Abstract
The MHC class II molecule DQ0602 confers strong susceptibility to narcolepsy but dominant protection against type 1 diabetes. The crystal structure of DQ0602 reveals the molecular features underlying these contrasting genetic properties. Structural comparisons to homologous DQ molecules with differential disease associations highlight a previously unrecognized interplay between the volume of the P6 pocket and the specificity of the P9 pocket, which implies that presentation of an expanded peptide repertoire is critical for dominant protection against type 1 diabetes. In narcolepsy, the volume of the P4 pocket appears central to the susceptibility, suggesting that the presentation of a specific peptide population plays a major role.
The human MHC class II molecule encoded by DQA1*0102/DQB1*0602 (termed DQ0602 hereafter) is uniquely capable of conferring dominant protection against type 1 diabetes (T1D) (1) and strong susceptibility to narcolepsy (2). These two associations, negative in T1D and positive in narcolepsy, are the strongest known for MHC-associated disorders.
T1D affects ≈1% of the population and is, in most patients, the result of an autoimmune-mediated specific destruction of insulin-secreting pancreatic β cells. Genetic susceptibility is mainly associated with the DQB1*0302 allele, whereas the DQB1*0602 allele confers almost absolute protection, even in the presence of the DQB1*0302 allele (3, 4).
Studies in the mouse as well as sequence comparison of human and mouse MHC class II molecules have revealed that, in general, the amino acid at β57 is critical in determining whether the allele predisposes to or protects against T1D. Alleles that encode Asp at 57β are protective, whereas alleles encoding Ala, Val, or Ser at 57β predispose (1). Animal studies have provided evidence that the predisposing MHC class II molecules mediate disease, at least in part, by presenting pancreatic β cell-derived peptides to diabetogenic T cells (5). In contrast, the data on MHC class II-associated protection are conflicting; i.e., it can be either dissociated from (6) or associated with (7) thymic deletion of diabetogenic T cells.
Narcolepsy is a chronic disabling sleep disorder of unknown origin that affects 0.02–0.06% of the population and is characterized mainly by excessive daytime sleepiness and cataplexy (8, 9). Narcolepsy is strongly positively associated with the DQB1*0602 allele, and 90–100% of patients with definite cataplexy carry this allele (2, 10).
In contrast, the closely related DQB1*0601 allele, which differs at only nine residues, protects against developing narcolepsy, suggesting that peptide binding differences between these two alleles determine whether they predispose to or protect against narcolepsy. Because of the strong DQB1*0602 association, narcolepsy has long been suspected to be an autoimmune disease (11, 12) like virtually all of the other MHC-associated disorders. Recent studies suggest that a deficiency in hypocretin neurotransmission (13–17), which is important for sleep and wakefulness (18), plays a central role in the development of narcolepsy. This deficiency appears to be due to the absence of hypocretin-secreting neurons in the hypothalamus while other prevalent adjacent neurons are unaffected (17). It has been suggested, therefore, that a DQ0602-restricted autoimmune attack on the hypocretin-secreting neurons is responsible for narcolepsy.
DQ0602 provides the opportunity in a single molecule to analyze the features conferring dominant protection against, and strong susceptibility to, two distinct diseases. The 1.8-Å resolution crystal structure of this molecule renders possible a detailed analysis in three dimensions and thus provides atomic level insights into mechanisms by which MHC class II molecules either predispose to or protect against disease.
Materials and Methods
Protein Expression and Purification. DQ0602 and DQ0602–hypocretin molecules were expressed in a soluble form fused to the leucine zipper dimerization motif of Fos and Jun (19) in Drosophila melanogaster S2 cells by using established protocols (20) and were affinity-purified on columns coupled with anti-DQ monoclonal antibody SPV-L3.
For crystallization purposes DQ0602–hypocretin was digested with V8 proteinase and purified through gel filtration and anion-exchange chromatography as described (30). Fractions containing DQ0602–hypocretin were pooled, buffer-exchanged to 10 mM Tris-HCl, and concentrated to ≈7 mg/ml.
