Abstract
Flaviviruses are human pathogens of world-wide medical importance. They have recently received much additional attention because of their spread to new regions (such as West Nile virus to North America), highlighting their potential as newly emerging disease agents. Using tick-borne encephalitis virus, we have developed and evaluated in mice a new genetic vaccine based on self-replicating but noninfectious RNA. This RNA contains all of the necessary genetic information for establishing its replication machinery in the host cell, thus mimicking a natural infection. However, genetic modifications in the region encoding the capsid protein simultaneously prevent the assembly of infectious virus particles and promote the secretion of noninfectious subviral particles that elicit neutralizing antibodies. These characteristics demonstrate that a new generation of flavivirus vaccines can be designed that stimulate the same spectrum of innate and specific immune responses as a live vaccine but have the safety features of an inactivated vaccine.
Flaviviruses are small, enveloped, positive-strand RNA viruses that include a number of important arthropod-borne human pathogens, such as West Nile virus, dengue viruses, yellow fever virus, Japanese encephalitis virus, and tick-borne encephalitis virus (TBEV) (1). The development of new vaccines against flaviviruses is an issue of ever-growing importance, because these pathogens continue to be a considerable medical problem in large areas of the world, new threats continually arise from the spread of flaviviruses to new geographic regions, and their potential abuse as bioterroristic agents persists.
Currently, conventional and experimental flavivirus vaccines can be divided into three major categories (2, 3): (i) Live vaccines, such as the widely used yellow fever 17D vaccine, are attenuated strains that initiate a productive infection in the vaccinee without causing disease symptoms. Live vaccines induce the most comprehensive immune response, because infection by an attenuated virus closely mimics a natural infection. In addition to virus particles, infected cells produce nonstructural proteins that are not incorporated into virus particles but are required for virus replication, and these proteins contribute to the targeting of infected cells by the immune system. Processing intermediates and RNA replicative forms (including double-stranded RNA) also stimulate various components of the innate and specific immune response (4). One type of live vaccine that has recently shown a great deal of promise involves the construction of attenuated “chimeras,” in which genes encoding structural proteins of one flavivirus are replaced by those of a different flavivirus (refs. 2 and 5 and references therein). (ii) Noninfectious vaccines are chemically inactivated whole virions or subunits of viruses that cannot spread in the host, an inherent safety feature of this approach. Protection in this case mainly depends on the ability of the surface proteins to induce antibodies that bind to and neutralize the virus, and their efficacy is greatly enhanced when the antigen is presented in particulate form (virions or subviral particles) (6, 7). (iii) Genetic vaccines involve direct immunization with RNA or DNA and have as one of their main advantages the simplicity and purity with which they can be produced (8). Application of in vitro synthesized infectious RNA or DNA, from which infectious RNA is transcribed in vivo, combines the advantages of genetic immunization with those of live virus vaccines, as demonstrated previously for TBEV (9) and Kunjin virus (10), a close relative of West Nile virus.
In this study, we use TBEV to introduce a new vaccine approach that merges the advantages of all three vaccination strategies discussed above. The procedure involves gene-gun-mediated application of self-replicating viral RNA with a genetic deletion preventing assembly of infectious virus particles and other modifications that promote secretion of immunogenic subviral particles. Because of the complete lack of infectivity, the safety profile of this RNA vaccine would be expected to resemble that of conventional noninfectious vaccines. Nevertheless, it exhibits important characteristics of a live virus vaccine, such as in vivo particle formation and release, RNA replication, and authentic nonstructural protein expression.
The flavivirus genome is a single, positive-stranded RNA molecule that encodes three structural proteins (capsid protein C, protein prM, which is a precursor to the small membrane protein M, and the large envelope protein E) and several nonstructural proteins in a single ORF (1). Previous investigations have revealed a remarkable functional flexibility of protein C, allowing the generation of infectious viral mutants carrying deletions that removed up to almost one-third of this protein (11, 12). We now demonstrate that a very large deletion (approximately two-thirds of the protein) results in an entirely noninfectious but RNA-replication-competent phenotype. The introduction of additional specific point mutations, which are known from studies in other flavivirus systems to override the mechanism that regulates the processing of the polyprotein precursor to the individual structural proteins (13, 14), caused an increase in the secretion of subviral particles containing the viral surface antigens. Immunization of mice with in vitro transcribed RNAs via gene-gun bombardment demonstrated that the ability to generate subviral particles correlated with the induction of neutralizing antibodies. The new vaccine is shown to reproducibly elicit a high-titered and highly protective immune response.
