Abstract
The mechanism to maintain homeostasis of the gut microbiota remains largely unknown despite its critical role in the body defense. In the intestines of mice with deficiency of activation-induced cytidine deaminase (AID), the absence of hypermutated IgA is partially compensated for by the presence of large amounts of unmutated IgM and normal expression levels of defensins and angiogenins. We show here a predominant and persistent expansion of segmented filamentous bacteria throughout the small intestine of AID–/– mice. Reconstitution of lamina propria IgA production in AID–/– mice recovered the normal composition of gut flora and abolished the local and systemic activation of the immune system. The results indicate that secretions of IgAs rather than innate defense peptides are critical to regulation of commensal bacterial flora and that the segmented filamentous bacteria antigens are strong stimuli of the mucosal immune system.
Mucosal epithelial surfaces represent points of continuous and intimate interactions between the immune system and the outside environment. Under a constant antigenic pressure from >400 species of commensal bacteria, the gut immune system has developed highly sophisticated and efficient defensive as well as symbiotic mechanisms (1, 2). Secretion of antibiotic peptides by epithelial cells represents an important component of the innate immune system in the gut. Bacteria or bacterial antigens are capable of stimulating secretion of large amounts of antimicrobial peptides by crypt Paneth cells (3, 4). Also, transgenic (Tg) mice expressing a human intestinal defensin are protected against enteric salmonellosis (5), whereas mice deficient in the metalloproteinase matrilysin MMP-7 and thus lacking mature cryptdins show decreased resistance to some intestinal infections (6). Therefore, antimicrobial peptides appear to be involved in the maintenance of the symbiotic environment in the gut and protection of crypt stem cells from infections.
Another front line body defense mechanism that provides protection against microbial agents at mucosal surfaces is production and secretion of IgA (7). Indeed, IgA is the most abundant Ig isotype in mucosal secretions, and at least 80% of all plasma cells in mice are located in the intestinal lamina propria (LP) (8). The gut IgA responses are initiated primarily in organized lymphoid structures present in the intestine, namely Peyer's patches (PP) (9) and the isolated lymphoid follicles (ILFs), which have architecture similar to PP (10, 11). These structures contain a large number of conventional B2 cells, which are derived from precursor cells generated in the bone marrow (BM) (9, 10). In addition, peritoneal cavity B1 cells contribute to intestinal IgA plasma cells. B1 cells are shown to generate large amounts of IgAs independent of T cells and germinal centers (GC) (12, 13). Both B1 and conventional B2 cells are most likely to switch in situ from IgM to IgA in the LP with the help of dendritic cells and factors secreted by stromal cells (14). However, it is unknown how these innate and humoral immune defenses participate for protection at the mucosal surfaces, especially their relative roles in regulation of gut flora homeostasis.
We recently reported that the lack of class-switch recombination and somatic hypermutation of the Ig genes due to deficiency of activation-induced cytidine deaminase (AID) (15) leads to excessive anaerobic expansion in the small intestine, which induces hypertrophic protrusion of ILFs and activation of the whole body immune system (11). These observations implicate critical roles of hypermutated IgAs that were generated in the LP outside PPs in regulation of gut flora.
In the present study we show that the segmented filamentous bacteria (SFB) expanded aberrantly in the intestine of AID–/– mice and that normal gut IgAs, rather than antimicrobial peptides produced by the innate immune defense system, rescued the anaerobic expansion in the small intestine of AID–/– mice. The reestablishment of the gut homeostasis prevents the B cell activation and accumulation in gut-associated lymphoid tissues as well as in the peripheral lymphoid organs.
Materials and Methods
Mice. AID–/–, AID+/+, RAG-2–/–, and GFP Tg mice, all on a C57BL/6 background were maintained in our animal facility in specific pathogen-free conditions. Parabiosis surgery was performed as described (16) and in accordance with the guidelines established by the Institute of Laboratory Animals of Kyoto University. For transfer experiments, 1 × 107 cells isolated from BM of AID–/– and AID+/+ were injected into RAG-2–/– mice through the retroorbital plexus. After the operation or cell transplantation, mice were given 500 mg/liter ampicillin (Sigma) and 1 g/liter neomycin (Nacalai Tesque, Kyoto) in drinking water for 6 days.
