh1, which peaked after 24–48 h, decreased over the next several days in wild-type photoreceptors and was still visible around day 8. Under similar conditions in flies expressing ceramidase, Rh1 levels peaked similar to wild-type flies; however, Rh1 disappeared rapidly thereafter, and none was visible after 4 days (Fig. 3B). These experiments indicate that ceramidase expression enhances the light-dependent turnover of Rh1.
Fig. 3.
Ceramidase expression facilitates the endocytic turnover of Rh1. (A) A pulse of 1D4-tagged Rh1 was induced in w1118 control flies by heat shock for 1 h at 37°C, and proteins were extracted from these fly retinas on days 1, 2, 4, 7, and 8 and were analyzed by Western blot for the tagged Rh1. The Rh1 level peaked between 1 and 2 days and decreased in amount thereafter, and a signal was still detected on day 8. (B) A pulse of 1D4-tagged Rh1 was induced as in A in flies expressing UAS-ceramidase under a GMR-GAL4 driver. In these flies the Rh1 peaked as in control retina, however, the tagged Rh1 was not visible beyond day 4. The blots were probed for IPP as loading controls (Lower).
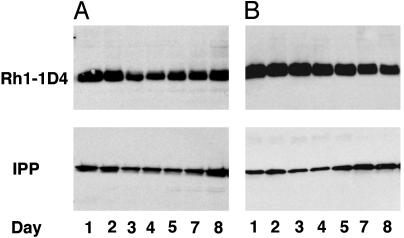
Ceramidase Facilitates Turnover of Ligand-Activated Rhodopsin. Drosophila phototransduction is a prototypic G protein-coupled receptor-signaling cascade (22–24). Like other G protein signaling cascades, invertebrate rhodopsin undergoes light-dependent turnover (16, 25, 26). In the blowfly, it has been demonstrated that rhodopsin has an extended half-life in flies maintained in the dark, whereas in flies maintained in light it has a short half-life (26). Although rhodopsins undergo photochemical interconversion, light-activated rhodopsins are eventually endocytosed and degraded (26–29). In Drosophila, visual arrestins act as clathrin adaptors and have been demonstrated to bind and internalize light-activated Rh1 (28). We hypothesized that if ceramidase facilitates a downstream process of endocytosis, although it can facilitate the turnover of light-activated rhodopsin it should have minimal effect on the half-life of an unactivated rhodopsin in flies maintained in dark. We therefore examined the effect of ceramidase on rhodopsin turnover in flies maintained in the dark. A pulse of rhodopsin in flies reared in the dark results in longer half-life, and a greater fraction of the pulsed rhodopsin is still visible 8 days after heat shock (Fig. 4A). Similar levels of rhodopsin were seen in ceramidase expressors maintained in the dark (Fig. 4B), thus suggesting that ceramidase does not accelerate the process of rhodopsin turnover in flies maintained in the dark (compare Figs. 3B and Fig. 4B). We therefore conclude that ceramidase enhances turnover of light-activated rhodopsin by facilitating endocytosis. Because ceramidase does not affect rhodopsin that is not light-activated, we believe it facilitates an existing normal mechanism for rhodopsin turnover. Although the molecular details of rhabdomere and rhodopsin turnover are yet to be elucidated, it has been known for several years that in flies the photoreceptor membrane is shed into the photoreceptor cell and cleared by endocytosis (17, 28–30). Ceramidase expression results in the clearing of ill-formed rhabdomeric elements in ninaEI17 mutants, whereas its expression in endocytic mutants such as arrestin and dynamin suppresses degeneration. Therefore, the consequence of ceramidase expression is determined by the underlying pathology of the phototransduction mutant.
Fig. 4.
