Abstract
Results from over a dozen prostate cancer susceptibility genome-wide scans, encompassing some 1,500 hereditary prostate cancer families, indicate that prostate cancer is an extremely heterogeneous disease with multiple loci contributing to overall susceptibility. In an attempt to reduce locus heterogeneity, we performed a genomewide linkage scan for prostate cancer susceptibility genes with 36 Jewish families, which represent a stratification of hereditary prostate cancer families with potentially increased locus homogeneity. The 36 Jewish families represent a combined dataset of 17 Jewish families from the Fred Hutchinson Cancer Research Center-based Prostate Cancer Genetic Research Study dataset and 19 Ashkenazi Jewish families collected at Johns Hopkins University. All available family members, including 94 affected men, were genotyped at markers distributed across the genome with an average interval of <10 centimorgans. Nonparametric multipoint linkage analyses were the primary approach, although parametric analyses were performed as well. Our strongest signal was a significant linkage peak at 7q11–21, with a nonparametric linkage (NPL) score of 3.01 (P = 0.0013). Simulations indicated that this corresponds to a genomewide empirical P = 0.006. All other regions had NPL P values ≥0.02. After genotyping additional markers within the 7q11–21 peak, the NPL score increased to 3.35 (P = 0.0004) at D7S634 with an allele-sharing logarithm of odds of 3.12 (P = 0.00007). These studies highlight the utility of analyzing defined sets of families with a common origin for reducing locus heterogeneity problems associated with studying complex traits.
In 2003, an estimated 220,900 men will be diagnosed with prostate cancer in the U.S., and 28,900 will die of the disease (1). Both epidemiological studies and segregation analyses confirm the existence of a genetic component to prostate cancer etiology. Two segregation analyses, both based on ascertainment of family history through probands treated with radical prostatectomy, show evidence for the dominant transmission of a rare high-risk allele (population prevalence of 0.3–0.6%), with carriers having an 88–89% risk of prostate cancer by 85 years of age, compared with 3–5% in noncarriers (2, 3). Carter et al. (2) suggest that the cumulative proportion of prostate cancer cases within the population attributable to high-risk susceptibility alleles is 43% for men ≤55 years of age, 34% for men ≤70 years of age, and 9% for men ≤85 years of age. By comparison, population-based studies from Sweden (4) and Australia (5) estimate a higher population prevalence of carriers (1.1–1.67%) and a lower lifetime incidence (63–79%). They suggest also that 23% of all prostate cancer cases diagnosed at <65 years of age may be due to inherited mutations in susceptibility genes (4).
These observations have motivated a large body of work aimed at finding susceptibility genes involved in hereditary prostate cancer (HPC). The scan of Smith et al. (6) in 1996 highlighted regions of chromosome 1q24–25, 4q27, and Xq27–28 as containing prostate cancer loci, with the result at 1q24–25 being statistically significant. A maximum multipoint logarithm of odds (LOD) score of 5.43 under the assumption of heterogeneity was observed, with 34% of families predicted to be linked. Several replication studies were published subsequently, with some confirming the initial findings (7–9), whereas others could not (10–14). A large metaanalysis of 772 families provided weak evidence overall, suggesting that about 6% of families could attribute their disease to HPC1 (15). Several studies focus on RNASEL as a candidate gene for the HPC1 locus (16), but attempts to demonstrate that mutations in RNASEL are solely responsible for the initial findings of linkage at 1q24–5 have been inconclusive (17–23).
Subsequent scans have identified loci on chromosomes 1p (CAPB) (24, 25), 1q (PCAP) (11), 20q (HPC20) (26), Xq (HCPX) (27), and HPC2/ELAC2 on chromosome 17p (28) and the MSR1 gene on chromosome 8p (29, 30). Other loci of interest have been reported on chromosomes 19p (31), 19q (32, 33), and 16q (34). As with HPC1, attempts at confirmation have proven inconclusive for HPC20 (35, 36), PCAP (14, 37, 38), HPCX (39, 40), and CAPB (14, 41), as well as for the HPC2/ELAC2 and MSR1 genes (42–46).
In late 2003, eight additional genomewide scans for prostate cancer susceptibility loci were reported, including our own studies of 254 Prostate Cancer Genetic Research Study (PROGRESS) and 188 Johns Hopkins University (JHU) families (47, 48). The aggregate results are summarized in a review by Easton et al. (49). The eight scans include 1,292 families with multiple cases of prostate cancer. Across all studies, 11 peaks with LOD scores >2 were observed, identifying regions on chromosomes 2–7, 9, 16, 17, 19, and 20. No chromosomal region was reported as significant at the LOD ≥2.0 level by more than one study, and only one LOD score ≥3.0 was reported (49).
Given the extreme locus heterogeneity associated with HPC, we have sought ways to define homogeneous subsets for refined analyses. One approach is to evaluate families from relatively isolated populations with a limited number of founders. One well defined population, which meets these criteria and has proven useful for genetic studies of other cancer susceptibility genes, is of Americans of Ashkenazi Jewish descent (50). For instance, founder mutations within the BRCA1, BRCA2, and MSH2 genes have been identified in studies of Ashkenazi Jewish individuals (51–55). Thus, we hypothesize that genomewide linkage analyses of HPC families of Jewish descent will increase locus homogeneity and our ability to find true susceptibility loci. Toward that end, we have analyzed a genomewide scan of 36 Jewish families by combining 17 families from the Fred Hutchinson Cancer Research Center-led PROGRESS and 19 families collected by JHU. These data highlight a region of chromosome 7q11–21 with significant results.