Cell Preparation. Lamina propria lymphocytes (LPL) were isolated as described (17). For intestinal epithelial cell (IEC) preparation, after a brisk rinse with RPMI medium 1640 (Sigma, Tokyo), small intestine fragments were mildly stirred for 15 min in RPMI medium 1640 (2% FCS/0.5 mM EDTA) at 37°C. After incubation, the fragments were collected and washed two times with RPMI medium 1640. All supernatants were pooled and passed through a glass-wool column, preequilibrated with RPMI medium 1640 (2% FCS). After centrifugation, cells were resuspended in 40% buffered Percoll (Sigma) and layered on 75% buffered Percoll. After spinning, IEC were collected from the surface of the 40% layer and washed twice with RPMI medium 1640. After quantification, IEC preparation was split into two fractions; one was used directly for RNA extraction, and the other was frozen as pellet (–70°C) for genomic DNA preparation.
Biopsy Samples and PCR of Gut Flora. A whole small intestine was cut into three equal parts. After removal of intestinal contents, the segments were longitudinally cut and washed vigorously three times in PBS. The supernatants from the first and third wash were used to evaluate the flora of the intestinal contents. Biopsies (three ≈1-mm3 pieces) were taken from the proximal and distal segments, DNA was extracted, and the 16S rRNA was amplified by using universal primers described (18). After purification, the PCR products were ligated into pGEM-T Easy Vector (Promega). After transfection and amplification in Escherichia coli, inserts were sequenced with ABI PRISM 3100 genetic analyzer (Perkin–Elmer). Sequences were identified by using the BLAST database. For each experiment, the intestinal content and biopsies were pooled from two or three age-matched mice. Bacterial cultures were performed as described (11).
RT-qPCR of Defensins. Total RNA was isolated from IEC by using the RNeasy mini kit from Qiagen (Tokyo). After DNase treatment, oligo(dT) primers were used for first-strand cDNA synthesis (reverse transcription) from 1 μg total RNA. All procedures were performed according to the manufacturer's instructions (Invitrogen, Tokyo). Quantitative PCR was done on an iCycler apparatus using SYBR Green Supermix according to instructions and analyzed by accompanying software (all from Bio-Rad). Except for GAPDH, all other primers were determined by beacon designer (v. 2.1, Premier Biosoft International, Palo Alto, CA). Sequences were as follows: GAPDH, 5′-TGTGTCCGTCGTGGATCTGA (forward) and 5′-CCTGCTTCACCACCTTCTTGAT (reverse); cryptdin 3–6 5′-TCCTCCTCTCTGCCCTYGTCCTG (forward) and 5′-AGACACAGCCTGGTCSTCTTCC (reverse); cryptdin 1, 5′-CTCTTCTCCTGGCTGCTCCTC (reverse); matrilysin, 5′-ACCCTGT TCTGCT T TGTGTGTC (forward) and 5′-TCTGAGCCTGTTCCCACTGATG (reverse); angiogenin 1, 5′-CCATGACGCCA AGCCA A AGG (forward) and 5′-GCTTCCATTCGCTCCACAGATG (reverse); angiogenin 4, 5′-GCTGGGTCTGGTTGTGATTCC (forward) and 5′-AGGCGAGGTTAGCTTTCTTTCC (reverse); fatty acid binding protein 2, 5′-AGCTCGGTGTAAACTTTCCCTAC (forward) and 5′-AAACCTCTCGGACAGCAATCAG (reverse).
Starting quantity of cDNA was 10 ng of RNA equivalent, optimized final primer concentrations ranged from 150 to 300 nM, and reactions were run for 4 min at 95°C, followed by 35 cycles of 20 sec at 60°C, 20 sec at 72°C, and 15 sec at 95°C. A serial diluted positive control (bulk IEC cDNA) and reverse transcription minus controls of each tested sample were included in each PCR. Specificity of PCR products was initially confirmed by sequence analysis and subsequently by melt-curve analysis.
SFB-qPCR. Genomic DNA (gDNA) was prepared by phenol extraction. Primers used for ribosomal DNA sequences from SFB were 5′-AGGAGGAGTCTGCGGCACATTAGC (forward) and 5′-TCCCCACTGCTGCCTCCCGTAG (reverse). Only the forward primer is specific for SFB. For the SYBR green-based PCR, 50 ng of heat-denatured (for 10 min) gDNA was used as template and primer concentrations were 300 nM. Reactions were run for 10 min at 95°C followed by 35 cycles of 20 sec at 65°C, 20 sec at 72°C, and 15 sec at 95°C. Each PCR included a serial diluted positive control (bulk gDNA from intestinal washings), an internal control (GAPDH), and cut-off control (Thy gDNA). Specificity of PCR products was initially confirmed by sequencing and subsequently by melt-curve analysis.