Ceramidase expression does not facilitate Rh1 turnover in flies maintained in the dark. (A) A pulse of 1D4-tagged Rh1 was induced in w1118 control flies by heat shock for 1 h at 37°C, and proteins were extracted from retina 1, 2, 3, 4, 5, 7, and 8 days after heat shock and were analyzed by Western blot for the tagged Rh1. The flies were maintained in dark during the whole procedure until homogenization. Rh1 turned over slowly in flies maintained in the dark and had a longer half-life, and a substantial fraction was seen even on day 8. (B) A pulse of 1D4-tagged Rh1 was induced in flies expressing UAS-ceramidase under GMR-GAL4 and was analyzed as described in A. The rate of turnover was not enhanced as in the case of light, and a significant fraction of Rh1 was seen even on day 8. The blots were probed for IPP as loading controls (Lower).
In this study we have used a sensitized background to reveal the effects of ceramidase on membrane turnover. By following the phenotypic changes from the long involuting rhabdomeric membranes seen in ninaEI17 mutant photoreceptor cells (Fig. 1 A and B) to cells almost devoid of rhabdomeres in ninaEI17 expressing ceramidase (Fig. 1 C and D), we have shown that ceramidase expression facilitates membrane turnover in these cells. The use of an inducible, tagged Rh1 (hs-Rh1–1D4) has allowed us to follow the turnover of rhodopsin in wild-type photoreceptors. Using the inducible rhodopsin, we have demonstrated that ceramidase specifically facilitated the turnover of light-activated receptor. Light is the ligand for rhodopsin, and these receptors are not engaged when photoreceptors are maintained in the dark. Ceramidase expression had no effect on the half-life of rhodopsin when maintained in the dark, and we therefore concluded that ceramidase-facilitated ligand induced endocytic turnover of rhodopsin. The study of rhodopsin turnover in wild-type photoreceptors permitted us to examine receptor endocytosis without complications of an underlying mutant phenotype. We have earlier demonstrated that ceramidase expression suppresses degeneration in endocytic mutants such as arr23 and mutant photoreceptors expressing a dominant-negative form of dynamin. Indeed, it would be very interesting to evaluate the turnover of rhodopsin in these mutants. We attempted to follow rhodopsin turnover in an inducible dominant-negative dynamin mutant background. However, the mutant photoreceptors degenerated, and the function of these photoreceptors was severely compromised. Because of the degeneration, heat-shock induction of 1D4-tagged Rh1 in these mutants did not result in synthesis of appreciable amounts of protein, and hence turnover could not be followed (data not shown). A similar degenerative pathology complicates analysis in arrestin mutant background, and the lack of these critical controls makes it difficult to evaluate effects of ceramidase on Rh1 turnover in these mutant backgrounds. Another approach is to investigate the interaction of ceramidase expressors with components of pathways that have been implicated in membrane transport and photoreceptor degeneration. A recent study addressed the probable dual role of phosphoinositides in activation and adaptation of phototransduction cascade (31). Arrestin 2 specifically bound phoshphoinositide/inositol phosphates. In this study, flies expressing engineered mutants of arrestin that were defective in phosphoinositide binding but not Rh1 binding were generated. The photoreceptors from these flies show delayed and decreased translocation of arrestin to the rhabdomere upon light activation, and those that were translocated were not internalized efficiently after binding Rh1. In a reverse approach, the authors also showed that light-dependent translocation of arrestin was defective in mutants that disrupt phosphoinositide metabolism. Earlier work on CDP-DAG synthase, an enzyme required for phosphoinositide biosynthesis, and RDGB, a phosphoinositide transfer protein, have implicated phosphoinositides in membrane turnover and signaling in Drosophila photoreceptors (6, 23, 32–34). Because phosphoinositides have been implicated in transport of phototransduction components to and from rhabdomeric membranes and sphingolipids are integral membrane components, specific interactions could influence phototransduction at multiple steps. It would thus be worthwhile to examine whether enzymes of the sphingolipid biosynthetic pathway, such as serine palmitoyltransferase and ceramidase, do interact with the phosphoinositide signaling pathway. It is also important to discern whether these interactions, if any, are mediated by specific protein–protein interactions or are caused by effects of changes in metabolite concentrations, such as ceramide and sphingosine, or rather are the result of a combined effect. Use of mass spectrometry and feeding experiments suggested that a decrease in steady-state levels of ceramide contributed to the beneficial effect of ceramidase in suppressing degenerations (4). We now believe that facilitation of endocytosis observed in ceramidase expressors is also consequent to its action on ceramide levels in photoreceptors. If so, then mutants such as lace that are defective in ceramide synthesis and upstream of neutral ceramidase in the de novo biosynthetic pathway should have a similar effect as ceramidase expression in ninaEI17mutant. Formation and breakdown of ceramide can affect the structure of membranes because of its topology, membrane sidedness, and limited flip-flop across membranes. We believe our results lend credence to suggestions that many of the actions of ceramide are caused by its role in membrane domain formation, membrane vesiculation, fusion and fission reactions, and trafficking (2, 35, 36). Sphingolipids are being increasingly implicated in yeast as important regulators of cell growth, heat stress response, and membrane trafficking (37, 38). In conclusion, whereas our earlier studies showed that expression of ceramidase suppresses degeneration in arr23, norpA, and dynamin mutant backgrounds, our current study has enabled us to propose that it does so by facilitating endocytosis and a decrease in ceramide contributes to these processes.
