Abstract
Drosophila melanogaster locomotor activity responds to different seasonal conditions by thermosensitive regulation of splicing of a 3′ intron in the period mRNA transcript. Here we demonstrate that the control of locomotor patterns by this mechanism is primarily light-dependent at low temperatures. At warmer temperatures, when it is vitally important for the fly to avoid midday desiccation, more stringent regulation of splicing is observed, requiring the light input received through the visual system during the day and the circadian clock at night. During the course of this study, we observed that a mutation in the no-receptor-potential-AP41 (norpAP41) gene, which encodes phospholipase-C, generated an extremely high level of 3′ splicing. This cannot be explained simply by the mutation's effect on the visual pathway and suggests that norpAP41 is directly involved in thermosensitivity.
Keywords: period gene, splicing, locomotor, entrainment, norpA
The period (per) gene in Drosophila melanogaster plays a central role in the circadian clock, acting as a negative regulator of its own transcription (reviewed in ref. 1). A number of other positive and negative transcriptional regulators are interwoven with PER to generate the circadian cycles observed in behavior, including TIMELESS (TIM) and the basic helix–loop–helix transcription factors dCLOCK and CYCLE. In addition, kinases such as DOUBLETIME regulate PER at the protein level, thereby contributing to the timing mechanism (1).
Under light/dark cycles (LD) 12:12, D. melanogaster behavior is bimodal, with a small morning locomotor activity peak at “lights on” [Zeitgeber time (ZT)0] and a larger evening activity peak around “lights off” (ZT12). Two classic per mutations, pers and perL, alter the free-running period, and under a 24-h Zeitgeber either advance (pers) or delay into the night phase (perL) the evening peak, while leaving morning locomotor activity largely unaffected (2). Thus per function is particularly relevant for this latter phase of locomotor behavior, and the importance of this response has been underscored by studies in which elevated temperatures delay the evening peak under LD cycles and cooler temperatures advance it (2–4). This is clearly an adaptive response in that under hot desiccating conditions, the locomotor activity of the fly moves toward the cooler later parts of the day, generating an apparent midday “siesta” (4).
Temperature changes regulate the splicing of an intron within the 3′ UTR of per, which then determines the position of the evening locomotor activity peak (4). It might be predicted that the regulation of this event would be tightly controlled, particularly at high temperatures when inappropriate locomotor behavior could prove fatal. We therefore examined the levels of per mRNA splicing in a number of different mutant backgrounds that either eliminate clock function (per01 and tim01) or impair light input [cryb, gl60j, and no-receptor-potential-AP41(norpAP41)], and combinations thereof.
The cryb mutation is considered a nearly null mutation in the cryptochrome gene, which encodes the dedicated circadian blue-light photoreceptor (5, 6). Mutations in norpA and glass affect the visual input pathway. The norpA gene encodes a phospholipase C (PLC) required for inositol signaling within the phototransduction pathway, in which NORPA is activated by photoactivated rhodopsin via a G protein complex (7, 8). norpA is primarily expressed within the rhabdomeres of the compound eye and the retina and axons of photoreceptor cells (9, 10) and even some tissues lacking phototransduction (11). The photoreceptors of severe norpA mutants such as norpAP41 show no electrophysiological response to light or PLC activity within the head (10, 12, 13). Thus in norpAP41 mutants, only CRY should be available for circadian entrainment. However, norpAP41; cryb mutants still entrain to LD cycles, suggesting the existence of some residual norpA-independent compound eye activity (14, 15).
glass (gl) encodes a transcription factor required for the development of photoreceptor cells, with the gl60j mutation eliminating the ocelli and retinal photoreceptor cells, the HB eyelet, and the DN1 dorsal neurons (15–17). Because CRY provides the only remaining photoreception in gl60j mutants, gl60j cryb double mutants are reported to be blind to light, with an unentrainable circadian clock (15). Thus gl60j is a more extreme visual mutant than norpAP41.
Although gl60j, norpAP41, and doubly mutant norpAP41; cryb flies entrain to LD cycles, they do so with altered patterns of locomotor activity. gl60j, cryb, and norpAP41 all alter the entrainment of the evening locomotor activity peak (6, 14, 15, 18); for example, norpAP41 mutants take longer to entrain to new LD cycles (18, 19) and do so with altered patterns of behavior (20). This raises the possibility that the light input alters the splicing of the per 3′ UTR intron, generating changes in evening activity. Our analysis of these mutants reveals that the canonical visual input pathway, as well as the clock and CRY, play important roles in the control of adaptive circadian behavior through regulation of per splicing. We also reveal that norpA may regulate per splicing independently of its role within the visual system.
Materials and Methods
RNA Analysis. RNA was extracted from collections of ≈50 adult heads at 4-h intervals by using Tri-reagent (Sigma) after entrainment to LD 12:12 at either 18 or 29°C. Levels of per splicing and overall per mRNA levels were determined by using a One-Step Reverse-Transcription PCR technique (Qiagen, Chatsworth, CA). To determine the proportion of per transcripts that were spliced, primers were designed 3′ and 5′ of the 3′ UTR intron, so that both the spliced and unspliced products were amplified in the same PCR reaction, allowing accurate quantification of their relative levels. The primers span the last intron in the per coding sequence to control for genomic contamination. To determine the overall per RNA levels, a multiplex PCR reaction was set up, amplifying per and the constitutively expressed gene rp49 simultaneously. Again, the primers span introns to control for genomic contamination.