Subjects and Methods
Prostate Cancer Family Collection. Seventeen families were collected as part of the Fred Hutchinson Cancer Research Centerled national PROGRESS. They derive from a larger dataset of 255 families ascertained from throughout North America and several other countries by advertising a toll-free number by means of public media, health-related publications, and the internet, as well as communications with urologists, other healthcare professionals, and prostate cancer support groups (10). To be eligible for the PROGRESS, families were required to meet at least one of the following criteria: (i) have three or more first-degree relatives with prostate cancer; (ii) have three generations with prostate cancer, through either paternal or maternal lineage; or (iii) have two first-degree relatives with prostate cancer diagnosed before age 65 or be African American. Families from the parent PROGRESS were eligible for this study if they self-identified as Jewish in response to a question on religious preference on a baseline questionnaire, which also collected information on each family member's country of origin. All 17 families are of Central or Eastern European descent and are likely to be Ashkenazi Jewish.
Medical records and death certificates were obtained to confirm the diagnosis of prostate cancer. Of the 48 medical records received on the putatively affected men who were genotyped, 100% confirmed the self-reported prostate cancer diagnosis. Death certificates confirmed an additional 6 of the 15 unsampled prostate cancer diagnoses in the 17 PROGRESS families. Only one sampled affected man's medical records were not available to confirm the diagnosis. Because of the high accuracy of prostate cancer self-reporting, the individual for whom records were not available was considered affected.
The 19 JHU Ashkenazi Jewish families are a subset of 188 HPC families that were collected and studied at the Brady Urology Institute at Johns Hopkins Hospital (48). A majority of cases were ascertained through referrals generated in response to a letter distributed to 8,000 urologists throughout the country. Families were also identified from family history records of patients seen at Johns Hopkins Hospital for treatment of prostate cancer. The remaining families were respondents to articles in a variety of lay publications describing JHU prostate cancer family studies. To qualify for the study, each family was required to have at least three first-degree relatives affected with prostate cancer. Medical records verified prostate cancer diagnosis for each affected male studied. Families were queried as to their religious preference to establish Jewish status. Individuals indicating Jewish were further asked to specify Ashkenazi or not, and all 19 families self-identified as Ashkenazi Jewish. Age at diagnosis of prostate cancer was confirmed either through medical records or from two other independent sources.
In the 36 PROGRESS and JHU Jewish families, all 45 genotyped affected men from JHU and 45 of 49 PROGRESS genotyped men self-identified as Jewish. The four genotyped affected men who did not self-report as Jewish are members of distinct families. None of the families were bilineal. In addition, no excess of breast or breast and ovarian cancer was observed when the Jewish datasets were compared, respectively, with the larger PROGRESS (n = 254) and JHU (n = 188) HPC datasets. It should be noted, however, that on average the Jewish families from both datasets were smaller and hence had fewer relatives at risk for breast or ovarian cancer than the non-Jewish families. For both the PROGRESS and JHU families, study forms and protocols were approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center and the Johns Hopkins Medical Institutions, respectively.
Genotyping. For both studies, genomic DNA was extracted from peripheral blood lymphocytes by using standard techniques (56). For the PROGRESS families, a total of 441 microsatellite markers were genotyped. Details of the PCR amplification, marker characteristics, and genotyping are described elsewhere (25, 47). Briefly, the average marker heterozygosity was 70%, and the average spacing between markers was 8.1 centimorgans. Genotyping data were checked for errors before analysis by using pedcheck (57), relpair (58), and prest (59). The JHU families were genotyped by using 406 microsatellite markers with an average intermarker spacing of 10 centimorgans and average heterozygosity of 80%. PCR conditions and genotyping methods are described elsewhere (48). For both datasets, all genotyping included the same Centre d'Étude du Polymorphisme Humain (CEPH) individual (1347-02) for quality control purposes.
To combine the two datasets, only markers present in the University of California, Santa Cruz genome browser April 2003 assembly (http://genome.ucsc.edu) were used (PROGRESS 421 markers, JHU 398 markers). Map order and distance were based on the University of California, Santa Cruz map. The markers from the other genomewide scan were given no genotypes for all individuals (0, 0). There were 26 markers in common in both scans. To avoid the problem of allele binning, these markers were treated as separate markers and given map distances <0.1.
Statistical Analysis. A genomewide linkage scan was performed by using nonparametric multipoint linkage analyses as the primary method of analysis because of the uncertainty regarding likely mode of inheritance (60). Parametric multipoint analyses were also performed. The computer program merlin, Version 0.9.8, was used to perform affected relative-pair linkage analyses (61). The estimated marker identical-by-descent sharing of alleles for the various affected relative-pairs was compared with the values expected under the null hypothesis of no linkage. Model-free allele sharing was evaluated by using the nonparametric linkage (NPL)all statistic. Allele-sharing LOD scores were then calculated based on the NPLall statistic, with equal weight assigned to all families (62). P values associated with LOD scores were calculated assuming the NPLall statistic was normally distributed and were not adjusted for multiple tests.
Empirical P values were calculated for the NPLall scores by performing simulations. The merlin program was used to generate and analyze 1,000 replicates of the entire genome from the original dataset of 36 Jewish families.
For the parametric analysis, a dominant two-liability class model was used. This model is based on the model used by Smith et al. (6) and assumes dominant inheritance of a disease allele with a frequency of 0.003. The two-liability class model is an affecteds-only analysis where the affection status of all unaffected men and women is assumed to be unknown. A maximum-likelihood approach was used to estimate the proportion of linked families by maximizing the admixed LOD score (heterogeneity LOD, HLOD), as implemented in the computer program genehunter, Version 2.0 (63, 64).
Results
The 36 Jewish HPC families have the following characteristics. There are 161 individuals genotyped, and, of the 149 affected men, 94 are genotyped. The mean age at diagnosis is 64.8 years, ranging from 38 to 81 years, and families contain from 2 to 10 genotyped individuals.