Results
Disregulation of Gut Flora in AID–/– Mice. We have found that 100-fold expansion of anaerobic bacteria in the small intestine of AID –/– mice causes ILF hyperplasia and GC enlargement in all secondary lymphoid tissues (11). Because the previous study depended on analyses of bacteria by culture methods, the results obtained may have been biased because of the presence of uncultured species. To obtain a better representation of the gut flora that triggered the excessive activation of lymphoid organs in AID–/– mice, we adopted a molecular approach in which bacterial 16S rRNA was amplified by using universal primers followed by sequence analyses of the PCR products. This approach enabled us to have a more “qualitative” view of the intestinal ecosystem by identifying uncultured bacteria present in intestinal secretions or strongly adherent to the mucosal epithelium.
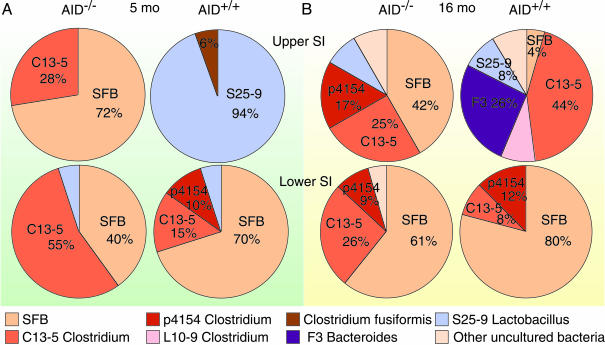
We found that SFB that cannot grow in vitro were the major components of the flora in biopsy samples from the upper part of the small intestine in 5-month-old AID–/– mice. In these mice, the lower segment of the small intestine was dominated by uncultured anaerobes, including SFB and those closely related to Clostridium species (Fig. 1A).
Fig. 1.
SFB expansion in AID–/– small intestine. Shown are intestinal flora in the biopsies from upper and lower segments of the small intestine of 5-month-old (A) and 16-month-old (B) AID–/– and AID+/+ mice. Biopsies were pooled from two littermates for each genotype, and the microflora was identified by sequence analyses of the 16S rRNA PCR products. Each color represents a specific sequence, and numbers indicate percentages in sequenced clones.
In sharp contrast, aerobic bacteria related to Lactobacillus constituted the predominant population of gut flora detected in biopsy samples from upper segment of the small intestine in age-matched AID+/+ mice (Fig. 1 A). As in AID–/– mice, anaerobes were the major population in lower part of the AID+/+ small intestine, and among them the most predominant were SFB (Fig. 1 A).
In 16-month-old AID–/– mice, SFB still represented a major bacterial population in the upper part of the small intestine (Fig. 1B). In contrast, SFB were seldom identified in the upper segments of aged AID+/+ mice, although the major bacterial population clearly shifted to anaerobes related to the genera Clostridium and Bacteroides (Fig. 1B). The complexity of bacterial species adherent to the gut mucosa increased with age, and this was more evident for the upper part of the small intestine.
These observations indicate that the gut microbial ecosystem is strongly disturbed in the absence of hypermutated intestinal IgA in AID–/– mice and has shifted to a more anaerobic environment dominated by SFB, especially in the upper part of the small intestine. In normal mice, SFB were not predominant even in aged mice, although changes toward an anaerobic predominance were seen with age.
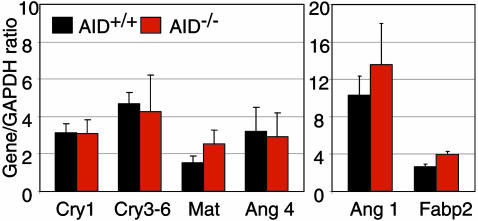
Anaerobe Expansion and Gut Innate Immune Defenses. It is possible that the anaerobic expansion in AID–/– mice stimulates the production of antimicrobial peptides from Paneth cells, which in turn may affect the composition of intestinal microflora. To investigate this possibility, we performed RT-qPCR of IEC using mRNA isolated from the whole length of the small intestine. We could not find any change in cryptdin expression in AID–/– mice as compared with AID+/+ mice (Fig. 2). We also measured the expression of matrilysin, the enzyme required for activation of cryptdins (6), but again we could not find any more differences than in unrelated mRNA variations (Fabp2 and angiogenin 1) between AID–/– mice and wild-type mice (Fig. 2). We measured the transcripts of angiogenin 4 gene, which was induced in germ-free mice after monoassociation with a commensal anaerobe (4). As shown in Fig. 2, there were no differences in expression levels of angiogenin 4 in IEC between AID–/– and AID+/+ mice. We thus conclude that the expression of antimicrobial peptides was not affected by the aberrant expansion of commensal anaerobes in the small intestine of AID–/– mice.