To ensure that quantification was accurate for both the per RNA cycle and the splice levels of the different products, the reaction was examined over a range of PCR cycles to ensure both products were amplifying within the exponential phase, and subsequent reactions were carried out only over this cycle range. To determine per splicing levels, the PCR was run for 29 cycles and, for overall per RNA levels, 23 cycles, for between 250 ng and 1 μg of initial total mRNA. RNA levels were quantified by running the PCR products on 2% agarose gels and band intensities quantified by using scion image software (Scion, Frederick, MD). Results show the average levels of splicing of per mRNA for at least three independent fly collections per mRNA extractions at each time point.
The primers for determining splicing levels were: 5′-AAG ACG GAG CCG GGC TCC AG (6,421–6,440 bp); and 3′-AAT TGG ATC CGT GGC GTT GGC TTT CG (from ref. 4; 7373–7358). For per: 5′-GGT AAT GAA GAA GGG TCA GAC (4,291–4,311); 3′-AGA CGG TCA ACT GCT TCT CG (4,761–4,781); and for rp49: 5′-GTT AGT CCG TGC GCC GTT CG (94–113); 3′-CCT GTA TGC TGG GCG GAT GG (368–387). per is numbered with respect to sequence X03636 and rp49, to Y13939.
Behavioral Analysis. Fly stocks were reared at 25°C in LD 12:12, and locomotor activity was recorded by using the Drosophix (Padova, Italy) locomotor monitoring system and software (21). One-day-old male flies were collected and placed in locomotor activity tubes inside incubators at either 18 or 29°C with an LD 12:12 light cycle. Flies were then allowed to entrain to this regime for 4 days before locomotor activity was monitored for at least 3 days in LD. To compare the behavioral profiles between flies in LD, the activities of successive days are superimposed, generating an average daily activity pattern (22). The time bin with the highest mean activity value for each fly was used as the metric for comparing evening peaks between genotypes by using ANOVA (statistica, Statsoft, Tulsa, OK).
Line numbers in the text refer to the results of statistical tests illustrated in Table 1.
Table 1. Results of ANOVAs.
| Comparison | Effect | Statistics | Ref. |
|---|---|---|---|
| per splicing | |||
| WT Canton-S, per01, tim01, cryb and per01; cryb at 18 and 29°C | Temperature | F1, 221 = 156.2, P << 0.0001 | 1 |
| Genotype × temperature | F4, 221 = 6.2, P < 0.0001 | 2 | |
| Temp. × time | F5, 221 = 3.73, P < 0.005 | 3 | |
| Genotype × time | F20, 221 = 0.97, ns | 4 | |
| Genotype × temp. × time | F20, 221 = 1.13, ns | 5 | |
| cryb, per01, tim01 and WT, 18°C | Genotype | F3, 112 = 4.7, P < 0.01 | 6 |
| per01; cryb | Temperature | F1, 41 = 8.1, P < 0.01 | 7 |
| per01; cryb compared to WT | Genotype | F1, 56 = 10.4, P < 0.01 | 8 |
| per01; cryb and WT at 29°C | Genotype | F1, 49 = 55.6, P << 0.0001 | 9 |
| per01, tim01 and cryb compared to WT at 29°C | Genotype | F3, 68 = 16.2, P < 0.0001 | 10 |
| Time | F5, 68 = 11.0, P << 0.0001 | 11 | |
| DD and LD 12:12 | Light × temp. × time | F5,85 = 2.8, P < 0.05 | 12 |
| LD 6:18 and LD 12:12 | Light regime | F1, 56 = 12.7, P < 0.001 | 13 |
| norpAP41, gl60j and WT | Genotype | F2, 127 = 191.9 P << 0.0001 | 14 |
| norpAP41; cryb and norpAP41 at 18°C | Genotype | F1, 36 = 5.2, P < 0.05 | 15 |
| WT, 29°C | Time | F1,5=3.2, P < 0.5 | 16 |
| norpAP41, 29°C | Time | F1,5=0.39, n.s. | 17 |
| gl60j, 29°C | Time | F1,5 = 1.22, n.s. | 18 |
| gl60j, gl60j cryb and norpAP41 | Genotype × temperature | F2, 89 = 9.5, P < 0.001 | 19 |
| Locomotor activity | |||
| norpAP41 and cryb at 18°C | Genotype | F1,25 = 0.21, ns | 20 |
| norpAP41, cryb and WT at 18°C | Genotype | F2,100 = 12.8, P << 0.0001 | 21 |
| norpAP41 and cryb at 29°C | Genotype | F1,77 = 9.2, P < 0.005 | 22 |
| cryb and WT at 29°C | Genotype | F1,140 = 22.6 P << 0.0001 | 23 |
| per mRNA levels | |||
| WT and norpAP41 | Time | F5,53 = 6.6, P << 0.0001 | 24 |
| Temperature | F1,53=7.7, P < 0.01 | 25 | |
| Genotype × temperature × time | F5,53=1.4, ns | 26 | |
| gl60j, WT, and norpAP41 | Genotype × temperature × time | F10,61 = 2.5, P < 0.05 | 27 |
n.s., not significant.