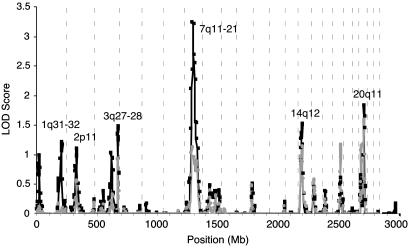
We performed a combined genomewide linkage analysis of the 36 families and identified a region of significant linkage on chromosome 7q11–21 (Fig. 1 and Table 1). The NPL P value was <0.01 for nine consecutive markers (D7S1818–D7S630) spanning 29.2 centimorgans (38.9 Mb). The maximum NPL score was 3.01 (P = 0.0013) at marker D7S502. Simulation studies find a genomewide probability of this NPL score of 0.006 in these data, thus, the genomewide P value is 0.006. The multipoint HLOD score was positive for the dominant two-liability model (HLOD = 1.14).
Fig. 1.
Initial genomewide scan results for the 36 Jewish families. Allele-sharing LOD scores implemented by merlin are indicated by black squares, and HLOD scores from multipoint parametric analysis using the two-liability class model analyzed with genehunter are indicated by gray circles. Vertical dashed lines separate the chromosomes. Mb, megabases.
Table 1. Initial linkage results with NPL P values ≤0.06.
| Nonparametric analysis
|
Parametric analysis*
|
|||||
|---|---|---|---|---|---|---|
| Chromosome | Peak marker | NPL | Position, Mb | P | HLOD | Position, Mb |
| 1q31-32 | D1S1660 | 1.53 | 195.1 | 0.06 | 0.10 | 211.8 |
| D1S413 | 195.1 | |||||
| 2p11 | D2S2333 | 1.60 | 85.5 | 0.06 | 0.57 | 88.3 |
| 3q27-28 | D3S1262 | 1.84 | 187.6 | 0.03 | 0.94 | 189.0 |
| D3S1580 | 189.9 | |||||
| 7q11-21 | D7S502 | 3.01 | 66.5 | 0.0013 | 1.14 | 51.5 |
| 14q12 | D14S1040 | 2.05 | 30.2 | 0.02 | 1.15 | 30.2 |
| D14S297 | 30.5 | |||||
| 20q11 | D20S195 | 1.80 | 32.5 | 0.04 | 1.65 | 30.8 |
Dominant parametric HLOD scores generated by using the two-liability class model.
Our analysis of 36 Jewish families also highlighted regions on chromosomes 1q31–32, 2p11, 3q27–28, 14q12, and 20q11 with P values of 0.02 to 0.06. The strongest of these was at 14q12 (P = 0.02). Other minor peaks with an NPL P value ≤0.06 include 1q31–32 (P = 0.06), 2p11 (P = 0.06), 3q27–28 (P = 0.03), and 20q11 (P = 0.04) (Table 1). The dominant multipoint HLOD scores at 14q12 and 20q11 were 1.15 and 1.59, respectively, when the two-liability class model was used. For all of the other minor peaks, the multipoint HLODs were <1.0.
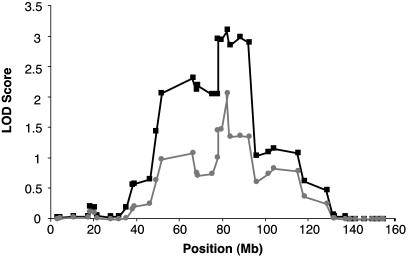
To refine the region of interest, we genotyped additional markers within the chromosome 7 peak and found support for the initial finding (Fig. 2 and Table 2). The final dataset included 11 markers across the 7q11–21 peak where genotypes are available from both JHU and PROGRESS datasets, including three markers not present in either original genomewide scan. With the inclusion of these markers, the NPL P values were <0.01 from D7S1818 to D7S630 as previously observed, and the maximum NPL score was 3.35 (P = 0.0004) at D7S634 with a corresponding allele-sharing LOD of 3.12 (P = 0.00007). Additionally, the multipoint HLOD generated by using the two-liability class model was 2.06. After stratifying by mean age at diagnosis, the NPL scores were 2.30 (P = 0.011) in 18 younger-age-at-diagnosis families (mean age <65) and 3.27 (P = 0.0005) in the 18 older-age-at-diagnosis families (mean age ≥65), indicating that although the older-age-at-diagnosis families account for most of the result, both younger- and older-age families contribute to the peak at 7q11–21. We had insufficient data from the older generations in many of the 36 families to accurately stratify by inheritance pattern (i.e., evidence of male-to-male transmission).
Fig. 2.
Fine mapping multipoint linkage results on chromosome 7. Allele-sharing LOD scores implemented by merlin are indicated by black squares, and HLOD scores from multipoint parametric analysis using the 2-liability class model analyzed with genehunter are indicated by gray circles.
Table 2. Chromosome 7 fine mapping linkage results.