Methods
Virus, Recombinant Subviral Particles (RSPs), and Infectious cDNA Clone. The prototypic strain Neudoerfl of Western subtype TBEV (genome sequence GenBank accession no. U27495) was used as the wild-type control. RSPs, all described mutants, and the commercial vaccine FSME IMMUN Inject (Baxter, Vienna, Austria) were derived from this same strain. Control preparations of purified virus and RSPs were prepared by following standardized procedures (15). The infectious cDNA clone for this virus, pTNd/c, and procedures to transcribe RNA in vitro and introduce it into BHK-21 cells by electroporation have been described in detail (11, 16).
Mutant Construction. Mutant C(▵28–89) was derived from the wild-type infectious cDNA clone by introducing an in-frame deletion into the genomic region coding for capsid protein C removing amino acid residues 28–89 of this protein. This was achieved by swapping the unique MluI–AgeI fragment of subclone pTNd/5′ (16) with a PCR-derived fragment synthesized with primers 5′-TTTACGCGTCAAAAACGTGGGAAAAGGAGGTCAGCGAC-3′ (sense) and 5′-AGCGTAAACCGGTGCCAAAC-3′ (antisense). Then the mutation was transferred into the full-length clone pTNd/c by taking advantage of unique SalI and SnaBI restriction sites. Similarly, mutant C(▵28–89)-S was generated with primer 5′-TTTACGCGTCAAAAACGTGGGA A A AGGAGGTCAGCGACGGACTGGATGAGCTGGTTGCTGGTCATCACTCTGTTGCCGCAGACGCAGGCTGCAACGGTGAGGAAAG-3′ (sense), which, in addition to the deletion, introduces three amino acid changes and the same antisense primer as before. Cloning procedures, plasmid preparations, and sequencing procedures were done according to standard protocols.
Protein Expression and Infectivity Assays. RNAs were transcribed in vitro and introduced into BHK-21 cells by electroporation as described (16). Protein expression was detected by indirect immunofluorescence staining of cells after acetone-methanol fixation 48 h after transfection (11). Protein E released into the supernatant was detected with a four-layer ELISA (17), and mean values were derived from three independent experiments. Passaging experiments in cell culture and infectivity determinations in suckling mice were performed as described (11).
Particle Characterization. Separation of viral and subviral particles was performed on discontinuous sucrose gradients (10%, 35%, and 50%) as described (11). The protein E concentration of individual fractions was measured by SDS/ELISA (18). Buoyant density was determined by equilibrium sucrose gradient centrifugation and density measurement of the particle-containing fractions as in previous studies (11, 15). The antigenic structure of particles was assessed and compared with control preparations with a set of 18 protein E-specific monoclonal antibodies in a standardized four-layer ELISA system (18–20). Purified particles were fractionated by SDS/PAGE (21) and individual proteins visualized by staining with PhastGel Blue R250 (Amersham Pharmacia).
Animal Experiments and Analysis of Immune Response. RNA was synthesized from the corresponding cDNA clones in vitro by T7-mediated transcription. Afterward, the DNA template was removed enzymatically, and RNA was coated onto gold microcarrier particles essentially as described (9). Groups of four adult mice (female BALB/c, ≈15-g body weight) were inoculated with two shots by the gene gun, delivering ≈1 μg of RNA per mouse. For booster immunizations, this procedure was repeated 4 weeks later. Four weeks after the booster, mice were bled and serum samples were tested for the presence of TBEV-specific antibody by ELISA (22). Neutralizing activity was tested in a newly developed focus reduction assay that will be described in more detail elsewhere. Briefly, aliquots of virus solutions containing 50 focus-forming units were mixed with a 2-fold dilution series of serum (starting with a 1:10 dilution) and used to infect BHK-21 cells. Cells were covered with a carboxymethylcellulose overlay. Two days later, foci were visualized by subsequent incubations with polyclonal rabbit anti-TBEV serum and anti-rabbit IgG conjugated with alkaline phosphatase and development with FAST solution (Sigma). Protection of mice was tested by an i.p. challenge–inoculation with a highly lethal dose (>1,000 LD50) of the virulent TBEV strain Hypr (23).