Crystallization and Data Collection. Purified DQ0602–hypocretin was crystallized in 100 mM glycine (pH 3.5), 16% (wt/vol) PEG 8000, and 100 mM magnesium acetate by using the sitting drop/vapor diffusion method. The crystals diffracted to 1.8-Å resolution after flash-freezing at 105 K. Before flash-freezing, DQ0602–hypocretin crystals were transferred for ≈1 min to a cryoprotectant solution of crystallization buffer plus 20% ethylene glycol.
Diffraction data were collected at the European Synchrotron Radiation Facility (Grenoble, France) on beamline ID29 by using a Quantum 210 charge-coupled device detector (Area Detector Systems, Poway, CA). The x-ray data were processed and scaled with the HKL suite (21) (Table 1).
Table 1. Crystallographic statistics.
| Data collection | |
| Beamline | ESRF-ID29 |
| Wavelength | 0.9564 |
| Resolution | 30.0 to 1.8 |
| Space group | P21212 |
| Cell dimensions, Å | 102.1;129.3;40.6 |
| Unique reflections | 49489 |
| Completeness, % | 97.3 (78.7) |
| Rmerge, % | 5.0 (29.6) |
| I/σI | 15.9 (3.6) |
| Average redundancy | 7.5 |
| Structure refinement | |
| Resolution range, Å | 25.0 to 1.8 |
| Number of reflections (Rfactor/Rfree) | 47,894/2,386 |
| R-factor, % | 18.9 (22.4) |
| Rfree, % | 20.5 (25.0) |
| rmsd. bonds/angles, Å | |
| Side chain | 2.1/3.2 |
| Main chain | 1.2/1.9 |
| Average B factors, Å2 | 16.4 |
| Average B factors hypocretin peptide, Å2 | 10.0 |
| Number of atoms (proteins/NAG/fucose/mannose/zinc/waters/glycine/acetate) | 3081/56/10/11/1/373/4/4 |
Numbers in parentheses refer to the indicated highest-resolution shell (1.85 to 1.80 Å). Rmerge = Σhkl Σi|I(hkl;i) - 〈I(hkl)〉|/Σhkl ΣiI(hkl;i), where I(hkl;i) is the intensity of an individual measurement and 〈I(hkl)〉 is the average intensity from multiple observations. Rfactor = Σhkl∥Fobs| - k|Fcalc∥/Σhkl Fobs| Rfree equals the R-factor against 5% of the data removed prior to refinement. rmsd, rms deviation from ideal geometry.
Structure Determination and Analysis. The structure was determined with the molecular replacement method. The search for DQ0602 was carried out within a data range of 25 to 3 Å with the program epmr (22) by using the crystal structure of DQ0302 (PDB ID code 1JK8) without the peptide as a search model. The calculations resulted in one clear solution with a correlation coefficient of 0.44, an R-factor of 0.50, and perfect crystal packing. The solution was confirmed by the presence of density consistent with the peptide in the initial 2Fo – Fc electron density map.
Refinement was carried out by using program cns (23) for rigid body refinement, followed by iterative cycles of simulated annealing, conjugate gradient minimization, individual B-factor refinement, and manual rebuilding by using program o (24). The final model (α-chain residues 2–183, β-chain residues 3–191, peptide plus linker residues 1–20, three N-linked sugars, and a divalent cation which on the basis of electron density and coordination geometry has been modeled as Zn; see Table 1) has an R-factor of 0.189 (Rfree = 0.205) using all data between 25 and 1.8 Å.
The stereochemical properties of the structure were assessed by procheck (25) and whatcheck (26) and showed only one residue (Asn-33β) in a disallowed region of the Ramachandran plot. The conformation of this residue is supported by unambiguous electron density. Structural superpositions were made by using program shp (27), and all figures were produced by using PyMOL (www.pymol.org). The program volumes (R.E.E., unpublished data) was used with a 1.4-Å radius probe to analyze cavities.