Results
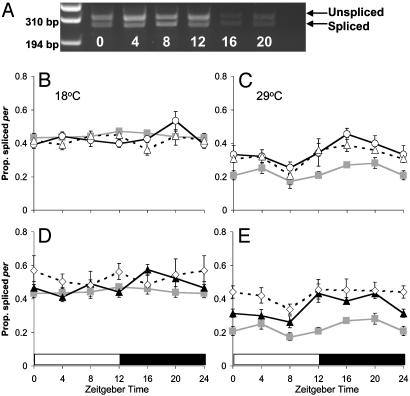
per Splicing in Clock and cry Mutants. Fig. 1 illustrates the proportion of per transcripts that were spliced at 18 and 29°C, averaged over several LD 12:12 cycles in Canton-S WT and per01, tim01, cryb, and per01; cryb mutant backgrounds. In all backgrounds splicing levels fall as the temperature rises, with 40–60% of transcripts spliced at 18°C and 20–45% at 29°C (Table 1, line 1; Fig. 1 B–E). However, not all genotypes react in the same way to temperature changes (Table 1, line 2).
Fig. 1.
Proportion of spliced per in clock mutants. Relative per mRNA splicing levels ± SEM are given for all ZTs. (A) Agarose gel of WT PCR products at 18°C, indicating the unspliced and spliced products. (B) 18°C; (C) 29°C. Canton-S ( ), per01 (○), and tim01 (▵). (D) 18°C; (E) 29°C. Canton-S (
), per01 (○), and tim01 (▵). (D) 18°C; (E) 29°C. Canton-S ( ), cryb (▴), and per01; cryb (⋄).
), cryb (▴), and per01; cryb (⋄).
The smallest but nevertheless significant effect of temperature on splicing levels is observed in per01; cryb (Fig. 1 D and E), suggesting that the temperature-sensing system for splicing may be compromised in the double mutant (Table 1, line 7). A significant temperature × time effect (Table 1, line 3) reveals that the temporal patterns of cycling differ among temperatures, and the absence of any other significant interactions (Table 1, lines 4, 5) suggests that all genotypes respond similarly. There is very little evidence for a significant day/night cycle in the proportion of per transcripts that are spliced at 18°C, but at 29°C, all genotypes reveal a higher level of splicing post lights off (ZT12) compared to the trough at ZT8. At 18°C, the per01 and tim01 mutations have no significant effect on the level of splicing of per mRNA compared to WT (Fig. 1B). However in cryb flies, splicing levels are significantly elevated (Table 1, line 6), particularly after lights off (Fig. 1D). This is also the case when per01; cryb is compared to WT (Table 1, line 8) (Fig. 1D). At 29°C, splicing levels are generally 5–10% higher in per01, tim01, and cryb mutants compared to WT in the light, but 15–20% higher after lights off at ZT12 (Table 1, lines 10, 11) (Fig. 1 C and E). This suggests that in the presence of light, splicing levels are reduced due largely to a clock-independent mechanism. In darkness, the clock and CRY become critical for maintaining this low splicing level at high temperatures.
The double mutant per01; cryb shows a highly significant increase in splicing of ≈20% throughout the day/night cycle compared to WT (Table 1, line 9) (Fig. 1E). Thus, at high temperature, either the presence of the circadian photoreceptor CRY or a functional circadian clock is sufficient to largely repress daytime splicing. With both eliminated, daytime splicing levels are elevated. In contrast, repression of splicing in the absence of light requires the circadian clock plus CRY. It seems somewhat counterintuitive that CRY, which is activated by light (5, 6, 23), plays a more prominent role in repressing splicing at night than it does during the day (Fig. 1E).
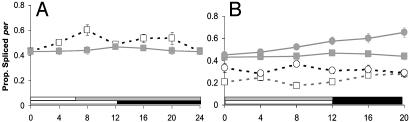
CRY is likely to be a dedicated circadian photoreceptor (5) yet at 29°C, splicing is repressed during the light phase even in cryb (Fig. 1E). This suggests that the light input to the splicing machinery cannot be primarily mediated by CRY. To confirm that light represses splicing, we investigated the effects that short photoperiods and constant darkness (DD) have on splicing levels in WT. Fig. 2 illustrates these results for LD 12:12, LD 6:18, and DD at 18°C. There is a significant effect of reducing photoperiod on the splicing level with an elevated level of splicing in DD compared to LD 12:12 (Table 1, line 12) (Fig. 2B), and similarly in LD 6:18, splicing levels are enhanced (Table 1, line 13) (Fig. 2A). Similar results have been observed by others (33). Because the repression of splicing by light in LD 12:12 at 29°C does not require the presence of CRY, we investigated whether the visual system played a role in setting the splicing level by examining the splicing of per transcripts in the mutants gl60j and norpAP41.
Fig. 2.
WT per splicing in altered photoperiods. Average relative per mRNA splicing levels ± SEM in Canton-S (A). LD 6:18 (□) and LD 12:12 ( ). (B) LD 12:12 18°C (
). (B) LD 12:12 18°C ( ) and 29°C (□) and during the first day of DD at 18°C (
) and 29°C (□) and during the first day of DD at 18°C ( ) and 29°C (○). LD 12:12 data reproduced from Fig. 1.
) and 29°C (○). LD 12:12 data reproduced from Fig. 1.