Results
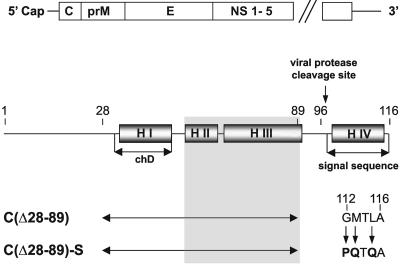
Generation of Capsid Deletion Mutants. The flavivirus protein C is a small protein with a large proportion of positively charged amino acid residues and a high percentage of α-helical secondary structures (11, 12, 24). A large deletion was engineered into the protein C-coding region of the full-length TBEV genome (Fig. 1). Previous studies had indicated that deletion of the hydrophobic helix I in conjunction with spontaneously occurring compensatory mutations in helix II or III was tolerated by the virus, yielding infectious progeny (11, 12). In mutant C(▵28–89), all three of these helices were removed by a deletion extending from residues 28–89. In a second mutant, C(▵28–89)-S, additional point mutations were introduced into helix IV, which serves as the internal signal sequence of the subsequently translated protein prM. It was shown previously that these mutations create an idealized signal sequence that no longer depends on coordinated cleavage by the viral protease, which regulates the subsequent processing steps of the surface proteins prM and E (13, 14).
Fig. 1.
Schematic of the TBEV genome and protein C. The genome (Upper) consists of a single long ORF encoding three structural proteins (C, prM, and E) and several nonstructural proteins and two flanking noncoding sequences (not drawn to scale). Protein C (Lower) is largely α-helical (four predicted helices, HI to H IV). H I coincides approximately with a stretch of hydrophobic amino acid residues (referred to as central hydrophobic domain, chD). The engineered deletion (double-headed arrow) removes all of H I/chD and an adjacent domain in which compensating mutations restoring viability of deletion mutants were previously observed to arise (shown as shaded area). H IV is an internal signal sequence of the subsequent component of the polyprotein, protein prM. It is cleaved off from mature protein C by the action of the viral protease NS2B/3. In mutant C(▵28–89)-S the signal sequence was modified by three point mutations as indicated in the figure, creating an idealized sequence. Numbers refer to amino acid positions in protein C.
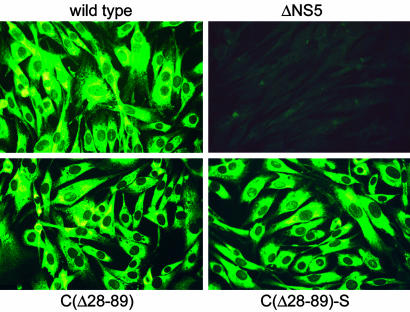
To assess the capability of the mutants to replicate and synthesize viral proteins, RNA from mutants C(▵28–89) and C(▵28–89)-S, wild-type TBEV, and the replication-deficient mutant ▵NS5 (used as a negative control) was transcribed in vitro and introduced into BHK-21 cells by electroporation. Protein E expression was detected for both of the capsid deletion mutants and the wild-type control, but not the replication-deficient negative control, demonstrating that neither the large capsid deletion nor the point mutations in helix IV significantly impaired RNA replication or translation (Fig. 2). ELISA analysis of cell culture supernatants (not shown) indicated that both mutants exported protein E, albeit in different quantities, as discussed in more detail below.
Fig. 2.
Expression of viral protein in BHK-21 cells. In vitro transcribed wild-type or mutant RNA (as indicated) was introduced by electroporation, and viral protein expression was determined by immunofluorescence staining with a polyclonal anti-TBEV serum 48 h after transfection. In the absence of RNA replication (replication-deficient mutant ▵NS5) no protein expression was detected.