Results
Peptide Binding. Soluble DQ0602 molecules were expressed in D. melanogaster S2 cells and were shown to be active in that they bound previously reported DQ0602 binding peptides (28) with similar apparent affinities (data not shown). Hypocretin was scanned for DQ0602 binding peptides at ScanProsite (www.expasy.org/tools/scanprosite) by using the proposed peptide binding motif for DQ0602 molecules (28). Two candidate peptides, hypocretin 1–13 (MNLPSTKVSWAAV) and hypocretin 45–57 (CSCRLYELLHGAG), were identified. Hypocretin 1–13 peptide bound the DQ0602 molecule with an apparent affinity that was 10-fold better than that of hypocretin 45–57. To facilitate complex formation, the hypocretin 1–13 peptide was linked through a 15-aa spacer to the encoded N terminus of the DQB1*0602 cDNA and expressed with DQA1*0102 in D. melanogaster S2 cells.
Crystal Structure. A soluble form of DQ0602 protein complexed with a peptide from human hypocretin (α-chain DQA0102 residues 1–196, β-chain DQB0602 residues 3–199, hypocretin 1–13; DQ0602–hypocretin) was produced as described previously for other human and murine MHC class II molecules (29, 30). DQ0602–hypocretin crystallized in an orthorhombic space group (P21212) with one molecule per asymmetric unit and was phased by molecular replacement. Its structure was determined to 1.8-Å resolution (see Materials and Methods and Table 1). The refined structure is consistent with the overall topology observed for other MHC class II molecules (31) and shows clear and unambiguous density for the peptide from the P1 to P11 residues plus seven residues from the linker (Fig. 1). The linker in total comprises 15 residues spanning from the C terminus of the peptide to the N terminus of the β-chain. In addition to the seven well ordered residues (GGGGSLV; Fig. 1D) included in the refined model, the course of the remainder of the linker main chain, although somewhat flexible, may be discerned from weak electron density. The position of the linker is stabilized by interactions with the exterior of the peptide binding groove and the N-linked sugar (N78α) and is oriented away from the putative T cell antigen receptor (TCR) recognition surface.
Fig. 1.
The crystal structure of DQ0602–hypocretin. (A) The solvent-accessible surface of the DQ0602 peptide binding groove is colored by electrostatic potential (blue, positive charge; red, negative charge) viewed onto the TCR recognition surface. The residues of the peptide are shown as ball-and-stick in atomic coloring (blue, nitrogen; red, oxygen; yellow, peptide carbon). The major pockets within the groove are labeled in the standard MHC class II nomenclature. (B) A composite omit map contoured at 1σ is shown in blue chicken wire. The peptide is depicted in ball-and-stick colored as in A and viewed through the β1-helix. (C) A superposition of the peptides from the DQ0602–hypocretin and DQ0302–insulinB structures. The peptides are shown with atomic coloring as in A except for the insulinB peptide carbon atoms (green). The view is as in B, and the residues are labeled in the standard MHC class II nomenclature. (D) Superposition of the DQ0602 and DQ0302 peptide binding grooves. The Cα traces for DQ0602 (gray) and for DQ0302 (green) are viewed as in A. The well ordered residues of the hypocretin peptide and linker are shown with atomic coloring (peptide coloring as in A; orange, linker carbon). Residues 46α to 55α and 85β to 91β show significant main chain and side chain conformational changes between the two MHC class II structures. The Cα positions of these residues in DQ0602 are indicated by spheres (blue, α-chain; magenta, β-chain). The concerted conformational changes impact on the P1 pocket (see text) and on the heterodimer interface. In particular, residue 48α changes from leucine in DQ0302 to tryptophan in DQ0602, and the side chain of Trp-48α (shown in blue sticks) is reoriented to form a tight, hydrophobic interaction at the interface with the β2 domain.
The hypocretin peptide is presented in the DQ0602 binding groove with peptide side chains P1, P4, P6, and P9 occupying the corresponding pockets (Fig. 1 A and C). The N-terminal portion of the peptide up to residue P1 and the C-terminal residues P10 and P11 extend out of the binding groove. Within the DQ0602 binding groove, the main chain of the hypocretin peptide adopts an extended conformation typical of MHC class II–peptide complexes. This conformation is stabilized by a set of conserved hydrogen bonding interactions between the MHC class II molecule and the backbone of the peptide (as reported in ref. 31) but in DQ0602 is supplemented with an extra bond between the main chain nitrogen of P9 and Asp-57β.