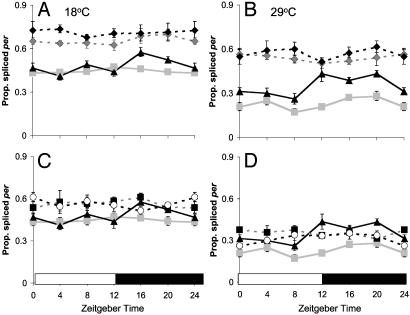
per Splicing in Visual Mutants. Fig. 3 shows that the proportion of per mRNA transcripts that are spliced at both temperatures is increased in both the norpAP41 and gl60j backgrounds compared to WT (Table 1, line 14). At 18°C, ≈65% of per transcripts are spliced in norpAP41 and ≈60% in gl60j, whereas at 29°C, these levels fall to ≈55% and ≈40%, respectively. Apart from a marginal difference between norpAP41; cryb, and norpAP41 at 18°C (Table 1, line 15), there are no significant effects for either norpAP41 or gl60j when combined with cryb (Fig. 3). These results indicate that the visual system rather than CRY is primarily responsible for the light-dependent repression of splicing. Unlike WT (Table 1, line 16), per splicing levels do not rise after lights off at 29°C in either norpAP41 (Table 1, line 17) or gl60j (Table 1, line 18).
Fig. 3.
Splicing in visual mutants. Relative per mRNA splicing levels ± SEM. (A) 18°C; (B) 29°C, Canton-S ( ), cryb (▴), norpAP41 (
), cryb (▴), norpAP41 ( ), and norpAP41; cryb (♦). (C) 18°C and (D) 29°C. Canton-S (
), and norpAP41; cryb (♦). (C) 18°C and (D) 29°C. Canton-S ( ), cryb (▴), gl60j (▪), and gl60j cryb (○). Canton-S and cryb data are reproduced from Fig. 1.
), cryb (▴), gl60j (▪), and gl60j cryb (○). Canton-S and cryb data are reproduced from Fig. 1.
Interestingly, in gl60j, there is a 20% difference between the splicing levels at different temperatures (60–40%), whereas in norpAP41, this difference is reduced to 10% (65–55%) (Table 1, line 19). The difference in gl60j is similar to that seen in WT (45–25%). Thus per splicing in norpAP41 is relatively insensitive to temperature changes. It is also clear that the level of splicing in norpAP41 is significantly higher at all times and temperatures than gl60j. Therefore, the effect of norpAP41 on splicing is greater than that of gl60j, despite gl60j being the more severe visual mutant.
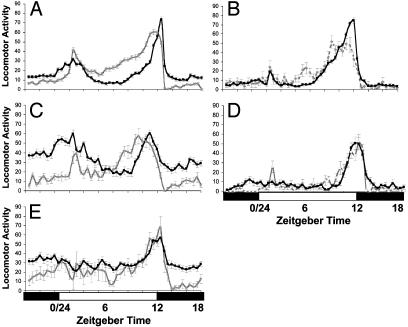
Locomotor Activity and Splicing Levels. Locomotor activity profiles of all genotypes were also monitored at 18 and 29°C (Fig. 4). Because each genotype shows a higher level of splicing at 18 than at 29°C, it would be predicted that this would generate an earlier evening activity peak at 18°C. As illustrated in Fig. 4 A–C, this is the case for WT, cryb and norpAP41, but not for norpAP41; cryb or gl60j (Fig. 4 D and E), where despite elevated splicing levels at higher temperatures (Fig. 3), there is no difference in the phase of activity. gl60j cryb does not entrain to LD cycles at 25°C (15), so was not included in this analysis.
Fig. 4.
Locomotor Activity profiles in LD 12:12. Activity is shown at 18°C (gray line) and 29°C (black line) ± SEM. (A) Canton-S. (B) cryb. (C) norpAP41. (D) norpAP41; cryb. (E) glass60j.
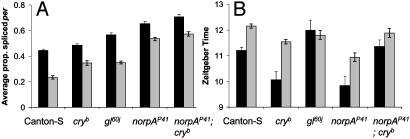
From Fig. 5A, the average proportion of per transcripts that are spliced at 18°C rises from WT (45%) to cryb (50%) to gl60j (60%) to norpAP41 (65%), and at 29°C from WT (25%) to gl60j and cryb (≈35%) to norpAP41 (55%). If the per splicing level is the only determinant of evening locomotor peak position, then a similar progression in the timing of this peak would be expected. The evening activity peaks of these different genotypes at 18 and 29°C are compared in Fig. 5B. It can be seen that for norpAP41, cryb, and WT, there is an inverse relationship between average splicing levels and the position of the activity peak at 18°C, with norpAP41 and cryb having similarly advanced activity peaks compared to WT (Table 1, lines 20, 21). At 29°C, the same inverse relationship holds, with norpAP41 advanced compared to cryb (Table 1, line 22), which is in turn earlier than WT (Table 1, line 23). Thus, those genotypes that show temperature-dependent changes in their evening activity generally display a correlation between average per splicing levels and the timing of the evening activity peak of the following day. Conversely, norpAP41; cryb and gl60j,, which show no significant differences in the phase of evening activity at different temperatures, have high splicing levels but relatively delayed evening activity peaks.
Fig. 5.
Splicing levels and evening activity peak positions. (A) Average per splicing levels across all time points. (B) Average evening peak position at 18 (▪) and 29°C ( ).
).
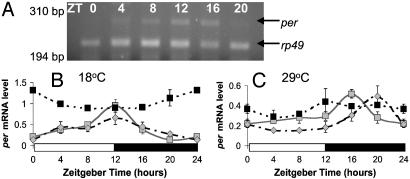
Underlying per mRNA Cycles. These observations raise the question of why the splicing level does not always relate to the timing of the evening locomotor activity peak, as in gl60j and norpAP41; cryb. Thus the per RNA profiles of gl60j and norpAP41 were compared to WT (Fig. 6). Because per does not cycle in cryb whole head homogenates (6), the underlying cycle in this background was not examined. Fig. 6 reveals that WT and norpAP41 show similar profiles (Table 1, lines 24–26), with an earlier per mRNA peak and higher overall level of per at the lower temperature. In contrast, there is no cycle in gl60j at either temperature, and levels of per are significantly different from WT and norpAP41 (Table 1, line 27).