Lack of Infectivity. Previously analyzed smaller capsid deletion mutants of TBEV were found to be infectious or were able to revert to an infectious phenotype after inoculation into suckling mouse brain (11, 12), the most sensitive growth system for this virus (25). Corresponding experiments performed with mutants C(▵28–89) and C(▵28–89)-S in BHK-21 cells and suckling mice, however, yielded no evidence of infectivity or genetic reversion events. The most stringent conditions applied involved the intracranial inoculation of suckling mice with concentrated samples of cell culture supernatants containing ≈70 ng of protein E per mouse, a dose that is equivalent to at least 106 infectious units in the case of wild-type virus. None of the mice developed signs of infection, and none of the mice seroconverted. To exclude the possibility of subclinical or abortive infections, selected brain samples were tested for the presence of viral RNA or protein by PCR and ELISA, respectively, and both tests consistently yielded negative results. Thus, both mutants were found to be competent for replication and translation but not found to yield infectious progeny or to regain infectivity through compensating mutations.
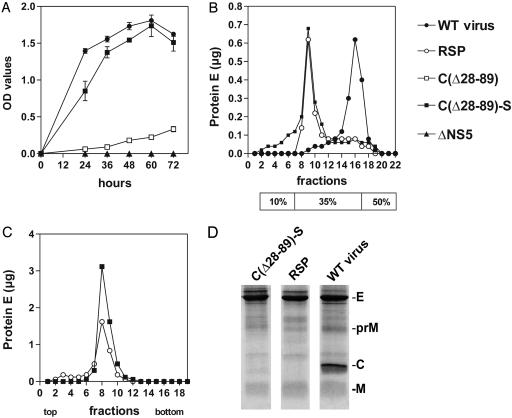
Characterization of Secreted Subviral Particles. The release of protein E into the supernatants of cultured BHK-21 cells transfected with the mutant and control RNAs was monitored by ELISA (Fig. 3A). This experiment revealed a major difference between mutant C(▵28–89) and mutant C(▵28–89)-S with regard to their capacity to export protein E. Whereas mutant C(▵28–89) released only small quantities of protein E, the modifications of the signal sequence engineered into mutant C(▵28–89)-S resulted in a substantial level of secretion. Both mutants, however, produced considerable levels of protein expression in the transfected cells. This finding suggests that the uncoupling of cleavage events at the C–prM junction is essential for achieving an efficient release of protein E in this experimental system.
Fig. 3.
Analysis of secreted particles. (A) The kinetics of the release of protein E from BHK-21 cells transfected with wild-type or mutant RNAs was monitored by ELISA. (B) Particles secreted by mutant C(▵28–89)-S were analyzed on a discontinuous (10%, 35%, and 50% as indicated below) sucrose gradient and compared with virus and RSP controls. (C) The buoyant density of particles secreted by mutant C(▵28–89)-S was determined by equilibrium sucrose gradient centrifugation in comparison with RSPs. (D) SDS/PAGE of viral and subviral particles. Particles secreted from cells transfected with C(▵28–89)-S were fractionated and compared with RSP and wild-type virus preparations. The positions of the structural proteins E, M (and its precursor, prM), and C are indicated.
In addition to virus particles, flavivirus-infected cells typically release noninfectious subviral particles (1), which can also be made by coexpression of the surface proteins prM and E in cell culture, and these are referred to as RSPs (15). RSPs are lipid-containing but capsidless particles that share relevant structural and functional similarity with infectious virions. They are excellent immunogens and noninfectious because they lack viral RNA (6, 15, 26, 27). As reported previously, flavivirus virions and RSPs differ with respect to size and density and therefore can be readily separated by density gradient centrifugation (15). A discontinuous sucrose gradient that yields a clear separation of the two species indicated that mutant C(▵28–89)-S exported particles matching the RSP control (Fig. 3B). RSPs have a buoyant density of 1.13–1.14 g/cm3 as opposed to the higher density of RNA-containing viral particles of 1.18–1.19 g/cm3 (15, 28). Particles obtained from mutant C(▵28–89)-S were subjected to equilibrium sucrose density centrifugation in parallel with conventionally produced RSPs. Both samples banded at the same position of the gradient corresponding to a density of 1.13 g/cm3 (Fig. 3C). The gradient-purified particles were then subjected to SDS/PAGE to visualize the individual protein components (Fig. 3D). The pattern obtained for the virus control exhibits distinct bands for the three structural proteins E, C, and M (and a minor band corresponding to its precursor protein, prM), whereas RSPs predominantly have proteins E and M (along with some prM). The pattern obtained for the C(▵28–89)-S sample was the same as that of RSPs.