The peptide binding motif for DQ0602 has been reported to show dependency on positions P4, P6, and P9 (28), which is consistent with the observed use of the corresponding pockets in the DQ0602–hypocretin crystal structure. The specificity of the P1 pocket is less well documented, but the impact of polymorphisms in this region on murine MHC class II stability has been discussed (32). A superposition of the DQ0602 structure with that of DQ0302 (ref. 30; PDB code 1JK8) reveals significant conformational changes for α45–53 and β85–90 (Fig. 1D). These changes may influence the stability of the heterodimer and certainly modulate the specificity of the P1 pocket. Sequence comparison shows significant polymorphism in the β85–93 region for the DQ alleles, whereas the DR alleles are relatively conserved.
Comparison of DQ0602 and DQ0302 and Implications for Diabetes. To address the possible molecular determinants of protection against versus susceptibility to T1D, the structure of DQ0602–hypocretin was compared to that of a previously reported structure of DQ0302 (DQ8) in complex with an insulin peptide (insulinB) (30). As discussed above, the main chains of the hypocretin and insulinB peptides are superimposable (Fig. 1C), with the exception of the flanking residues (P1, P10–P11). In both complexes the peptide anchor residues are accommodated in pockets P1, P4, P6, and P9. In Fig. 2, residues potentially implicated in the development of T1D by comparison of positively, negatively, and neutrally associated DQ sequences are listed and mapped onto the DQ0602 structure. Functional and epidemiological data have focused attention on polymorphisms at 57β (1, 4), which contributes to pocket P9. However, the sequence comparisons also indicate a potential role for residues 9β and 66α at pocket P6 and 37β at pocket P4.
Fig. 2.
Disease-associated polymorphisms. (A) Disease-associated residues as highlighted by sequence alignment of selected DQ chains. Residues associated with T1D are red, and those associated with narcolepsy are green. The binding groove pockets to which these residues contribute are indicated. (B) The DQ0602 binding groove is shown in Cα trace, with the hypocretin peptide depicted in the standard format (Fig. 1) and the Cα positions of residues highlighted in A indicated by spheres (red, T1D; green, narcolepsy).
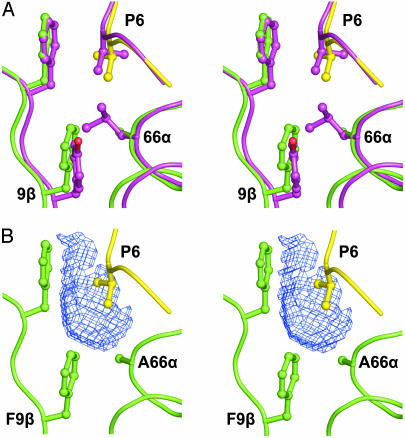
For both the DQ0602 and DQ0302 crystal structures a valine residue serves as the peptide anchor residue in the P6 pocket. The main chain conformations of the MHC class II residues contributing to this pocket are conserved between the two structures, but polymorphisms at 66α and 9β contribute two changes in side chains that combine to significantly increase the volume of the pocket in DQ0602 compared with that in DQ0302. This difference in pocket size is ≈42 Å3, which is sufficient to accommodate an additional two to three nonhydrogen atoms (Fig. 3). The change at 9β from tyrosine to phenylalanine is relatively subtle but is compounded by the more substantial change of leucine to alanine at 66α. The smaller volume in DQ0302 constrains the orientation of the valine P6 anchor and limits the ability of the pocket to accommodate larger hydrophobic or polar side chains. DQ0604, which is neutral for T1D development, shares the same α-chain as DQ0602 and contains a very limited number of β-chain polymorphisms as compared with DQ0602. The P6 pocket in DQ0604 contains tyrosine at 9β but alanine at 66α. Thus, there is a progression in P6 pocket size with DQ0602 possessing a large cavity, the pocket in DQ0604 having a slightly reduced volume and that in DQ0302 showing a very significant reduction. This variation in