Our approach of genetically modulating sphingolipid biosynthetic pathway in Drosophila phototransduction mutants, a prototypic G protein-coupled receptor signaling cascade, will help in integrating signaling, lipid metabolism, membrane turnover, and degenerative pathways.
Acknowledgments
We thank Dr. Charles Zuker for fly stocks and reagents, Dr. Robert Molday for antibody, and Dr. Shyam Sharan for comments on the manuscript.
Abbreviation: IPP antibody, polyclonal rabbit anti-inositol polyphosphate 1-phosphatase antibody.
References
- 1.Merrill, A. H., Jr., Schmelz, E. M., Dillehay, D. L., Spiegel, S., Shayman, J. A., Schroeder, J. J., Riley, R. T., Voss, K. A. & Wang, E. (1997) Toxicol. Appl. Pharmacol. 142, 208–225. [DOI] [PubMed] [Google Scholar]
- 2.van Blitterswijk, W. J., van der Luit, A. H., Veldman, R. J., Verheij, M. & Borst, J. (2003) Biochem. J. 369, 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hannun, Y. A., Luberto, C. & Argraves, K. M. (2001) Biochemistry 40, 4893–4903. [DOI] [PubMed] [Google Scholar]
- 4.Acharya, U., Patel, S., Koundakjian, E., Nagashima, K., Han, X. & Acharya, J. K. (2003) Science 299, 1740–1743. [DOI] [PubMed] [Google Scholar]
- 5.Zuker, C. S., Cowman, A. F. & Rubin, G. M. (1985) Cell 40, 851–858. [DOI] [PubMed] [Google Scholar]
- 6.Zuker, C. S. (1996) Proc. Natl. Acad. Sci. USA 93, 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar, J. P. & Ready, D. F. (1995) Development (Cambridge, U.K.) 121, 4359–4370. [DOI] [PubMed] [Google Scholar]
- 8.Chang, H. Y. & Ready, D. F. (2000) Science 290, 1978–1980. [DOI] [PubMed] [Google Scholar]
- 9.Leonard, D. S., Bowman, V. D., Ready, D. F. & Pak, W. L. (1992) J. Neurobiol. 23, 605–626. [DOI] [PubMed] [Google Scholar]
- 10.O'Tousa, J. E., Leonard, D. S. & Pak, W. L. (1989) J. Neurogenet. 6, 41–52. [DOI] [PubMed] [Google Scholar]
- 11.Acharya, J. K., Labarca, P., Delgado, R., Jalink, K. & Zuker, C. S. (1998) Neuron 20, 1219–1229. [DOI] [PubMed] [Google Scholar]
- 12.Kumar, J. P., Bowman, J., O'Tousa, J. E. & Ready, D. F. (1997) Dev. Biol. 188, 43–47. [DOI] [PubMed] [Google Scholar]
- 13.Freeman, M. (1996) Cell 87, 651–660. [DOI] [PubMed] [Google Scholar]
- 14.Hay, B. A., Wolff, T. & Rubin, G. M. (1994) Development (Cambridge, U.K.) 120, 2121–2129. [DOI] [PubMed] [Google Scholar]
- 15.Van Vactor, D., Jr., Krantz, D. E., Reinke, R. & Zipursky, S. L. (1988) Cell 52, 281–290. [DOI] [PubMed] [Google Scholar]