The antigenic structure of viral and subviral particles of TBEV can be characterized by their reactivity with a set of monoclonal antibodies (15, 19, 20). The reactivity pattern obtained for the C(▵28–89)-S particles was found to be identical to the RSP control (data not shown) and thus corroborated the results from the physical characterizations, namely that the particles generated by mutant C(▵28–89)-S, were indistinguishable from RSPs.
RNA Immunization. The feasibility of using noninfectious, replicating RNA for immunization was assessed in the established adult mouse model (9, 23, 25). Groups of four mice were inoculated by gene-gun bombardment with gold particles coated with in vitro-transcribed RNAs from mutant C(▵28–89), C(▵28–89)-S, ▵NS5 (replication-deficient control), or untreated gold particles (mock control). In addition, four mice were immunized with a commercial TBEV vaccine (formalin-inactivated whole-virus), FSME IMMUN Inject. A booster immunization was administered 4 weeks later and serum samples were then drawn to analyze the specific immune response (Table 1). Every mouse inoculated with the replicating RNA mutants had seroconverted. The titers showed very little variation among individual mice and were no more than an order of magnitude lower than those observed when mice were immunized with the inactivated whole-virus vaccine, despite the fact that the dose used in these controls was very large relative to body weight. Neutralization tests revealed the presence of significant amounts of neutralizing antibody in mice immunized with mutant C(▵28–89)-S and the whole-virus vaccine but not in those inoculated with mutant C(▵28–89), which fails to secrete a substantial amount of protein E, or the negative controls. Immunization of an additional group of mice (nos. 5–8) with mutant C(▵28–89)-S yielded results that were essentially identical to those with the first group (Table 1). Finally, all of the mice were challenged by injecting them i.p. with 1,000 times the LD50 of the highly virulent TBEV strain Hypr. All of the mice inoculated with either C(▵28–89), C(▵28–89)-S or FSME IMMUN Inject survived these very stringent challenge conditions without any clinical sign of disease, whereas the challenge was lethal for the mice of the two control groups.
Table 1. Antibody response and protection of immunized adult mice.
| Inoculum* | Mouse no.† | IgG titer‡ | FRNT50 titer§ | Protection¶ |
|---|---|---|---|---|
| Mock | 1-4 | negative | negative | - |
| ΔNS5 | 1-4 | negative | negative | - |
| FSME IMMUN | ||||
| Inject | 1-4 | 30,000 | 80 | |
| 1 | 100,000 | + | ||
| 2 | 30,000 | + | ||
| 3 | 100,000 | + | ||
| 4 | 30,000 | + | ||
| C(Δ28-89) | 1-4 | 10,000 | negative | |
| 1 | 10,000 | + | ||
| 2 | 10,000 | + | ||
| 3 | 10,000 | + | ||
| 4 | 3,000 | + | ||
| C(Δ28-89)-S | 1-4 | 10,000 | 20 | |
| 1 | 10,000 | + | ||
| 2 | 10,000 | + | ||
| 3 | 10,000 | + | ||
| 4 | 10,000 | + | ||
| 5-8 | 10,000 | 20 | ||
| 5 | 10,000 | + | ||
| 6 | 10,000 | + | ||
| 7 | 10,000 | + | ||
| 8 | 10,000 | + |
Mice were inoculated by two gene-gun applications into the abdominal epidermis, delivering a total of ≈ 1 μg of RNA; Mock, inoculation with empty microcarrier; FSME IMMUN Inject, subcutaneous inoculation with half a human dose (0.25 ml containing 1 μg of protein E) of the commercial vaccine. All inoculations were repeated after 4 weeks.
Individual mice or pools from the sera of four mice were investigated.
Determined by testing a 0.5 log dilution series of serum by ELISA. The titer represents the largest dilution, yielding an absorbance value > 0.1; negative, titer < 100.