| Position, Mb
|
Gap, Mb*
|
Nonparametric analysis
|
Parametric analysis†
|
||
|---|---|---|---|---|---|
| Marker | NPL | P | HLOD | ||
| D7S510 | 38.90 | 1.06 | 1.15 | 0.12 | 0.26 |
| D7S519 | 45.82 | 3.28 | 2.03 | 0.02 | 0.65 |
| D7S1818 | 49.10 | 2.36 | 2.48 | 0.007 | 0.99 |
| D7S1830 | 51.46 | 15.00 | 2.62 | 0.004 | 1.09 |
| D7S502‡ | 66.46 | 1.49 | 2.75 | 0.003 | 0.76 |
| D7S3046‡ | 67.95 | 0.51 | 2.78 | 0.003 | 0.71 |
| D7S2435‡ | 68.46 | 6.52 | 2.75 | 0.003 | 0.74 |
| D7S2518‡ | 74.98 | 2.49 | 2.74 | 0.003 | 1.01 |
| D7S669‡ | 77.47 | 0.26 | 3.07 | 0.0011 | 1.46 |
| D7S2204‡ | 77.73 | 1.72 | 3.08 | 0.001 | 1.48 |
| D7S634‡ | 79.45 | 2.95 | 3.35 | 0.0004 | 2.06 |
| D7S2212‡ | 82.40 | 0.99 | 3.26 | 0.0006 | 1.36 |
| D7S820‡ | 83.39 | 4.65 | 3.35 | 0.0004 | 1.36 |
| D7S630‡ | 88.04 | 4.36 | 3.3 | 0.0005 | 1.36 |
| D7S657‡ | 92.40 | 3.26 | 2.02 | 0.02 | 0.61 |
| D7S821 | 95.66 | 5.59 | 1.93 | 0.03 | 0.75 |
Distance from previous marker.
Dominant parametric HLOD scores generated by using the two-liability class model.
Markers with genotypes available from both JHU and PROGRESS families.
Finally, we considered the degree to which Jewish families accounted for results on chromosome 7q reported in our most recent genomewide scans (47, 48). In the case of the 254 PROGRESS families, we reported an HLOD of 2.25 (LOD = 1.55) at marker D7S2212 on 7q21 using a recessive parametric model. Furthermore, we noted an NPL result of 1.79 (P = 0.038) in this same region. Analysis of the 237 non-Jewish families from the PROGRESS dataset yielded an NPL score of 1.11 (P = 0.134), suggesting that the majority of the initial NPL result in the genomewide scan for the PROGRESS families can be accounted for by the Jewish families. In the genomewide study of 188 JHU families by Xu et al. (48), the strongest result on 7q was an allele-sharing LOD of 1.63 with marker D7S486. This result is at 7q22, which is ≈27 Mb from the region at 7q21 defined here. When 17 of the 19 JHU Ashkenazi Jewish families were analyzed by using D7S486, the allele-sharing LOD was only 0.04, suggesting that the JHU Ashkenazi Jewish families do not contribute significantly to the 7q22 result reported previously (48).
Discussion
We present here a genomewide scan for prostate cancer susceptibility loci in Jewish families, representing a well defined, isolated population. Only one significant linkage result was identified, on 7q11–21, although there were five other nonsignificant linkage peaks. The clear indication of only one susceptibility locus in these 36 Jewish families highlights the utility of this approach when investigating a disease with extreme locus heterogeneity, such as prostate cancer. Indeed, at this point in time, susceptibility loci have been identified on nearly every human chromosome (49).
The initial linkage result at 7q11–21 was a maximum NPL = 3.01 (P = 0.0013). Simulations indicated that this corresponds to a genomewide empirical P = 0.006. This initial finding was supported (NPL = 3.35, P = 0.0004) when additional markers in the region were genotyped. For both the initial and fine mapping linkage results, the multipoint HLOD scores (1.14 and 2.06, respectively) are less significant than the NPL results, presumably because of misspecification of the genetic models used in the parametric linkage analysis. For this heterogeneous disease, the parametric models were developed by using segregation analyses of populations that were not specifically Jewish (2–5). Therefore, these models are unlikely to accurately define prostate cancer genetics in the Jewish families.
Other studies have reported results of interest adjacent to 7q11–21, although none has been statistically significant. In an analysis of 326 affected sib pairs, Goddard et al. (65) modeled age as a covariate and observed a LOD score of 1.68 with markers overlapping the peak defined here at 7q11–21, with markers D7S3046 and D7S2204. In a previous study of the same affected sib-pair dataset, Witte et al. (32) identified a prostate cancer aggressiveness loci ≈44 Mb away at 7q31–32, by using Gleason score as an outcome variable. Their result at 7q31–32 (P = 0.0007) was one of the three strongest signals in the entire study. Fine-scale mapping by the same group (66) and a separate analysis of 100 German HPC families (67) support the finding at 7q31–32. We believe the locus defined by studies at 7q31–32 is distinct from the result reported here at 7q11–21. In 254 PROGRESS HPC families, we noted an NPL score of 1.77 (P = 0.028) with marker D7S1824 at 7q31–32 (47). However, the NPL score in the 237 PROGRESS families that did not self-identify as Jewish was 1.64 (P = 0.05), suggesting that the Jewish families do not contribute to the aggressiveness locus at 7q31–32. Furthermore, 7q31–32 is ≈44 Mb from the locus defined here at 7q11–21.
The strongest result reported previously at 7q11–21 was that described by our own analyses of 254 PROGRESS HPC families. Janer et al. (47) observed a peak LOD score at marker D7S2212 on 7q21 by using a recessive parametric model, with an HLOD of 2.25 (LOD = 1.55). Stratification of the dataset by age at diagnosis only slightly improved this result. In 214 families with a median age at diagnosis of 56–72 years, the HLOD was 2.41 at D7S2212 (LOD = 1.68). Analysis of the 31 HPC families with multiple breast and/or ovarian cancer identified an HLOD of 2.21 at D7S2204 (LOD = 2.15), where both the LOD and HLOD were 1.96 in the 15 HPC ovarian cancer families alone. In addition, analysis of the 237 families from the PROGRESS that did not self-identify as Jewish yielded an NPL score of 1.11 (P = 0.134) compared with 1.79 (P = 0.038) in the entire dataset, suggesting that the small number (n = 17) of PROGRESS Jewish families contribute disproportionately to the NPL result in the full dataset (n = 254). The co-occurrence of the multiple breast and/or ovarian cancer linkage result with the 7q11–21 result in the Jewish HPC families suggests that the 7q11–21 locus is important for a larger subset of HPC families than those who self-identify as Jewish.