pocket volume suggests that DQ0602 can accommodate a broad range of peptide side chains at P6, that DQ0604 can accommodate a somewhat reduced number, and that DQ0302 can accommodate a substantially limited set of amino acid types. In addition to the polymorphism at 9β, DQ0604 and DQ0602 contain sequence differences in the binding groove at 57β (in the P9 pocket), 70β (at the putative TCR recognition surface), and 30β. The latter difference is positioned between the P6 and P9 pockets and changes 30β from tyrosine in DQ0602 to histidine in DQ0604. This polymorphism could potentially have an indirect influence on the character of either pocket but is a relatively subtle change. In contrast, the polymorphism at 57β is within the P9 pocket and changes the nature of this residue from a negatively charged aspartic acid in DQ0602 to a hydrophobic valine in DQ0604. The difference between DQ0602 and DQ0604 is paralleled for DQ0602 and DQ0302 by a change from aspartic acid to alanine, again a hydrophobic residue. Therefore, the polymorphism at 57β implies that the specificity of the P9 pocket is similar in DQ0604 and DQ0302 but significantly different in DQ0602. In the DQ0602 structure the P9 pocket is relatively shallow and polar in character. Asp-57β points toward the cavity and is coordinated by a network of hydrogen bonds involving Tyr-37β, Arg-76α, the main chain nitrogen of Ala-P9, and a water molecule (Fig. 4A). In particular, the carboxylate group of Asp-57β forms salt bridges to the side chain of Arg-76α. The hydrogen bond between Asp-57β and the main chain nitrogen at peptide position P9 is not generally conserved in other MHC class II molecules, and this, plus the salt bridges, may account for the reported enhancement in stability of the DQ0602 heterodimer (33). The water molecule is well ordered and, in the absence of a small polar residue at peptide position P9 (e.g., serine), serves to fill the cavity. Comparison of the DQ0602 and DQ0302 crystal structures (Fig. 4) reveals that, in DQ0302, the replacement of Asp-57β by alanine allows Arg-76α to form hydrogen bonds to the carboxylate group of the glutamic acid side chain at peptide position P9. Indeed, the carboxylate group of the P9 residue occupies positions essentially equivalent to those of Asp-57β.
Fig. 3.
The P6 pocket and T1D. (A) Stereoview of a superposition of selected residues from DQ0602–hypocretin with the equivalent residues from DQ0302–insulinB. Main chain is shown as coil, and selected residue side chains are depicted as ball-and-stick (yellow, hypocretin peptide; green, DQ0602; magenta, DQ0302–insulinB). The P6 residue in both the hypocretin and insulinB peptides is valine. (B) Stereoview of the DQ0602–hypocretin complex as in A. The volume bound by the blue chicken wire shows the increase in the P6 pocket size of DQ0602 when compared with DQ0302. The volume was defined by the program volumes as local difference between the solvent-accessible surfaces of the two binding grooves after superposition.
Fig. 4.
The P9 pocket and T1D. (A) Stereoview of selected residues contributing to the P9 pocket of DQ0602. Main chain and selected residue side chains are depicted as sticks and color-coded as in Fig. 3. A tightly bound water molecule is shown as a red sphere, and hydrogen bonds are represented as orange dashed lines. (B) Stereoview of the equivalent residues at the P9 pocket of the DQ0302–insulinB complex. The color coding is as in A but with green substituted by magenta. (C) Superposition of the residues shown in A and B. The color coding is conserved, with the exception of the peptide in the DQ0302–insulinB complex (magenta, carbon). For clarity, hydrogen bonds and water are omitted.