Fifty percent focus reduction titer determined with a 2-fold dilution series of serum pools; negative, titer < 10.
+, mouse survived without any sign of disease during the 28-day observation period; -, mice died 8-12 d after challenge.
Discussion
Noninfectious, self-replicating RNAs generated by deletion mutagenesis of the capsid protein C represent a promising new type of vaccine against flaviviruses. Our study was performed with TBEV, for which a useful and well-established small animal model is available (9, 25). The high degree of structural and functional similarity among all members of the genus Flavivirus (1, 24, 29), however, strongly suggests that this approach could be applied equally well to viruses, such as West Nile virus or dengue virus, for which no vaccines are currently available (2). It is possible that this approach could be extended to members of the other two genera (Pestivirus and Hepacivirus) of the family Flaviviridae, such as hepatitis C virus. We expect that this kind of vaccination will achieve a safety profile similar to inactivated or subunit vaccines, because there is no spread of virus in the body. On the other hand, the in vivo replication of the subgenomic RNA at the inoculation site and the expression of all of the nonstructural proteins should induce a much more comprehensive immune response than an inactivated or subunit vaccine and would be expected to resemble that elicited by a live virus vaccine (2). Ongoing investigations indeed indicate that cytotoxic T cells are induced at levels similar to those observed with attenuated infectious virus (unpublished observation).
Specific antibody titers achieved with doses of ≈1 μg of RNA were 3–10 times lower than those observed when mice were immunized with a widely used commercial TBEV vaccine. This vaccine (FSME IMMUN Inject) contains formalin-inactivated whole-virus adsorbed to aluminum hydroxide as an adjuvant, and mice were immunized with a dose of 1 μg of protein E, which corresponds to half the dose used for immunizing adult humans. Thus, FSME IMMUN Inject was applied in our mouse experiments in an ≈1,000-fold higher ratio of antigen to body-weight than is recommended for human vaccinations. Although the titers achieved with the RNA vaccine were lower (the difference was statistically significant; P = 0.03), the achieved immune responses were sufficient to provide complete protection in all cases. Remarkably, the titers obtained for individual RNA-immunized mice showed virtually no variation, indicating a high reproducibility and reliability of the method. Clearly, more extensive studies involving larger numbers of animals and applying various inoculation doses will be necessary to compare the protective efficiency of the RNA vaccine with the currently used vaccine in more detail.
The induction of neutralizing antibodies, which are mainly directed against the large envelope protein E, is of paramount importance for achieving a solid protective immune response against flaviviruses (2, 30). The efficacy of inactivated vaccines, such as the one used as a control in our experiments, mainly depends on this component. Therefore, it is an important characteristic of the mutant C(▵28–89)-S that it is able to elicit a significant neutralizing antibody response. The data show that the proportion of antibodies possessing neutralizing activity was roughly equivalent in the cases of C(▵28–89)-S RNA and FSME IMMUN Inject. This finding is in good agreement with earlier work that had shown that the presentation of protein E in particulate form is essential for inducing neutralizing antibody (6, 7). Significant secretion of particles was achieved by introducing “uncoupling” mutations in the C-prM signal sequence (13, 14). Although the exact molecular mechanisms remain unclear, we hypothesize that the mutated protein C interferes with the proteolytic liberation of protein prM, which consequently prevents proper processing and export of protein E. This block was apparently circumvented by the mutations. Results obtained with yellow fever virus indicate that this uncoupling of the proteolytic events severely interferes with the formation of infectious virus particles (14) and probably increases the formation of subviral particles. Both of these effects are potentially useful for enhancing the safety and efficacy of the capsid-deletion vaccine.
Neutralizing antibody, however, clearly is not the only protective mechanism that can be used for vaccination against flaviviruses (30, 31). The role of antibodies against nonstructural protein NS1 (32–34) and cytotoxic T cells against various antigens, in particular protein NS3, are well documented (30, 35). The solid protection achieved with mutant C(▵28–89) despite the lack of measurable neutralizing antibody, suggests a significant contribution of various other components of the immune response and supports the view that the presentation of homologous nonstructural proteins is a favorable property of a replicating vaccine. Other genetic modifications may be possible to further enhance the formation and export of subviral particles. The relative importance of individual components of the immune response elicited with this type of vaccine remains uncertain, and it will be necessary to study the correlation between particle generation, induction of neutralizing antibodies, and protection in more detail in the future.