The joint genomewide scan reported here also highlighted regions on chromosomes 1q31–32, 2p11, 3q27–28, 14q12, and 20q11 with P values of 0.02–0.06. Other studies have reported linkage peaks in some of these regions. On chromosome 20, we observed a minor peak at 20q11, which is >10 Mb from the HPC20 locus defined by Berry and colleagues (26). At 2p11, Gibbs et al. (25), in an analysis of 94 PROGRESS families, observed a LOD score of 1.58 in families with a mean age at diagnosis of <65 years. In the study of all 254 PROGRESS families, an HLOD of 1.47 is reported at D14S1280 near the 14q12 peak observed in the 36 Jewish families described here (allele-sharing LOD = 1.38 at D14S1280) (47). Finally, our joint result at 1q31–32 is very near the HPC1 locus, located at 1q24–25. HPC1 was initially reported by Smith et al. (6) (HLOD = 5.43) when analyzing a set of 91 families, 79 of which were from JHU and 12 of which were from Sweden. Recently, Xu et al. (48) reported the analysis of 17 of the 19 JHU Ashkenazi Jewish families described here at HPC1, demonstrating that the highest allele-sharing LOD score was 1.70 at 1q31–32 (D1S413). By comparison, in the combined analysis of 36 Jewish families from PROGRESS and JHU, the maximum NPL score was 1.53 (P = 0.06) and the maximum allele-sharing LOD was 1.22.
RNASEL has been proposed as a candidate gene for the HPC1 locus (16), and as with other candidate genes, replication studies have proven inconclusive (17–23). Although the RNASEL 471delAAAG mutation was found to be associated with prostate cancer in Ashkenazi Jews in one study (18), this was not confirmed in another similar study (21). Only 1 individual of 161 in the 36 Jewish families described here carries the 471delAAAG mutation (Avi Orr-Urtreger and Mariela Langlois, personal communication). Thus, the 471delAAAG mutation does not appear to be responsible for prostate cancer susceptibility in the 36 Jewish families described here. However, we cannot exclude the possibility that other RNASEL mutations could be responsible for the linkage result we observe at the HPC1 locus.
Previous studies of the JHU Ashkenazi Jewish families have highlighted three additional loci of interest at 8p22, 10p15, and 20p13 (48, 68). None of these loci is supported in this joint analysis. The most provocative was the initial linkage report for 8p22, where a small number of Ashkenazi Jewish families (n = 11) contributed disproportionately to the linkage result with a maximum allele-sharing LOD of 1.31 compared with 1.39 for the remaining 133 non-Jewish Caucasian families (68). However, in this combined analysis of 36 Jewish families, no evidence for linkage was seen at 8p22 (allele-sharing LOD = 0.01). The differing results could reflect the small number of families available before the combined analysis.
Analysis of HPC families after stratifying by, or adjusting for, age at diagnosis has proven to be informative in previous linkage studies (reviewed in refs. 49 and 69–71). Stratification of the 36 Jewish families by mean age at diagnosis (<65 vs. ≥65 years) indicated that the older-age-at-diagnosis families contribute disproportionately to the chromosome 7 result (NPL = 3.27, P = 0.0005). However, the younger families also have a peak at 7q11–21 with an NPL = 2.3 (P = 0.011), suggesting that these families contribute to the result as well. Overall, these observations support the notion that the result reported here at 7q11–21 was not due to the contribution of only one or two families, and that replication of this result is likely in other datasets of Jewish HPC families. In addition, the differences in the younger- vs. older-age-at-diagnosis families is not surprising, as the median number of affected individuals with genotypes is 2.0 in the younger families and 3.0 in the older families. We note, however, that the median number of affected individuals reported per family is 4.0 for both the younger and older age at diagnosis families. Thus, the weaker linkage result obtained in the younger families probably reflects the reduced number of affected men who were genotyped in these families, and accordingly the reduced power of the analysis.
In summary, we performed a prostate cancer genomewide screen in Jewish families and identified a strong linkage result on chromosome 7q with a relatively small number of families. Our result suggests that reducing locus heterogeneity by grouping Jewish families of similar background is a useful strategy for linkage studies of complex diseases. Previously identified cancer susceptibility genes for which Ashkenazi Jewish founder mutations have been described include the BRCA1, BRCA2, and MSH2 genes (51–55). Mutations in these genes are also strongly relevant for disease susceptibility in non-Jewish populations. Therefore, identification of the 7q11–21 gene is likely to be important in advancing our understanding of the etiology of this complex and common disease.
Acknowledgments
We especially thank the PROGRESS and JHU families for participating in this research. We thank Dr. Avi Orr-Urtreger and Mariela Langlois for providing the RNASEL sequencing information and Drs. Kyriacos Markianos and Joshua Akey for helpful comments on the PROGRESS linkage results. We also thank Toby Karyadi for perl programming assistance. This research was supported by National Institutes of Health Grants RO1 CA80122 and RO1 CA089600 (to J.L.S.), RO1 CA78836 and K05CA90754 (to E.A.O.), and RO1 CA58236 (to W.B.I.); Fred Hutchinson Cancer Research Center Interdisciplinary Training Grant CA80416 (to D.M.F.); the Pew Biomedical Scholars Program (G.P.J.); and Department of Defense Grants DAMD17-98-1-8469 (to W.B.I.) and DAMD17-00-1-0087 (to J.X.).
Abbreviations: HPC, hereditary prostate cancer; JHU, Johns Hopkins University; LOD, logarithm of odds; HLOD, heterogeneity LOD; Mb, megabases; NPL, nonparametric linkage; PROGRESS, Prostate Cancer Genetic Research Study.