Fig. 2.
Innate immune defenses are not perturbed by commensal anaerobe expansion. Data are cryptdin, matrilysin, and angiogenin 4 mRNA expression by using RT-qPCR in the IEC preparations from small intestines of 4- to 5-month-old AID–/– and AID+/+ mice (n = 3, duplicate determinations; mean gene/GAPDH ratio ± SD plotted). Expression of angiogenin 1 and fatty acid binding protein (Fabp2) are showed as controls for IEC.
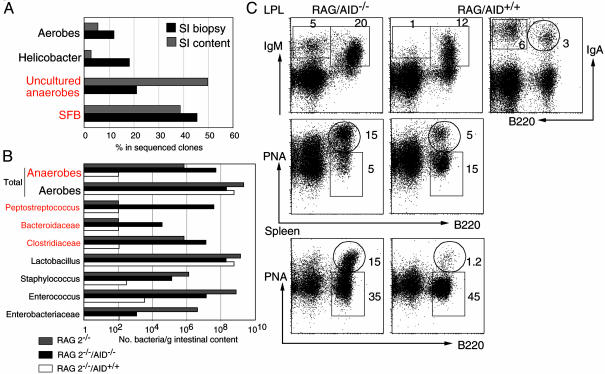
Transfer of AID–/– Activation Phenotype to RAG-2–/– Mice. We examined whether the aberrant expansion of SFB is a common feature of gut IgA deficiency, such as in RAG-2–/– mice. We found that the sequence of SFB was among the most frequently identified in biopsy samples and in the intestinal content of the small intestine of RAG-2–/– mice (Fig. 3A). The expansion of anaerobic bacteria of the genera Clostridium was also observed by microbiological methods (Fig. 3B, gray bars). To explore more about the importance of IgA in regulation of gut flora homeostasis and how changes in the bacterial ecosystem influenced the immune system, RAG-2–/– mice were then transplanted with BM cells from AID–/– and AID+/+ mice. Significantly, the total number of anaerobes drastically decreased 10 weeks after transfer of BM from AID+/+ mice (Fig. 3B, white bars). In contrast, the gut anaerobic expansion persisted after transfer of BM from AID–/– mice, and the major anaerobes cultured were Peptostreptococcus, Clostridium, and Bacteroides sp. (Fig. 3B, black bars). Strikingly, the anaerobic predominance in RAG-2–/– transferred with AID–/– BM was associated with accumulation of IgM-producing B cells and plasma cells and induction of B220+PNA+ GC B cells in the gut LP (Fig. 3C). As expected, RAG-2–/– mice transferred with AID+/+ BM developed B220–IgA+ plasma cells and had normal population of B220+IgA+ B cells as well as B220+IgM+ B cells in LP of the small intestine (Fig. 3C). Consistent with our previous observations (11, 15), we found more activated B220+PNA+ GC B cells in spleen of RAG-2–/– injected with AID–/– BM but not in those injected with BM from normal mice (Fig. 3C). Thus, the immunodeficiency caused by either the complete absence of mature B cells or the lack of hypermutated IgA led to a profound disregulation of the gut flora with excessive anaerobic expansion, which correlated with generalized activation of the immune system.
Fig. 3.
Anaerobe expansion in RAG-2–/– mice is reversed by AID+/+ but not AID–/– BM transplantation. (A) Small intestinal microflora in biopsy samples (black bars) and intestinal content (gray bars) of RAG-2–/– mice identified by using bacterial 16S rRNA universal primers. Samples were from 10-week-old mice (n = 4). (B) Microflora identified by microbiological methods in the small intestinal contents of RAG-2–/– mice (gray bars), RAG-2–/– mice injected with BM from AID+/+ mice (white bars), or RAG-2–/– mice injected with BM from AID–/– mice (black bars). (C) Fluorescence-activated cell sorting profiles of LP and spleen stained for B220, IgM, IgA, and peanut agglutinin (PNA) from the RAG-2–/– mice transferred with BM from AID–/– or AID+/+ mice. Numbers indicate the lymphocyte percentages in the gates. Data showed in B and C are from the same animals and are representative profiles from two sets of mice.