Application of naked, self-replicating RNA derived from a positive-stranded RNA virus and expressing a heterologous gene has successfully been used to elicit a specific humoral and cellular immune response (36–38). However, the same principle can also be applied in the form of a DNA vaccine in which the viral sequence is placed under the control of a eukaryotic promoter to drive the transcription of the noninfectious, replicating RNA in vivo (36, 37). Similarly, application of DNA from which infectious RNA corresponding to an attenuated flavivirus is transcribed in vivo has recently been introduced as an experimental vaccine against West Nile virus (10). The ideal flavivirus replicon vaccine should ultimately induce a broad immune response by mimicking a natural infection without actually initiating a productive infection in the vaccinee.
Acknowledgments
We thank Angela Dohnal, Agnes Leitner, and Gabriel O'Riordain for their excellent technical assistance and Stefan Kiermayr for help and advice.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TBEV, tick-borne encephalitis virus; RSP, recombinant subviral particles.
References
- 1.Lindenbach, B. D. & Rice, C. M. (2001) in Fields Virology, eds. Knipe, D. M., Howley, P. M., et al. (Lippincott Williams & Wilkins, Philadelphia), pp. 991–1041.
- 2.Pugachev, K. V., Guirakhoo, F., Trent, D. W. & Monath, T. P. (2003) Int. J. Parasitol. 33, 567–582. [DOI] [PubMed] [Google Scholar]
- 3.Murphy, B. R. & Chanock, R. M. (2001) in Fields Virology, eds. Knipe, D. M., Howley, P. M., et al. (Lippincott Williams & Wilkins, Philadelphia), pp. 435–467.
- 4.Leitner, W. W., Hwang, L. N., deVeer, M. J., Zhou, A., Silverman, R. H., Williams, B. R., Dubensky, T. W., Ying, H. & Restifo, N. P. (2003) Nat. Med. 9, 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monath, T. P., Guirakhoo, F., Nichols, R., Yoksan, S., Schrader, R., Murphy, C., Blum, P., Woodward, S., McCarthy, K., Mathis, D. et al. (2003) J. Infect. Dis. 188, 1213–1230. [DOI] [PubMed] [Google Scholar]
- 6.Heinz, F. X., Allison, S. L., Stiasny, K., Schalich, J., Holzmann, H., Mandl, C. W. & Kunz, C. (1995) Vaccine 13, 1636–1642. [DOI] [PubMed] [Google Scholar]
- 7.Aberle, J. H., Aberle, S. W., Allison, S. L., Stiasny, K., Ecker, M., Mandl, C. W., Berger, R. & Heinz, F. X. (1999) J. Immunol. 163, 6756–6761. [PubMed] [Google Scholar]
- 8.Liu, M. A. (2003) J. Intern. Med. 253, 402–410. [DOI] [PubMed] [Google Scholar]
- 9.Mandl, C. W., Aberle, J. H., Aberle, S. W., Holzmann, H., Allison, S. L. & Heinz, F. X. (1998) Nat. Med. 4, 1438–1440. [DOI] [PubMed] [Google Scholar]
- 10.Hall, R. A., Nisbet, D. J., Pham, K. B., Pyke, A. T., Smith, G. A. & Khromykh, A. A. (2003) Proc. Natl. Acad. Sci. USA 100, 10460–10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kofler, R. M., Heinz, F. X. & Mandl, C. W. (2002) J. Virol. 76, 3534–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kofler, R. M., Leitner, A., O'Riordain, G., Heinz, F. X. & Mandl, C. W. (2003) J. Virol. 77, 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stocks, C. E. & Lobigs, M. (1998) J. Virol. 72, 2141–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, E., Stocks, C. E., Amberg, S. M., Rice, C. M. & Lobigs, M. (2000) J. Virol. 74, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schalich, J., Allison, S. L., Stiasny, K., Mandl, C. W., Kunz, C. & Heinz, F. X. (1996) J. Virol. 70, 4549–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandl, C. W., Ecker, M., Holzmann, H., Kunz, C. & Heinz, F. X. (1997) J. Gen. Virol. 78, 1049–1057. [DOI] [PubMed] [Google Scholar]