References
- 1.Jemal, A., Murray, T., Samuels, A., Ghafoor, A., Ward, E. & Thun, M. J. (2003) CA Cancer J. Clin. 53, 5–26. [DOI] [PubMed] [Google Scholar]
- 2.Carter, B. S., Beaty, T. H., Steinberg, G. D., Childs, B. & Walsh, P. C. (1992) Proc. Natl. Acad. Sci. USA 89, 3367–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaid, D. J., McDonnell, S. K., Blute, M. L. & Thibodeau, S. N. (1998) Am. J. Hum. Genet. 62, 1425–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grönberg, H., Damber, L., Damber, J.-E. & Iselius, L. (1997) Am. J. Epidemiol. 146, 552–557. [DOI] [PubMed] [Google Scholar]
- 5.Cui, J., Staples, M. P., Hopper, J. L., English, D. R., McCredie, M. R. & Giles, G. G. (2001) Am. J. Hum. Genet. 68, 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith, J. R., Freije, D., Carpten, J. D., Grönberg, H., Xu, J., Isaacs, S., Brownstein, M. J., Bova, G. S., Guo, H., Bujnovszky, P., et al. (1996) Science 274, 1371–1374. [DOI] [PubMed] [Google Scholar]
- 7.Cooney, K. A., McCarthy, J. D., Lange, E., Huang, L., Miesfeldt, S., Montie, J. E., Oesterling, J. E., Sandler, H. M. & Lange, K. (1997) J. Natl. Cancer Inst. 89, 955–959. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh, C.-L., Oakley-Girvan, I., Gallagher, R. P., Wu, A. H., Kolonel, L. N., The, C. Z., Halpern, J., West, D. W., Paffenberger, R. S., Jr., & Whittemore, A. S. (1997) J. Natl. Cancer Inst. 89, 1893–1894. [DOI] [PubMed] [Google Scholar]
- 9.Neuhausen, S. L., Farnham, J. M., Kort, E., Tavtigian, S. V., Skolnick, M. H. & Cannon-Albright, L. A. (1999) Hum. Mol. Genet. 8, 2437–2442. [DOI] [PubMed] [Google Scholar]
- 10.McIndoe, R. A., Stanford, J. L., Gibbs, M., Jarvik, G. P., Brandzel, S., Neal, C. L., Li, S., Gammack, J. T., Gay, A. A., Goode, E. L., et al. (1997) Am. J. Hum. Genet. 61, 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berthon, P., Valeri, A., Cohen-Akenine, A., Drelon, E., Paiss, T., Wohr, G., Latil, A., Millasseau, P., Mellah, I., Cohen, N., et al. (1998) Am. J. Hum. Genet. 62, 1416–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eeles, R. A., Durocher, F., Edwards, S., Teare, D., Badzioch, M., Hamoudi, R., Gill, S., Biggs, P., Dearnaley, D., Ardern-Jones, A., et al. (1998) Am. J. Hum. Genet. 62, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goode, E., Stanford, J. L., Chakrabarti, L., Gibbs, M., Kolb, S., McIndoe, R. A., Buckley, V. A., Schuster, E. F., Neal, C. L., Miller, E. L., et al. (2000) Genet. Epidemiol. 18, 251–275. [DOI] [PubMed] [Google Scholar]
- 14.Berry, R., Schaid, D. J., Smith, J. R., French, A. J., Schroeder, J. J., McDonnell, S. K., Peterson, B. J., Wang, Z. Y., Carpten, J. D., Roberts, S. G., et al. (2000) Am. J. Hum. Genet. 66, 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu, J. & International Consortium for Prostate Cancer Genetics (2000) Am. J. Hum. Genet. 66, 945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpten, J., Nupponen, N., Isaacs, S., Sood, R., Robbins, C., Xu, J., Faruque, M., Moses, T., Ewing, C., Gillanders, E., et al. (2002) Nat. Genet. 30, 181–184. [DOI] [PubMed] [Google Scholar]
- 17.Rokman, A., Ikonen, T., Seppala, E. H., Nupponen, N., Autio, V., Mononen, N., Bailey-Wilson, J., Trent, J., Carpten, J., Matikainen, M. P., et al. (2002) Am. J. Hum. Genet. 70, 1299–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rennert, H., Bercovich, D., Hubert, A., Abeliovich, D., Rozovsky, U., Bar-Shira, A., Soloviov, S., Schreiber, L., Matzkin, H., Rennert, G., et al. (2002) Am. J. Hum. Genet. 71, 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, L., McDonnell, S. K., Elkins, D. A., Slager, S. L., Christensen, E., Marks, A. F., Cunningham, J. M., Peterson, B. J., Jacobsen, S. J., Cerhan, J. R., et al. (2002) Am. J. Hum. Genet. 71, 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casey, G., Neville, P. J., Plummer, S. J., Xiang, Y., Krumroy, L. M., Klein, E. A., Catalona, W. J., Nupponen, N., Carpten, J. D., Trent, J. M., et al. (2002) Nat. Genet. 32, 581–583. [DOI] [PubMed] [Google Scholar]