Fig. 6.
per mRNA cycles. (A) Agarose gel showing Canton-S per and rp49 PCR products at 29°C. per mRNA levels are shown relative to rp49 ± SEM in Canton-S ( ), norpAP41 (
), norpAP41 ( ), and gl60j (▪) at 18°C (B) and 29°C (C). Results for Canton-S confirm those of ref. 4; note different y axis scales for A and B.
), and gl60j (▪) at 18°C (B) and 29°C (C). Results for Canton-S confirm those of ref. 4; note different y axis scales for A and B.
Discussion
Setting the Splicing Level. To entrain locomotor behavior to different seasons, the fly's clock must respond to changes in both light and temperature. This is mediated through a molecular switch, whereby increases in temperature repress the splicing of an intron within the 3′ UTR of per, delaying the onset of evening locomotor activity (4). Light also represses splicing, with higher splicing levels seen in shorter photoperiods (Fig. 2A), allowing locomotor activity to be fine-tuned to any given set of photoperiodic and temperature conditions. During the first day of DD, the level of splicing rises continuously (Fig. 2B). This is presumably because at the beginning of DD, the level of splicing is set low from the previous day's light input. Normally the light from the next day maintains this repressed level of splicing, but because this light input is absent, the repression of splicing is lifted, leading to a gradual rise in splicing levels.
The most obvious source for light input into the splicing machinery is the circadian photoreceptor CRY. However, analysis of the splicing levels in cryb shows that, although this mutation has an effect on splicing levels at 18°C, this effect is marginal and is seen only after lights off. This implies first, that any function of CRY in the repression of splicing is not via the activation of this molecule by light (24); second, because CRY is relatively dispensable for circadian locomotor rhythmicity per se (6, 23), it also suggests that any minor role in splicing at low temperature is unrelated to the functioning of the clock. As the temperature rises, PER, TIM, and CRY all become involved in the regulation of per mRNA splicing. At 29°C, all three mutants show the same splicing phenotype, with ≈30% of transcripts spliced during the day, but at night splicing is enhanced to ≈45%. Although PER, TIM, and CRY are known to associate in light conditions (6, 23, 25, 26), CRY and TIM can also associate in darkness (25), so it is not unexpected that the elimination of any one of the three proteins has a similar effect. Night time is also when the levels of these proteins are at their highest (1), and therefore any effects would be maximal.
At 29°C and in the presence of light, the levels of splicing in per01; cryb are elevated above those of either single mutant, which are themselves similar to WT. This suggests that the presence of either PER or CRY is required for light to repress splicing at 29°C. After lights off, the elevated levels of splicing of per are very similar in per01, tim01, cryb, and per01; cryb (Fig. 1 C and E). Therefore PER, TIM, and CRY probably work together to repress splicing in the dark at 29°C. An alternative view for the virtually identical per01, tim01, and cryb splicing levels at 29°C is that this reflects a masking effect of light, so that exogenous LD cycles have a greater effect on splicing at night compared to WT, which shows a modest but significant day–night rhythm. Such stronger masking effects on locomotor behavior have also been observed in cryb mutants (27), but any mechanism that might relate or explain these observations remains obscure.
The examination of whole head homogenates means that the majority of biological material is derived from the eyes so may not represent exactly what occurs in the pacemaker neurons. The eyes are peripheral clocks, and the cryb mutation stops the cycling of the clock in whole head homogenates, although cycling continues in the pacemaker cells (6). One possibility is that the splicing observed in cryb does not truly reflect the role of CRY in setting splicing levels but is instead a consequence of the clock having stopped in the eyes, thus explaining why per01, tim01, and cryb all show the same splicing phenotype. However, if this splicing phenotype is simply what happens when the clock stops, then per01; cryb should show the same splicing phenotype as either single mutant. This is not the case, because the daytime splicing in per01; cryb at 29°C is dramatically elevated compared to either single mutant. Thus the splicing phenotypes of per01, tim01, and cryb cannot simply be a result of the clock having stopped. This means that it is the presence of these proteins, rather than their clock-dependent cycling, which is important to the regulation of per splicing levels.
In gl60j, there is no per mRNA cycle in whole head homogenates (Fig. 6). This means that in the majority of cells in the gl60j head, the clock has either stopped or cells have become desynchronized. If the former is true, then splicing levels of gl60j should resemble those of per01 or tim01, and this is clearly not the case. If the latter is true, this could prevent the observation of any splicing rhythm, but the level of splicing observed should still represent the average level of splicing in this mutant background, which is clearly significantly different from WT. In any case, splicing levels observed in all visual mutants are likely to represent the effect of removing visual photoreception, because these elevated levels are similar to those observed in WT in DD.
norpAP41 and gl60j have considerably higher splicing levels than WT and cryb mutants at both temperatures, indicating that information received via the visual system rather than CRY drives this repression of splicing, which is borne out by analysis of gl60j cryb and norpAP41; cryb double mutants. The splicing levels of gl60j and gl60j cryb are similar at both temperatures, which is also true of norpA and norpAP41; cryb at 29°C. At 18°C, there is slightly more spliced per RNA in norpAP41; cryb than in the norpAP41 single mutant, reflecting the earlier result where cryb showed a marginal enhancement of splicing at cooler temperatures. These results also demonstrate that unspecific genetic background effects are not responsible for this marginal effect of cryb, because the double mutant background should make any interacting loci heterozygous. This lack of significant background effects in determining overall splicing levels has been confirmed by examining several natural European D. melanogaster lines. All mutants studied here show the same significantly enhanced splicing patterns when compared to any of the wild-caught isolates (B.H.C. and C.P.K., unpublished observations).