- 17.Heinz, F. X., Tuma, W., Guirakhoo, F. & Kunz, C. (1986) J. Biol. Stand. 14, 133–141. [DOI] [PubMed] [Google Scholar]
- 18.Heinz, F. X., Stiasny, K., Puschner-Auer, G., Holzmann, H., Allison, S. L., Mandl, C. W. & Kunz, C. (1994) Virology 198, 109–117. [DOI] [PubMed] [Google Scholar]
- 19.Guirakhoo, F., Heinz, F. X. & Kunz, C. (1989) Virology 169, 90–99. [DOI] [PubMed] [Google Scholar]
- 20.Holzmann, H., Stiasny, K., York, H., Dorner, F., Kunz, C. & Heinz, F. X. (1995) Arch. Virol. 140, 213–221. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. & Favre, M. (1973) J. Mol. Biol. 80, 575–599. [DOI] [PubMed] [Google Scholar]
- 22.Heinz, F. X., Berger, R., Tuma, W. & Kunz, C. (1983) Virology 126, 525–537. [DOI] [PubMed] [Google Scholar]
- 23.Wallner, G., Mandl, C. W., Ecker, M., Holzmann, H., Stiasny, K., Kunz, C. & Heinz, F. X. (1996) J. Gen. Virol. 1035–1042. [DOI] [PubMed]
- 24.Jones, C. T., Ma, L., Burgner, J. W., Groesch, T. D., Post, C. B. & Kuhn, R. J. (2003) J. Virol. 77, 7143–7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandl, C. W., Holzmann, H., Meixner, T., Rauscher, S., Stadler, P. F., Allison, S. L. & Heinz, F. X. (1998) J. Virol. 72, 2132–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferlenghi, I., Clarke, M., Ruttan, T., Allison, S. L., Schalich, J., Heinz, F. X., Harrison, S. C., Rey, F. A. & Fuller, S. D. (2001) Mol. Cell 7, 593–602. [DOI] [PubMed] [Google Scholar]
- 27.Gehrke, R., Ecker, M., Aberle, S. W., Allison, S. L., Heinz, F. X. & Mandl, C. W. (2003) J. Virol. 77, 8924–8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinz, F. X. & Allison, S. L. (2000) Adv. Virus Res. 55, 231–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, Y., Corver, J., Chipman, P. R., Zhang, W., Pletnev, S. V., Sedlak, D., Baker, T. S., Strauss, J. H., Kuhn, R. J. & Rossmann, M. G. (2003) EMBO J. 22, 2604–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konishi, E., Ajiro, N., Nukuzuma, C., Mason, P. W. & Kurane, I. (2003) Vaccine 21, 3675–3683. [DOI] [PubMed] [Google Scholar]
- 31.Burton, D. R. (2002) Nat. Rev. Immunol. 2, 706–713. [DOI] [PubMed] [Google Scholar]
- 32.Kreil, T. R., Maier, E., Fraiss, S. & Eibl, M. M. (1998) J. Virol. 72, 3076–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs, S. C., Stephenson, J. R. & Wilkinson, G. W. (1992) J. Virol. 66, 2086–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlesinger, J. J., Brandriss, M. W., Cropp, C. B. & Monath, T. P. (1986) J. Virol. 60, 1153–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobigs, M., Arthur, C. E., Mullbacher, A. & Blanden, R. V. (1994) Virology 202, 195–201. [DOI] [PubMed] [Google Scholar]
- 36.Anraku, I., Harvey, T. J., Linedale, R., Gardner, J., Harrich, D., Suhrbier, A. & Khromykh, A. A. (2002) J. Virol. 76, 3791–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smerdou, C. & Liljestrom, P. (1999) Curr. Opin. Mol. Ther. 1, 244–251. [PubMed] [Google Scholar]
- 38.Ying, H., Zaks, T. Z., Wang, R. F., Irvine, K. R., Kammula, U. S., Marincola, F. M., Leitner, W. W. & Restifo, N. P. (1999) Nat. Med. 5, 823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]