- 21.Kotar, K., Hamel, N., Thiffault, I. & Foulkes, W. D. (2003) J. Med. Genet. 40, e22 (lett.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, H., Griffin, A. R., Wu, Y. Q., Tomsho, L. P., Zuhlke, K. A., Lange, E. M., Gruber, S. B. & Cooney, K. A. (2003) J. Med. Genet. 40, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakazato, H., Suzuki, K., Matsui, H., Ohtake, N., Nakata, S. & Yamanaka, H. (2003) Br. J. Cancer 89, 691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbs, M., Stanford, J. L., McIndoe, R. A., Jarvik, G. P., Kolb, S., Goode, E. L., Chakrabarti, L., Schuster, E. F., Buckley, V. A., Miller, E. L., et al. (1999) Am. J. Hum. Genet. 64, 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibbs, M., Stanford, J. L., Jarvik, G. P., Janer, M., Badzioch, M., Peters, M. A., Goode, E. L., Kolb, S., Chakrabarti, L., Shook, M., et al. (2000) Am. J. Hum. Genet. 67, 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry, R., Schroeder, J. J., French, A. J., McDonnell, S. K., Peterson, B. J., Cunningham, J. M., Thibodeau, S. N. & Schaid, D. J. (2000) Am. J. Hum. Genet. 67, 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu, J., Meyers, D., Freije, D., Isaacs, S., Wiley, K., Nusskern, D., Ewing, C., Wilkens, E., Bujnovszky, P., Bova, G. S., et al. (1998) Nat. Genet. 20, 175–179. [DOI] [PubMed] [Google Scholar]
- 28.Tavtigian, S. V., Simard, J., Teng, D. H., Abtin, V., Baumgard, M., Beck, A., Camp, N. J., Carillo, A. R., Chen, Y., Dayananth, P., et al. (2001) Nat. Genet. 27, 172–180. [DOI] [PubMed] [Google Scholar]
- 29.Xu, J., Zheng, S. L., Komiya, A., Mychaleckyj, J. C., Isaacs, S. D., Hu, J. J., Sterling, D., Lange, E. M., Hawkins, G. A., Turner, A., et al. (2002) Nat. Genet. 32, 321–325. [DOI] [PubMed] [Google Scholar]
- 30.Xu, J., Zheng, S. L., Komiya, A., Mychaleckyj, J. C., Isaacs, S. D., Chang, B., Turner, A. R., Ewing, C. M., Wiley, K. E., Hawkins, G. A., et al. (2003) Am. J. Hum. Genet. 72, 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh, C. L., Oakley-Girvan, I., Balise, R. R., Halpern, J., Gallagher, R. P., Wu, A. H., Kolonel, L. N., O'Brien, L. E., Lin, I. G., Van Den Berg, D. J., et al. (2001) Am. J. Hum. Genet. 69, 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witte, J. S., Goddard, K. A., Conti, D. V., Elston, R. C., Lin, J., Suarez, B. K., Broman, K. W., Burmester, J. K., Weber, J. L. & Catalona, W. J. (2000) Am. J. Hum. Genet. 67, 92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neville, P. J., Conti, D. V., Krumroy, L. M., Catalona, W. J., Suarez, B. K., Witte, J. S. & Casey, G. (2003) Genes Chromosomes Cancer 36, 332–339. [DOI] [PubMed] [Google Scholar]
- 34.Suarez, B. K., Lin, J., Burmester, J. K., Broman, K. W., Weber, J. L., Banerjee, T. K., Goddard, K. A. B., Witte, J. S., Elston, R. C. & Catalona, W. J. (2000) Am. J. Hum. Genet. 66, 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng, S. L., Xu, J., Isaacs, S. D., Wiley, K., Chang, B., Bleecker, E. R., Walsh, P. C., Trent, J. M., Meyers, D. A. & Isaacs, W. B. (2001) Hum. Genet. 108, 430–435. [DOI] [PubMed] [Google Scholar]
- 36.Cancel-Tassin, G., Latil, A., Valeri, A., Guillaume, E., Mangin, P., Fournier, G., Berthon, P. & Cussenot, O. (2001) Int. J. Cancer 93, 455–456. [DOI] [PubMed] [Google Scholar]
- 37.Gibbs, M., Chakrabarti, L., Stanford, J. L., Goode, E. L., Kolb, S., Schuster, E. F., Buckley, V. A., Shook, M., Hood, L., Jarvik, G. P., et al. (1999) Am. J. Hum. Genet. 64, 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittemore, A. S., Lin, I. G., Oakley-Girvan, I., Gallagher, R. P., Halpern, J., Kolonel, L. N., Wu, A. H. & Hsieh, C. L. (1999) Am. J. Hum. Genet. 65, 254–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lange, E. M., Chen, H., Brierley, K., Perrone, E. E., Bock, C. H., Gillanders, E., Ray, M. E. & Cooney, K. A. (1999) Clin. Cancer Res. 5, 4013–4020. [PubMed] [Google Scholar]
- 40.Peters, M. A., Jarvik, G. P., Janer, M., Chakrabarti, L., Kolb, S., Goode, E. L., Gibbs, M., DuBois, C. C., Schuster, E. F., Hood, L., et al. (2001) Hum. Hered. 51, 107–113. [DOI] [PubMed] [Google Scholar]
- 41.Badzioch, M., Eeles, R., Leblanc, G., Foulkes, W. D., Giles, G., Edwards, S., Goldgar, D., Hopper, J. L., Bishop, D. T., Moller, P., et al. (2000) J. Med. Genet. 37, 947–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rebbeck, T. R., Walker, A. H., Zeigler-Johnson, C., Weisburg, S., Martin, A. M., Nathanson, K. L., Wein, A. J. & Malkowicz, S. B. (2000) Am. J. Hum. Genet. 67, 1014–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vesprini, D., Nam, R. K., Trachtenberg, J., Jewett, M. A., Tavtigian, S. V., Emami, M., Ho, M., Toi, A. & Narod, S. A. (2001) Am. J. Hum. Genet. 68, 912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, J., Zheng, S. L., Carpten, J. D., Nupponen, N. N., Robbins, C. M., Mestre, J., Moses, T. Y., Faith, D. A., Kelly, B. D., Isaacs, S. D., et al. (2001) Am. J. Hum. Genet. 68, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller, D. C., Zheng, S. L., Dunn, R. L., Sarma, A. V., Montie, J. E., Lange, E. M., Meyers, D. A., Xu, J. & Cooney, K. A. (2003) Cancer Res. 63, 3486–3489. [PubMed] [Google Scholar]