Unlike the clock and cryb mutants, there is no day–night difference in splicing levels at 29°C in either gl60j or norpAP41. One possibility is that visual system structures are required for the repression of splicing even in the dark, hence the overall elevated splicing levels in norpAP41 and gl60j at all times. This would be surprising, because such a role would obviously have to be light independent. More likely, the light input received through the eyes sets the splicing level during the day, and the clock maintains this repression at night. Thus, if the visual input is removed or reduced, as in DD, gl60j, or norpAP41 mutants, or in shorter photoperiods, then the subsequent splicing level is set higher. The difference in roles between cry and the visual system on per splicing levels may also partly explain recent observations that cryb mutants are able to adapt the timing of locomotor activity to long and short photoperiods, whereas flies with defective visual photoreception, including gl60j, are not (27).
norpA and the Thermal Signal for per Splicing. Interestingly, although gl60j is the more severe visual mutant (15), norpAP41 has significantly higher per splicing levels than gl60j at both 18 and 29°C. Additionally, whereas the difference between splicing levels at 18 and 29°C is maintained in gl mutants (≈65% and ≈45% of transcripts spliced vs. ≈45% and 25% in WT at 18 and 29°C, respectively), this is greatly reduced in norpAP41 (65% and 55%). One possible explanation for this is that norpA may be a signaling molecule in the temperature-sensing pathway for the clock. The patterns of locomotor activity support a role for norpA in temperature sensing, with the norpAP41 fly's locomotor patterns seemingly more sensitive to high temperatures than WT (Fig. 4). Additionally, norpAP41 evening locomotor activity peaks early at both 18 and 29°C, and per mRNA splicing shows a corresponding elevation compared to WT. These are responses associated with low temperatures in WT D. melanogaster, and therefore norpAP41 mutants behave as if they have an impaired ability to detect high temperatures. norpAP41 flies still detect temperature changes (witness the altered evening peaks and splicing levels); they just react as if the temperature is colder than it actually is.
Thus, the enhanced per splicing seen in norpAP41 may reflect a direct link between norpA-encoded PLC signaling and the temperature sensitivity of the splicing mechanism, independent of norpA visual function. In the phototransduction cascade, rhodopsin activates a G-protein isoform that in turn activates the PLC encoded by norpA. As a result of this activation, Ca2+ permeable light-sensitive channels are opened, including members of the transient receptor potential (TRP) class (reviewed in ref. 28). Recently it has been demonstrated that dANKTM1, a D. melanogaster TRP channel, is activated by temperatures from 24 to 29°C (29). In addition, D. melanogaster painless mutant larvae have a disrupted TRP channel and display defective responses to thermal stimuli (30). Because several TRP family members act as thermal sensors in mammals, TRP channels appear to have an ancient heat-sensing function that is retained in both vertebrates and invertebrates (29, 30). Given that we have identified a heat-sensing role for norpA, and norpA is known to activate TRP channels in photoreception, it is not unreasonable to suppose that norpA plays a general role in responses to temperature stimuli.
per splicing levels may also impact on aspects of behavior other than the timing of evening locomotor activity. For instance, the free-running period of norpAP41 is ≈1 h shorter than WT (20). The splicing levels of per mRNA are greatly elevated in this background, and elevated splicing is predicted to advance the PER protein cycle (4) and thus speed up the clock. In fact, the splicing mechanism should have the effect of speeding up the clock at colder temperatures and slowing it down at high temperatures, thereby providing a potential basis for temperature compensation.
The Relationship Between Splicing and Evening Activity. From Fig. 5B, it can be seen that the position of the evening activity peak at different temperatures moves in different mutant backgrounds. For WT, norpAP41, and cryb, the level of splicing appears to correlate with the position of the evening activity peak at different temperatures. At 18°C, there is a small but significantly greater relative amount of spliced per RNA in cryb than in WT, resulting in the earlier evening activity peak seen in cryb flies. This difference in per splicing is greatest after lights off at both temperatures (Fig. 1 D and E). This is when PER levels will be rising, because TIM is present for PER stabilization (31), so enhancement of PER accumulation by elevated per splicing is likely to have its most noticeable effect around dusk or early evening. A similarly consistent situation is seen in norpAP41: there is more spliced per mRNA present at 18°C (65%) than 29°C (55%), accounting for the earlier peak of evening activity at 18°C. Additionally these levels are higher than those seen in either WT (45% and 25% per transcripts spliced at each temperature) or cryb (55% and 40%) and relates to the earlier phases of locomotor activity seen in norpAP41 compared to the other genotypes. However, at 18°C there is more spliced per in norpAP41 than in cryb, but the evening activity peak occurs at the same time. The simplest explanation is that there is a limit to how early the evening activity peak can occur, no matter what the per splicing level, because splicing alters the accumulation of PER protein; this is limited by the light-dependent degradation of TIM (4). Therefore, in general, the level of splicing determines when the peak level of locomotor activity will occur, as previously reported (4).