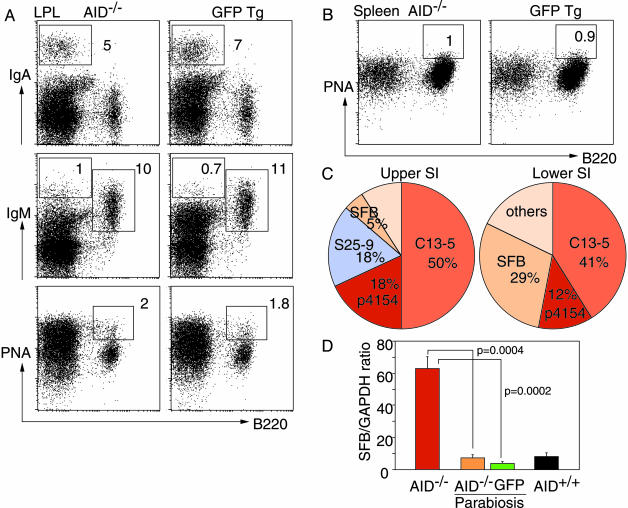
Rescue of AID–/– Mice by Anastomosis with Normal Mice. To determine whether the chronic activation processes in AID–/– mice induced by gut microflora and leading to B cell hyperplasia could be extinguished (reversed) by the presence of normal, hypermutated IgA, we made parabiosis between AID–/– and GFP Tg mice at the age of 2 months. This procedure does not require irradiation, which potentially induces gut inflammation, and allows a physiological exchange of gut-primed IgA B cells from GFP Tg to the AID–/– partner. Six months after anastomosis, AID–/– mice no more exhibited protruding ILF or hypertrophy of PPs in the small intestine, and the size of lymph nodes and spleen was also decreased (data not shown). Analyses of LPL from the small intestine revealed the presence of normal percentages and numbers of B220–IgA+ plasma cells and B220+IgM+ B cells in both parabiotic partners (Fig. 4A and data not shown). B220–IgM+ plasma cells were present in LPL preparations from both mice, and their percentages and numbers were comparable with those seen in normal mice (Fig. 4A and data not shown). Significantly, the GC B cells were drastically decreased not only in LPL but also in spleen of parabiotic AID–/– mice and reached the same levels as in the GFP Tg partner (Fig. 4 A and B).
Fig. 4.
IgA reconstitution in AID–/– mice rescue the gut microbiota and B cell activation phenotype. (A and B) Representative fluorescence-activated cell sorting profiles of LPL and spleen stained for B220, IgM, IgA, and peanut agglutinin (PNA) from AID–/– and GFP Tg mice 6 months after anastomosis. Numbers indicate the percentages of lymphocytes in the gates. (C) Intestinal flora in the biopsies from upper and lower segments of the small intestine of AID–/– mice after parabiosis with GFP Tg mice. Results shown in A–C are from the same animals and are representative profiles from four sets of mice. (D) Quantitative PCR for SFB in IEC preparations from AID–/– mice (red bar), AID+/+ mice (black bar), and AID–/– mice after parabiosis with GFP Tg mice (orange and green bars, respectively). Data are mean ± SE; n = 4; P values (unpaired Student t test) are shown.
Bacterial flora in gut biopsies from upper and lower segments of the small intestine was very similar in parabiotic pairs (Fig. 4C and data not shown). As a consequence of the presence of normal gut IgA in AID–/– mice with parabiosis, the population of SFB was drastically decreased, with a paralleled increase of Lactobacillus sp., especially in the upper part of the small intestine. The amount of SFB sequences that could be detected in the IEC preparations by qPCR was much less in AID–/– mice after anastomosis with GFP Tg mice than in AID–/– mice, and the relative expression of SFB was very similar to that seen in IEC from AID+/+ or GFP Tg mice (Fig. 4D). This finding indicates that SFB must be strongly anchored to the epithelial layer and that IgAs but not IgMs are capable of preventing this attachment.