- 46.Wang, L., McDonnell, S. K., Cunningham, J. M., Hebbring, S., Jacobsen, S. J., Cerhan, J. R., Slager, S. L., Blute, M. L., Schaid, D. J. & Thibodeau, S. N. (2003) Nat. Genet. 35, 128–129. [DOI] [PubMed] [Google Scholar]
- 47.Janer, M., Friedrichsen, D. M., Stanford, J. L., Badzioch, M. D., Kolb, S., Deutsch, K., Peters, M. A., Goode, E. L., Welti, R., DeFrance, H. B., et al. (2003) Prostate 57, 309–319. [DOI] [PubMed] [Google Scholar]
- 48.Xu, J., Gillanders, E. M., Wiley, K. E., Chang, B.-l., Isaacs, S. D., Zheng, S. L., Jones, M., Gildea, D., Riedesel, E., Albertus, J., et al. (2003) Prostate 57, 320–325. [DOI] [PubMed] [Google Scholar]
- 49.Easton, D. F., Schaid, D. J., Whittemore, A. S. & Isaacs, W. J. (2003) Prostate 57, 261–269. [DOI] [PubMed] [Google Scholar]
- 50.Ostrer, H. (2001) Nat. Rev. Genet. 2, 891–898. [DOI] [PubMed] [Google Scholar]
- 51.Struewing, J. P., Abeliovich, D., Peretz, T., Avishai, N., Kaback, M. M., Collins, F. S. & Brody, L. C. (1995) Nat. Genet. 11, 198–200. [DOI] [PubMed] [Google Scholar]
- 52.Roa, B. B., Boyd, A. A., Volcik, K. & Richards, C. S. (1996) Nat. Genet. 14, 185–187. [DOI] [PubMed] [Google Scholar]
- 53.Neuhausen, S. L., Mazoyer, S., Friedman, L., Stratton, M., Offit, K., Caligo, A., Tomlinson, G., Cannon-Albright, L., Bishop, T., Kelsell, D., et al. (1996) Am. J. Hum. Genet. 58, 271–280. [PMC free article] [PubMed] [Google Scholar]
- 54.Neuhausen, S. L., Godwin, A. K., Gershoni-Baruch, R., Schubert, E., Garber, J., Stoppa-Lyonnet, D., Olah, E., Csokay, B., Serova, O., Lalloo, F., et al. (1998) Am. J. Hum. Genet. 62, 1381–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foulkes, W. D., Thiffault, I., Gruber, S. B., Horwitz, M., Hamel, N., Lee, C., Shia, J., Markowitz, A., Figer, A., Friedman, E., et al. (2002) Am. J. Hum. Genet. 71, 1395–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) in Molecular Cloning: A Laboratory Manual, ed. Nolan, C. (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed., Vol. 2, pp. 9.16–9.19. [Google Scholar]
- 57.O'Connell, J. R. & Weeks, D. E. (1998) Am. J. Hum. Genet. 63, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boehnke, M. & Cox, N. J. (1997) Am. J. Hum. Genet. 61, 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun, L., Wilder, K. & McPeek, M. S. (2002) Hum. Hered. 54, 99–110. [DOI] [PubMed] [Google Scholar]
- 60.Isaacs, W. B., Xu, J. & Walsh, P. (2001) in Prostate Cancer, Biology, Genetics, and the New Therapeutics, eds. Chung, L. W. K., Isaacs, W. B. & Simons, J. W. (Humana, Clifton, NJ).
- 61.Abecasis, G. R., Cherny, S. S., Cookson, W. O. & Cardon, L. R. (2002) Nat. Genet. 30, 97–101. [DOI] [PubMed] [Google Scholar]
- 62.Kong, A. & Cox, N. (1997) Am. J. Hum. Genet. 61, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kruglyak, L., Daly, M. J., Reeve-Daly, M. P. & Lander, E. S. (1996) Am. J. Hum. Genet. 58, 1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 64.Markianos, K., Daly, M. J. & Kruglyak, L. (2001) Am. J. Hum. Genet. 68, 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goddard, K. A., Witte, J. S., Suarez, B. K., Catalona, W. J. & Olson, J. M. (2001) Am. J. Hum. Genet. 68, 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neville, P. J., Conti, D. V., Paris, P. L., Levin, H., Catalona, W. J., Suarez, B. K., Witte, J. S. & Casey, G. (2002) Neoplasia 4, 424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paiss, T., Worner, S., Kurtz, F., Haeussler, J., Hautmann, R. E., Gschwend, J. E., Herkommer, K. & Vogel, W. (2003) Eur. J. Hum. Genet. 11, 17–22. [DOI] [PubMed] [Google Scholar]
- 68.Xu, J., Zheng, S. L., Hawkins, G. A., Faith, D. A., Kelly, B., Isaacs, S. D., Wiley, K. E., Chang, B., Ewing, C. M., Bujnovszky, P., et al. (2001) Am. J. Hum. Genet. 69, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ostrander, E. A. & Stanford, J. L. (2000) Am. J. Hum. Genet. 67, 1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh, R., Eeles, R. A., Durocher, F., Simard, J., Edwards, S., Badzioch, M., Kote-Jarai, Z., Teare, D., Ford, D., Dearnaley, D., et al. (2000) Prostate Cancer Prostatic Dis. 3, 241–247. [DOI] [PubMed] [Google Scholar]
- 71.Nupponen, N. N. & Carpten, J. D. (2001) Cancer Metastasis Rev. 20, 155–164. [DOI] [PubMed] [Google Scholar]