The level of splicing of the per intron cannot be the only determinant of evening peak position, because the relationship between the per splicing level and evening activity peak position breaks down in norpAP41; cryb and gl60j, where there are different levels of splicing at the two temperatures but no corresponding difference in the evening peak position. When we analyzed the underlying per mRNA cycles of gl60j, norpAP41, and WT flies at 18°C and 29°C (Fig. 6), it was found that whereas per levels cycle in norpAp41 and WT, this cycle is lost in gl60j. If there is no underlying per RNA cycle, then there is no mRNA peak to be advanced or delayed by splicing.
At the cellular level, although gl is not a clock component, when mutated, it eliminates a number of clock-expressing cells within the head, including the eyes, ocelli, H-B eyelet, and the DN1s (15). Despite this, the primary effect on the clock is to remove most of the visual entrainment pathway, but the clock in the key pacemaker cells of gl60j mutants must still be functional, because behavior still entrains to LD cycles and remains rhythmic in DD. It is significant that the crosstalk between different classes of clock cells is essential for the generation of robust behavioral rhythms (32). Thus loss of the overall per mRNA rhythm may be a consequence of disrupting this network in gl60j, and, while leaving the basic system intact, this affects the more subtle temperature-sensitive aspects of entrainment. A similar argument based on an interruption of the entrainment network can also be proposed to explain the corresponding results with norpAP41; cryb double mutants, because in this case per mRNA is assumed to be noncycling because of the cryb background. However, the locomotor behavior of cryb single mutants remains thermosensitive even though overall per mRNA is noncycling. Thus, only when the photoreceptive pathway and mRNA cycle are both compromised (as in gl and norpAP41; cryb) is locomotor behavior insensitive to temperature-dependent changes in per splicing levels.
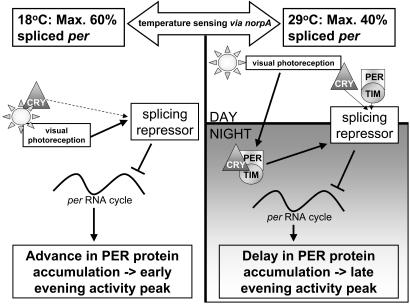
A Model as to How Light and Temperature Set the Splicing Level. Fig. 7 shows a model of how light and temperature may set the splicing level of the clock. How temperature is detected by the splicing machinery is not yet clear, but there is compelling evidence that norpA plays a role. At low temperatures, the splicing level is primarily set by light via the visual system rather than CRY, which is then remembered during the night. In longer periods of darkness such as in DD, this memory decays, and splicing levels begin to rise. Thus the visual system represses splicing by enhancing the effects of an unknown repressor molecule(s) that is sensitive to temperature change and the norpA PLC. At high temperatures, the regulation of splicing is more stringent and complex and recruits the circadian clock. Again, the light input received through the visual system sets the low splicing level during the day. This appears to also depend on the presence of at least two of the three molecules, PER, TIM, or CRY, because elimination of any one of these gives a barely detectable daytime rise in splicing, reflecting the very low levels of PER, TIM, and CRY at this time (23, 31). However, elimination of both PER and CRY in the per01; cryb double mutant lifts all light-dependent repression during the day.
Fig. 7.
A model for how the clock and visual system set the splicing level. For description, see text.
At night, the level of splicing set during the day by the visual system is again remembered and maintained by the clock at night. If per, tim, or cry is eliminated, then this repression of splicing is lost at night, generating the day/night difference in splicing levels. In gl60j cryb or norpAP41; cryb, because there is no visual light input during the day, there is no splicing level for the clock to remember, and therefore there is no day/night difference in splicing levels. Thus at high temperature, the visual system activates the repressor molecule during the day, and the clock maintains this activation at night. We assume that recruiting the clock at high temperature to inhibit per splicing is required to ensure that the fly's locomotor/foraging behavior is adaptive and does not encroach on those times of the day when there would be a significant risk of desiccation.
Acknowledgments
We thank Jeff Hall (Brandeis University) for the various mutant stocks and Isaac Edery (Rutgers University) for sharing unpublished information. We thank two anonymous reviewers for very helpful comments on an earlier draft.This work was supported by a postgraduate studentship from the Medical Research Council (to B.H.C.), a Royal Society Wolfson Research Merit Award (to C.P.K.), a Biotechnology and Biological Sciences Research Council David Phillips Fellowship (to E.R.), and a Biotechnology and Biological Sciences Research Council grant (to C.P.K. and E.R.).
Abbreviations: LD, light/dark cycles; DD, constant darkness; ZT, Zeitgeber time; PLC, phospholipase C; TRP, transient receptor potential.
Note Added in Proof. We have learned that a similar paper to ours is in press (33).