DQ0602 Binding Pockets and Narcolepsy. DQ0602 confers strong susceptibility to narcolepsy, whereas DQ06011 protects. These molecules share the same α-chain and differ at only nine residues in their β-chains. Of these nine polymorphic residues, 3β is distant from the binding groove and three residues (66β, 67β, and 70β on the β1-helix) are at the putative TCR recognition surface. The remaining five polymorphic residues (9β, 13β, 26β, 37β, and 38β; Fig. 2) contribute to binding pockets. Residue 9β changes from phenylalanine in DQ0602 to leucine in DQ06011. This change conserves the hydrophobic nature and has minimal effect on the shape of the P6 pocket. The effects of the polymorphisms on the P4 and P9 pockets are more pronounced. At the P9 pocket the change of 38β from alanine in DQ0602 to valine in DQ06011 is very conservative, but the change from tyrosine to aspartic acid at 37β introduces significantly more negative charge and may be expected to impact on the hydrogen bonding network within the pocket. Overall, these changes retain the polar character of the P9 pocket but may subtly modulate anchor specificity. The largest pocket in the DQ0602 structure is the hydrophobic P4 pocket with a volume of ≈100 Å3. For the DQ0602–hypocretin complex the threonine side chain of the P4 peptide residue only partially fills this volume (Fig. 5A). On the basis of the structure the pocket specificity would be predicted to include large hydrophobic residues, and this is, indeed, in agreement with observed anchor preferences (28). In contrast, the P4 pocket in DQ06011 is likely to have a very significantly smaller volume (Fig. 5B). This difference results from the polymorphisms at residues 13β and 26β, which, respectively, change glycine to alanine and leucine to tyrosine in DQ06011 compared with DQ0602. The impact of these changes on peptide specificity is predicted to be very substantial. For example, the relatively small threonine, which acts as the P4 anchor residue in the DQ0602–hypocretin complex, can no longer be accommodated because of steric clashes with the larger side chains at 13β and 26β (Fig. 5B).
Fig. 5.
The P4 pocket and narcolepsy. (A) Stereoview of selected residues contributing to the P4 pocket of DQ0602. Main chain and selected residue side chains are depicted as sticks and color-coded as in Fig. 3. (B) Superposition with polymorphic residues modeled for DQ06011 colored in magenta.
Discussion
The MHC class II molecule DQ0602 represents a paradox. This molecule is implicated in dominant protection against T1D and, conversely, strong susceptibility to narcolepsy. The current study of DQ0602 at 1.8-Å resolution has generated suggestions for the functional mechanisms by which this molecule exerts its effect on T cells in the disease process. For T1D this analysis reveals a previously unsuspected contribution by the P6 pocket and interplay between the volume of this pocket and the specificity of the P9 pocket. In particular, the increased volume of the P6 pocket will accommodate a more diverse set of amino acid side chains. This implies that presentation of an expanded peptide repertoire may be critical for dominant protection against T1D. In narcolepsy, the analysis highlights the volume of the P4 pocket as central to the susceptibility, suggesting that the presentation of a specific peptide population plays a major role in the disease development.
An expanded peptide repertoire presented by DQ0602 molecules could mediate its protective effect against T1D by means of at least two mechanisms, thymic deletion of T cells and/or thymic selection of regulatory T cells. Some evidence to distinguish between these possible mechanisms may be derived from the observation that protection from T1D also occurs in DQB1*0602-positive first-degree relatives of T1D patients that have antibodies against GAD65 (34). Such autoantibodies normally appear as the first sign of disease in prediabetic patients (35). This observation argues against thymic deletion of autoreactive T cells as the main mechanism for DQ0602-mediated dominant protection, because, if this were the case, an immune response against even a single β cell antigen would not have been expected to occur. In contrast, indirect lines of evidence suggest that DQ0602-mediated thymic selection of regulatory T cells might be of importance in the protection against human T1D. Firstly, such regulatory T cells have in mice been shown to prevent the spontaneous development of various autoimmune disorders, including T1D (36). Second, thymic selection of regulatory T cells appears to depend on a TCR affinity for the selecting self-peptide–MHC complex that is higher than that required for positive selection but lower than that leading to deletion (37). This high-affinity interaction might in humans be facilitated by the unique stability of the DQ0602 molecule (33), combined with its ability to bind an expanded peptide repertoire, allowing optimal presentation of β cell antigens expressed in the thymus. Such regulatory T cells in the periphery would be highly reactive to and specific for the β cell peptides. Accordingly, these cells could respond swiftly to even minor β cell insults. As a result, an incipient immune-mediated attack on β cells could be controlled quickly by the regulatory T cells, leaving behind a limited antibody response to a β cell antigen as seen in DQB1*0602-positive first-degree relatives of T1D patients.