Thus, the presence of normal IgAs reestablished the bacterial homeostasis in the small intestine of AID–/– mice with parabiosis. This was partly due to the exclusive capacity of IgA to prevent or limit the access of SFB to the mucosal epithelium, thereby preventing the activation process in the gut LP.
Discussion
Although the importance of mucosal IgA-mediated protection is unquestionable, the role of intestinal IgA in regulation of the gut flora and how disregulation of gut bacteria affects the immune system are only partially understood (1). We have shown here that hypermutated mucosal IgAs are critical for regulation of bacterial composition and represent the main element of the immune system involved in maintenance of gut flora homeostasis. Unmutated IgMs, although secreted into the gut lumen in the small intestine of AID–/– mice, could not prevent the excessive and aberrant expansion of anaerobes.
In vitro and in vivo studies revealed that crypt Paneth cells responded to bacterial challenge by expression and secretion of large amounts of microbicidal peptides (3, 4). However, the anaerobic expansion in AID–/– small intestine did not induce a significant response from Paneth cells, and, apparently, the basal secretion of defensins could not maintain the bacterial homeostasis. It is possible that the adherence of commensal anaerobes remained restricted to the epithelium that covers only the upper part of the villi, leaving the crypts relatively unaffected. Under severe inflammatory conditions with destruction of gut architecture or in the presence of highly invasive pathogens, Paneth cell peptides might be critical for maintenance of crypt sterility and thus protection of the epithelial stem cells (4–6).
What are the main bacteria that constantly activate the immune system of AID–/– mice, and what are the mechanisms that regulate them? Before trying to answer these questions, it should be mentioned that there is no satisfactory method available to date that can allow identification of such an enormous diversity of bacterial species, like those present in the gut. Molecular analyses revealed a diverse bacterial composition in the small intestinal contents (data not shown), yet the repertoire of bacteria adherent to the gut epithelium was rather limited and dominated by uncultured anaerobes. Among these, SFB seem to be a major population that activates the immune system because (i) SFB strongly attach to the gut epithelium (19, 20) and are the main (if not the only) bacterial species that could be detected in IEC suspension (data not shown). (ii) SFB was the major commensal anaerobe present in the upper segments of small intestine in AID–/– mice but not in normal mice. Indeed, in AID–/– mice we observed that ILF hypertrophy began and was more prominent in the upper segments of the small intestine (11). (iii) SFB retreated to the lower segments of AID–/– small intestine after reconstitution of normal gut IgAs, and this correlated with decrease of activation status in the gut-associated lymphoid tissues and in the peripheral lymphoid organs.
A delay in colonization of the small intestine with SFB and the retreat of SFB to the large intestine were known to occur in young mice in relation with the presence of IgAs, acquired by passive transfer and later secreted by IgA plasma cells located in the LP (21).
Thus, IgAs but not unmutated IgMs appear to prevent the attachment of SFB to the gut epithelium. Once attached, SFB may trigger a series of events that begin with uptake and antigen presentation to intraepithelial immune cells (19), activation of local B and T cells (21–23), production of cytokines and chemokines responsible for accumulation of B cells, hyperplasia of ILF, and GC induction.
Our studies revealed the importance of IgA in regulation of commensal gut flora homeostasis by demonstrating that the absence of normal IgAs leads to a significant shift in anaerobe populations in the small intestine. The compensatory mechanisms in response to the anaerobic expansion involve adaptive immune responses rather than the local antimicrobial defenses.
The elucidation of contribution of organized (PP and ILF) versus diffuse (LP) gut tissues in induction of mucosal IgA, the role of nonmutated versus hypermutated IgA in control of anaerobe expansion, and exactly how disregulation of the gut ecosystem affects the systemic immune system represent challenges for future studies.
Acknowledgments
We thank Dr. M. Okabe (Osaka University, Osaka) for kindly providing GFP Tg mice and Dr. H. Nagaoka for helpful discussions. This study was supported in part by a Center of Excellence Grant from the Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviations: SFB, segmented filamentous bacteria; AID, activation-induced cytidine deaminase; Tg, transgenic; PP, Peyer's patch; ILF, isolated lymphoid follicle; LP, lamina propria; LPL, LP lymphocyte; IEC, intestinal epithelial cell; BM, bone marrow; GC, germinal center.