- 16.Stein, P. J., Brammer, J. D. & Ostroy, S. E. (1979) J. Gen. Physiol. 74, 565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwemer, J. & Henning, U. (1984) Cell Tissue Res. 236, 293–303. [DOI] [PubMed] [Google Scholar]
- 18.Stark, W. S., Sapp, R. & Schilly, D. (1988) J. Neurocytol. 17, 499–509. [DOI] [PubMed] [Google Scholar]
- 19.MacKenzie, D., Arendt, A., Hargrave, P., McDowell, J. H. & Molday, R. S. (1984) Biochemistry 23, 6544–6549. [DOI] [PubMed] [Google Scholar]
- 20.Baker, E. K., Colley, N. J. & Zuker, C. S. (1994) EMBO J. 13, 4886–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colley, N. J., Cassill, J. A., Baker, E. K. & Zuker, C. S. (1995) Proc. Natl. Acad. Sci. USA 92, 3070–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardie, R. C., Martin, F., Cochrane, G. W., Juusola, M., Georgiev, P. & Raghu, P. (2002) Neuron 36, 689–701. [DOI] [PubMed] [Google Scholar]
- 23.Montell, C. (1999) Annu. Rev. Cell Dev. Biol. 15, 231–268. [DOI] [PubMed] [Google Scholar]
- 24.Ranganathan, R., Harris, W. A. & Zuker, C. S. (1991) Trends Neurosci. 14, 486–493. [DOI] [PubMed] [Google Scholar]
- 25.Claing, A., Laporte, S. A., Caron, M. G. & Lefkowitz, R. J. (2002) Prog. Neurobiol. 66, 61–79. [DOI] [PubMed] [Google Scholar]
- 26.Schwemer, J. (1984) J. Comp. Physiol. A 154, 535–547. [Google Scholar]
- 27.Ranganathan, R. & Stevens, C. F. (1995) Cell 81, 841–848. [DOI] [PubMed] [Google Scholar]
- 28.Kiselev, A., Socolich, M., Vinos, J., Hardy, R. W., Zuker, C. S. & Ranganathan, R. (2000) Neuron 28, 139–152. [DOI] [PubMed] [Google Scholar]
- 29.Orem, N. R. & Dolph, P. J. (2002) Mol. Vis. 8, 455–461. [PubMed] [Google Scholar]
- 30.Sapp, R. J., Christianson, J. & Stark, W. S. (1991) J. Neurocytol. 20, 597–608. [DOI] [PubMed] [Google Scholar]
- 31.Lee, S. J., Xu, H., Kang, L. W., Amzel, L. M. & Montell, C. (2003) Neuron 39, 121–132. [DOI] [PubMed] [Google Scholar]
- 32.Wu, L., Niemeyer, B., Colley, N., Socolich, M. & Zuker, C. S. (1995) Nature 373, 216–222. [DOI] [PubMed] [Google Scholar]
- 33.O'Tousa, J. E. (2002) Adv. Exp. Med. Biol. 514, 493–505. [PubMed] [Google Scholar]
- 34.Pak, W. L. & Leung, H. T. (2003) Recept. Channels 9, 149–167. [PubMed] [Google Scholar]
- 35.Ait Slimane, T. & Hoekstra, D. (2002) FEBS Lett. 529, 54–59. [DOI] [PubMed] [Google Scholar]
- 36.van Meer, G. & Lisman, Q. (2002) J. Biol. Chem. 277, 25855–25858. [DOI] [PubMed] [Google Scholar]
- 37.Obeid, L. M., Okamoto, Y. & Mao, C. (2002) Biochim. Biophys. Acta. 1585, 163–171. [DOI] [PubMed] [Google Scholar]
- 38.D'Hondt, K., Heese-Peck, A. & Riezman, H. (2000) Annu. Rev. Genet. 34, 255–295. [DOI] [PubMed] [Google Scholar]