The mechanism for DQB1*0602-associated susceptibility to narcolepsy is also an active area of research. The current analysis provides clear evidence that the P4 pocket and, to a certain extent, the P9 pocket differ significantly between the DQ0602 molecule and the narcolepsy protecting DQ06011 molecule. This strongly suggests that differential peptide binding between these two (otherwise closely related) molecules is central to their positive or negative association with narcolepsy. The most obvious explanation is that narcolepsy is the result of a DQ0602-restricted T cell-mediated autoimmune attack, e.g., on hypocretin-producing cells in the hypothalamus. Autoimmune deviations have not as yet been documented in patients with narcolepsy, but they clearly represent a potentially important area for study. If narcolepsy is indeed a T cell-mediated autoimmune disorder, the role of DQ0602 molecules in this context would then be to positively select autoreactive T cells in the thymus and to present self-peptides to these T cells in the hypothalamus.
Acknowledgments
We are grateful to the staff of the European Synchrotron Radiation Facility and European Molecular Biology Laboratory outstation (Grenoble, France) for assistance with data collection. We thank Andrew McMichael, Angela Vincent, and Nick Willcox for discussion. This work was supported by the Danish and British Medical Research Councils, the Karen Elise Jensen Foundation, the Lundbeck Foundation, the Danish Multiple Sclerosis Society, the European Commission Integrated Program SPINE (QLG2-CT-2002-00988), and the European Commission Descartes Prize. E.Y.J. is a Cancer Research UK Principal Research Fellow.
Abbreviations: T1D, type 1 diabetes; TCR, T cell antigen receptor.
Data deposition: Coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1UVQ).
References
- 1.Todd, J. A., Bell, J. I. & McDevitt, H. O. (1987) Nature 329, 599–604. [DOI] [PubMed] [Google Scholar]
- 2.Matsuki, K., Grumet, F. C., Lin, X., Gelb, M., Guilleminault, C., Dement, W. C. & Mignot, E. (1992) Lancet 339, 1052. [DOI] [PubMed] [Google Scholar]
- 3.Todd, J. A. & Wicker, L. S. (2001) Immunity 15, 387–395. [DOI] [PubMed] [Google Scholar]
- 4.Baisch, J. M., Weeks, T., Giles, R., Hoover, M., Stastny, P. & Capra, J. D. (1990) N. Engl. J. Med. 322, 1836–1841. [DOI] [PubMed] [Google Scholar]
- 5.Tisch, R. & McDevitt, H. (1996) Cell 85, 291–297. [DOI] [PubMed] [Google Scholar]
- 6.Luhder, F., Katz, J., Benoist, C. & Mathis, D. (1998) J. Exp. Med. 187, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt, D., Verdaguer, J., Averill, N. & Santamaria, P. (1997) J. Exp. Med. 186, 1059–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldrich, M. S. (1990) N. Engl. J. Med. 323, 389–394. [DOI] [PubMed] [Google Scholar]
- 9.Overeem, S., Mignot, E., van Dijk, J. G. & Lammers, G. J. (2001) J. Clin. Neurophysiol. 18, 78–105. [DOI] [PubMed] [Google Scholar]
- 10.Rogers, A. E., Meehan, J., Guilleminault, C., Grumet, F. C. & Mignot, E. (1997) Neurology 48, 1550–1556. [DOI] [PubMed] [Google Scholar]
- 11.Carlander, B., Eliaou, J. F. & Billiard, M. (1993) Neurophysiol. Clin. 23, 15–22. [DOI] [PubMed] [Google Scholar]
- 12.Mignot, E., Tafti, M., Dement, W. C. & Grumet, F. C. (1995) Adv. Neuroimmunol. 5, 23–37. [DOI] [PubMed] [Google Scholar]
- 13.Ripley, B., Overeem, S., Fujiki, N., Nevsimalova, S., Uchino, M., Yesavage, J., Di Monte, D., Dohi, K., Melberg, A., Lammers, G. J., et al. (2001) Neurology 57, 2253–2258. [DOI] [PubMed] [Google Scholar]