References
- 1.Cebra, J. J. (1999) Am. J. Clin. Nutr. 69, 1046S–1051S. [DOI] [PubMed] [Google Scholar]
- 2.Hooper, L. V. & Gordon, J. I. (2001) Science 292, 1115–1118. [DOI] [PubMed] [Google Scholar]
- 3.Ayabe, T., Satchell, D. P., Wilson, C. L., Parks, W. C., Selsted, M. E. & Ouellette, A. J. (2000) Nat. Immunol. 1, 113–118. [DOI] [PubMed] [Google Scholar]
- 4.Hooper, L. V., Stappenbeck, T. S., Hong, C. V. & Gordon, J. I. (2003) Nat. Immunol. 4, 269–273. [DOI] [PubMed] [Google Scholar]
- 5.Salzman, N. H., Ghosh, D., Huttner, K. M., Paterson, Y. & Bevins, C. L. (2003) Nature 422, 522–526. [DOI] [PubMed] [Google Scholar]
- 6.Wilson, C. L., Ouellette, A. J., Satchell, D. P., Ayabe, T., Lopez-Boado, Y. S., Stratman, J. L., Hultgren, S. J., Matrisian, L. M. & Parks, W. C. (1999) Science 286, 113–117. [DOI] [PubMed] [Google Scholar]
- 7.Fagarasan, S. & Honjo, T. (2003) Nat. Rev. Immunol. 3, 63–72. [DOI] [PubMed] [Google Scholar]
- 8.van der Heijden, P. J., Stok, W. & Bianchi, A. T. (1987) Immunology 62, 551–555. [PMC free article] [PubMed] [Google Scholar]
- 9.Cebra, J. J. & Shroff, K. E. (1994) in Handbook of Mucosal Immunology, eds. Ogra, P., Mestecky, J., Lamm, M., Strobel, W., McGhee, J. & Bienenstock, J. (Academic, San Diego), pp. 151–157.
- 10.Hamada, H., Hiroi, T., Nishiyama, Y., Takahashi, H., Masunaga, Y., Hachimura, S., Kaminogawa, S., Takahashi-Iwanaga, H., Iwanaga, T., Kiyono, H., et al. (2002) J. Immunol. 168, 57–64. [DOI] [PubMed] [Google Scholar]
- 11.Fagarasan, S., Muramatsu, M., Suzuki, K., Nagaoka, H., Hiai, H. & Honjo, T. (2002) Science 298, 1424–1427. [DOI] [PubMed] [Google Scholar]
- 12.Kroese, F. G., Butcher, E. C., Stall, A. M., Lalor, P. A., Adams, S. & Herzenberg, L. A. (1989) Int. Immunol. 1, 75–84. [DOI] [PubMed] [Google Scholar]
- 13.Macpherson, A. J., Gatto, D., Sainsbury, E., Harriman, G. R., Hengartner, H. & Zinkernagel, R. M. (2000) Science 288, 2222–2226. [DOI] [PubMed] [Google Scholar]
- 14.Fagarasan, S., Kinoshita, K., Muramatsu, M., Ikuta, K. & Honjo, T. (2001) Nature 413, 639–643. [DOI] [PubMed] [Google Scholar]
- 15.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102, 553–563. [DOI] [PubMed] [Google Scholar]
- 16.Donskoy, E. & Goldschneider, I. (1992) J. Immunol. 148, 1604–1612. [PubMed] [Google Scholar]
- 17.Kamata, T., Nogaki, F., Fagarasan, S., Sakiyama, T., Kobayashi, I., Miyawaki, S., Ikuta, K., Muso, E., Yoshida, H., Sasayama, S. & Honjo, T. (2000) J. Immunol. 165, 1387–1394. [DOI] [PubMed] [Google Scholar]
- 18.Swidsinski, A., Khilkin, M., Kerjaschki, D., Schreiber, S., Ortner, M., Weber, J. & Lochs, H. (1998) Gastroenterology 115, 281–286. [DOI] [PubMed] [Google Scholar]
- 19.Meyerholz, D. K., Stabel, T. J. & Cheville, N. F. (2002) Infect. Immun. 70, 3277–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamauchi, K. E. & Snel, J. (2000) Infect. Immun. 68, 6496–6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, H. Q., Bos, N. A. & Cebra, J. J. (2001) Infect. Immun. 69, 3611–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talham, G. L., Jiang, H. Q., Bos, N. A. & Cebra, J. J. (1999) Infect. Immun. 67, 1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umesaki, Y., Setoyama, H., Matsumoto, S., Imaoka, A. & Itoh, K. (1999) Infect. Immun. 67, 3504–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]