References
- 1.Stanewsky, R. (2003) J. Neurobiol. 54, 111–147. [DOI] [PubMed] [Google Scholar]
- 2.Hamblen-Coyle, M. J., Wheeler, D. A., Rutila, J. E., Rosbash, M. & Hall, J. C. (1992) J. Insect Behav. 5, 417–446. [Google Scholar]
- 3.Ewer, J., Hamblen-Coyle, M., Rosbash, M. & Hall, J. C. (1990) J. Neurogenet. 7, 31–73. [DOI] [PubMed] [Google Scholar]
- 4.Majercak, J., Sidote, D., Hardin, P. E. & Edery, I. (1999) Neuron 24, 219–230. [DOI] [PubMed] [Google Scholar]
- 5.Emery, P., Stanewsky, R., Hall, J. C. & Rosbash, M. (2000) Nature 404, 456–457. [DOI] [PubMed] [Google Scholar]
- 6.Stanewsky, R., Kaneko, M., Emery, P., Beretta, B., Wager-Smith, K., Kay, S. A., Rosbash, M. & Hall, J. C. (1998) Cell 95, 681–692. [DOI] [PubMed] [Google Scholar]
- 7.Bloomquist, B. T., Shortridge, R. D., Schneuwly, S., Perdew, M., Montell, C., Steller, H., Rubin, G. & Pak, W. L. (1988) Cell 54, 723–733. [DOI] [PubMed] [Google Scholar]
- 8.Bacigalupo, J., Bautista, D. M., Brink, D. L., Hetzer, J. F. & O'Day, P. M. (1995) J. Neurosci. 15, 7196–7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneuwly, S., Burg, M. G., Lending, C., Perdew, M. H. & Pak, W. L. (1991) J. Biol. Chem. 266, 24314–24319. [PubMed] [Google Scholar]
- 10.McKay, R. R., Chen, D. M., Miller, K., Kim, S., Stark, W. S. & Shortridge, R. D. (1995) J. Biol. Chem. 270, 13271–13276. [DOI] [PubMed] [Google Scholar]
- 11.Kim, S., McKay, R. R., Miller, K. & Shortridge, R. D. (1995) J. Biol. Chem. 270, 14376–14382. [DOI] [PubMed] [Google Scholar]
- 12.Inoue, H., Yoshioka, T. & Hotta, Y. (1985) Biochem. Biophys. Res. Commun. 132, 513–519. [DOI] [PubMed] [Google Scholar]
- 13.Yoshioka, T., Inoue, H. & Hotta, Y. (1985) J. Biochem. 97, 1251–1254. [DOI] [PubMed] [Google Scholar]
- 14.Malpel, S., Klarsfeld, A. & Rouyer, F. (2002) Development (Cambridge, U.K.) 129, 1443–1453. [DOI] [PubMed] [Google Scholar]
- 15.Helfrich-Forster, C., Winter, C., Hofbauer, A., Hall, J. C. & Stanewsky, R. (2001) Neuron 30, 249–261. [DOI] [PubMed] [Google Scholar]
- 16.Lindsley, D. L. & Zimm, G. G. (1992) Genetic Variations of Drosophila melanogaster (Academic, San Diego).
- 17.Moses, K., Ellis, M. C. & Rubin, G. M. (1989) Nature 340, 531–536. [DOI] [PubMed] [Google Scholar]
- 18.Emery, P., Stanewsky, R., Helfrich-Forster, C., Emery-Le, M., Hall, J. C. & Rosbash, M. (2000) Neuron 26, 493–504. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler, D. A., Hamblen-Coyle, M. J., Dushay, M. S. & Hall, J. C. (1993) J. Biol. Rhythms 8, 67–94. [DOI] [PubMed] [Google Scholar]
- 20.Dushay, M. S., Rosbash, M. & Hall, J. C. (1989) J. Biol. Rhythms 4, 1–27. [DOI] [PubMed] [Google Scholar]
- 21.Peixoto, A. A., Hennessy, J. M., Townson, I., Hasan, G., Rosbash, M., Costa, R. & Kyriacou, C. P. (1998) Proc. Natl. Acad. Sci. USA 95, 4475–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tauber, E., Roe, H., Costa, R., Hennessy, J. M. & Kyriacou, C. P. (2003) Curr. Biol. 13, 140–145. [DOI] [PubMed] [Google Scholar]
- 23.Emery, P., So, W. V., Kaneko, M., Hall, J. C. & Rosbash, M. (1998) Cell 95, 669–679. [DOI] [PubMed] [Google Scholar]
- 24.Lin, F. J., Song, W., Meyer-Bernstein, E., Naidoo, N. & Sehgal, A. (2001) Mol. Cell. Biol. 21, 7287–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ceriani, M. F., Darlington, T. K., Staknis, D., Mas, P., Petti, A. A., Weitz, C. J. & Kay, S. A. (1999) Science 285, 553–556. [DOI] [PubMed] [Google Scholar]
- 26.Rosato, E., Codd, V., Mazzotta, G., Piccin, A., Zordan, M., Costa, R. & Kyriacou, C. P. (2001) Curr. Biol. 11, 909–917. [DOI] [PubMed] [Google Scholar]
- 27.Rieger, D., Stanewsky, R. & Helfrich-Forster, C. (2003) J. Biol. Rhythms 18, 377–391. [DOI] [PubMed] [Google Scholar]
- 28.Hardie, R. C. & Raghu, P. (2001) Nature 413, 186–193. [DOI] [PubMed] [Google Scholar]
- 29.Viswanath, V., Story, G. M., Peier, A. M., Petrus, M. J., Lee, V. M., Hwang, S. W., Patapoutian, A. & Jegla, T. (2003) Nature 423, 822–823. [DOI] [PubMed] [Google Scholar]
- 30.Tracey, W. D., Jr., Wilson, R. I., Laurent, G. & Benzer, S. (2003) Cell 113, 261–273. [DOI] [PubMed] [Google Scholar]
- 31.Young, M. W., Wager-Smith, K., Vosshall, L., Saez, L. & Myers, M. P. (1996) Cold Spring Harbor Symp. Quant. Biol. 61, 279–284. [PubMed] [Google Scholar]
- 32.Peng, Y., Stoleru, D., Levine, J. D., Hall, J. C. & Rosbash, M. (2003) PLoS Biol. 1, 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majercak, J., Chen, W.-F. & Edery, I. (2004) Mol. Cell Biol., in press. [DOI] [PMC free article] [PubMed]