- 14.Chemelli, R. M., Willie, J. T., Sinton, C. M., Elmquist, J. K., Scammell, T., Lee, C., Richardson, J. A., Williams, S. C., Xiong, Y., Kisanuki, Y., et al. (1999) Cell 98, 437–451. [DOI] [PubMed] [Google Scholar]
- 15.Hara, J., Beuckmann, C. T., Nambu, T., Willie, J. T., Chemelli, R. M., Sinton, C. M., Sugiyama, F., Yagami, K., Goto, K., Yanagisawa, M. & Sakurai, T. (2001) Neuron 30, 345–354. [DOI] [PubMed] [Google Scholar]
- 16.Lin, L., Faraco, J., Li, R., Kadotani, H., Rogers, W., Lin, X., Qiu, X., de Jong, P. J., Nishino, S. & Mignot, E. (1999) Cell 98, 365–376. [DOI] [PubMed] [Google Scholar]
- 17.Peyron, C., Faraco, J., Rogers, W., Ripley, B., Overeem, S., Charnay, Y., Nevsimalova, S., Aldrich, M., Reynolds, D., Albin, R., et al. (2000) Nat. Med. 6, 991–997. [DOI] [PubMed] [Google Scholar]
- 18.Taheri, S., Zeitzer, J. M. & Mignot, E. (2002) Annu. Rev. Neurosci. 25, 283–313. [DOI] [PubMed] [Google Scholar]
- 19.Kalandadze, A., Galleno, M., Foncerrada, L., Strominger, J. L. & Wucherpfennig, K. W. (1996) J. Biol. Chem. 271, 20156–20162. [DOI] [PubMed] [Google Scholar]
- 20.Hansen, B. E., Andersson, E. C., Madsen, L. S., Engberg, J., Sondergaard, L., Svejgaard, A. & Fugger, L. (1998) Tissue Antigens 51, 119–128. [PubMed] [Google Scholar]
- 21.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 22.Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. (1999) Acta Crystallogr. D 55, 484–491. [DOI] [PubMed] [Google Scholar]
- 23.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M. & Pannu, N. S., (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 24.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 26.Hooft, R. W., Vriend, G., Sander, C. & Abola, E. E. (1996) Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 27.Stuart, D. I., Levine, M., Muirhead, H. & Stammers, D. K. (1979) J. Mol. Biol. 134, 109–142. [DOI] [PubMed] [Google Scholar]
- 28.Ettinger, R. A. & Kwok, W. W. (1998) J. Immunol. 160, 2365–2373. [PubMed] [Google Scholar]
- 29.Kozono, H., White, J., Clements, J., Marrack, P. & Kappler, J. (1994) Nature 369, 151–154. [DOI] [PubMed] [Google Scholar]
- 30.Lee, K. H., Wucherpfennig, K. W. & Wiley, D. C. (2001) Nat. Immunol. 2, 501–507. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh, P., Amaya, M., Mellins, E. & Wiley, D. C. (1995) Nature 378, 457–462. [DOI] [PubMed] [Google Scholar]
- 32.Latek, R. R., Suri, A., Petzold, S. J., Nelson, C. A., Kanagawa, O., Unanue, E. R. & Fremont, D. H. (2000) Immunity 12, 699–710. [DOI] [PubMed] [Google Scholar]
- 33.Ettinger, R. A., Liu, A. W., Nepom, G. T. & Kwok, W. W. (1998) J. Immunol. 161, 6439–6445. [PubMed] [Google Scholar]
- 34.Gianani, R., Verge, C. F., Moromisato-Gianani, R. I., Yu, L., Zhang, Y. J., Pugliese, A. & Eisenbarth, G. S. (1996) J. Autoimmun. 9, 423–425. [DOI] [PubMed] [Google Scholar]
- 35.Yu, L., Rewers, M., Gianani, R., Kawasaki, E., Zhang, Y., Verge, C., Chase, P., Klingensmith, G., Erlich, H., Norris, J. & Eisenbarth, G. S. (1996) J. Clin. Endocrinol. Metab. 81, 4264–4267. [DOI] [PubMed] [Google Scholar]
- 36.Sakaguchi, S. (2000) Cell 101, 455–458. [DOI] [PubMed] [Google Scholar]
- 37.Jordan, M. S., Boesteanu, A., Reed, A. J., Petrone, A. L., Holenbeck, A. E., Lerman, M. A., Naji, A. & Caton, A. J. (2001) Nat. Immunol. 2, 301–306. [DOI] [PubMed] [Google Scholar]