Abstract
The Arabidopsis thaliana genome contains 20 CNGCs, which are proposed to encode cyclic nucleotide gated, non-selective, Ca2+-permeable ion channels. CNGC7 and CNGC8 are the two most similar with 74% protein sequence identity, and both genes are preferentially expressed in pollen. Two independent loss-of-function T-DNA insertions were identified for both genes and used to generate plant lines in which only one of the two alleles was segregating (e.g., cngc7-1+/−/cngc8-2−/− and cngc7-3−/−/cngc8-1+/−). While normal pollen transmission was observed for single gene mutations, pollen harboring mutations in both cngc7 and 8 were found to be male sterile (transmission efficiency reduced by more than 3000-fold). Pollen grains harboring T-DNA disruptions of both cngc7 and 8 displayed a high frequency of bursting when germinated in vitro. The male sterile defect could be rescued through pollen expression of a CNGC7 or 8 transgene including a CNGC7 with an N-terminal GFP-tag. However, rescue efficiencies were reduced ∼10-fold when the CNGC7 or 8 included an F to W substitution (F589W and F624W, respectively) at the junction between the putative cyclic nucleotide binding-site and the calmodulin binding-site, identifying this junction as important for proper functioning of a plant CNGC. Using confocal microscopy, GFP-CNGC7 was found to preferentially localize to the plasma membrane at the flanks of the growing tip. Together these results indicate that CNGC7 and 8 are at least partially redundant and provide an essential function at the initiation of pollen tube tip growth.
Introduction
Fertilization in flowering plants requires a series of carefully coordinated events, including pollen grain germination, pollen tube growth, and directional changes in pollen tube tip growth that guide pollen tubes into the micropyle of an ovule [1]–[3]. When pollen tubes reach a synergid, they burst and discharge sperm cells [4]–[7]. These series of events involve signaling processes that coordinate dynamic changes in the cytoskeleton, ion homeostasis, and membrane trafficking.
Ca2+ signals are thought to play a central role in pollen tube tip growth and fertilization [8]–[12]. Evidence from pharmacological and genetic approaches support an important role for at least two different types of Ca2+-permeable channels, cyclic nucleotide gated channels (CNGCs) and glutamate receptor-like proteins (GLRs) [13]–[18]. In addition, a knockout of a plasma membrane Ca2+-pump ACA9 results in pollen defects that include slow tube growth and a reduced ability to discharge sperm cells to synergids [19]. A double knockout of two pollen-expressed Ca2+-dependent protein kinases CPKs 17 and 34 results in tubes that are slow, short and impaired in their ability to find ovules [20]. Moreover, Ca2+ signals have been implicated in regulating the dynamics of the actin cytoskeleton [21], [22] and the activity of Rops, which are small GTPases that can regulate cytoskeletal and secretory processes [22]–[24].
In Arabidopsis thaliana, 6 of the 20 CNGCs show detectable expression in pollen [25], [26] and CNGC18 was shown to be essential for pollen tube tip growth [13], [14]. This is consistent with pharmacological evidence that cyclic nucleotide monophosphate (cNMP) signals can trigger growth-altering Ca2+ signals [27]–[29]. While it is possible that cNMP triggered Ca2+ signals are a direct result of Ca2+ conductance through a CNGC, these channels are also permeable to K+, and could be functioning in a way that indirectly triggers a Ca2+ release from an internal store [9], [30], [31]. Regardless, a GFP-tagged CNGC18 was found to localize to the growing apical region [13], [14], supporting a model in which cNMP signals have a specific role in regulating signaling and tip growth.
Here we show that two additional pollen-expressed CNGCs (7 and 8) are essential to pollen tube growth. A double knockout of CNGC7 and 8 results in pollen grains that burst when germinated in vitro. A GFP-tagged CNGC7 was found to localize to the plasma membrane, with the strongest GFP signal at the flanks of the pollen tube tip. This favors a model in which the formation and maintenance of pollen tube tip growth requires multiple CNGCs, including CNGC18 and either CNGC7 or 8.
Results
CNGC7 and 8 have Redundant Functions Required for Pollen Transmission
Among the six CNGCs that are most highly expressed in A. thaliana pollen (Figure 1), CNGC7 (At1g15990) and 8 (At1g19780) are the two most closely related (74% aa identity). To determine if these genes have redundant functions in pollen development, two independent T-DNA gene disruptions for each gene were obtained from publically available T-DNA insertion collections: cngc7-1, 7-3, 8-1, and 8-2 [32]–[34] (Figure 2). The cngc7-3 and 8-2 alleles have insertions located in exons that encode essential features for a CNGC. As individual mutations, all four insertions showed normal Mendelian segregation when heterozygous plants were self-fertilized or tested for pollen transmission in a manual cross (Table 1).
Figure 1. Expression profiles showing preferential pollen expression for six CNCGs in Arabidopsis thaliana.
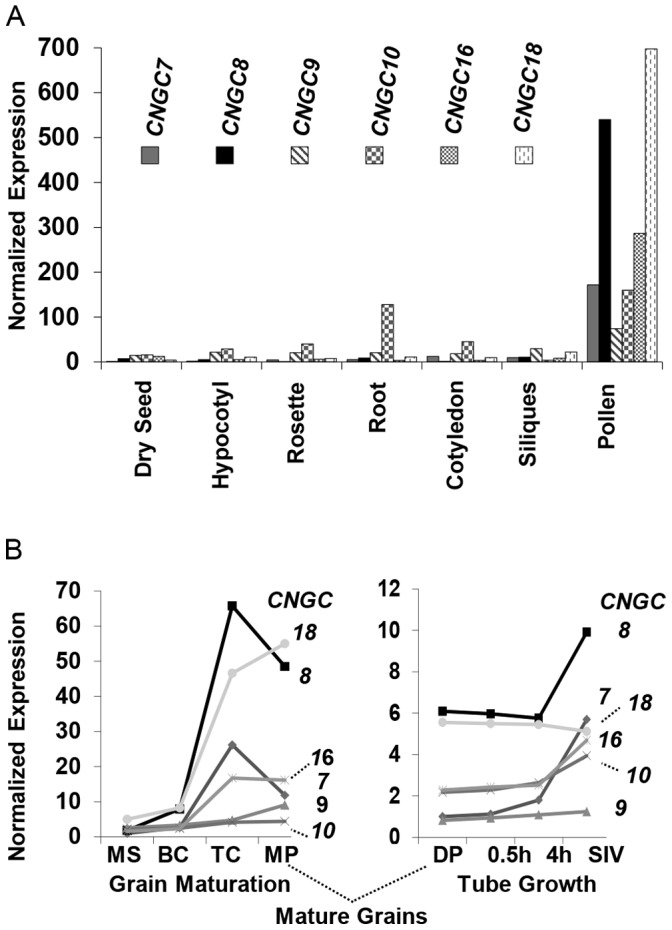
A) Relative expression levels in different tissues are shown for CNGC7, 8, 9, 10, 16, and 18 (AT1G15990, AT1G19780, AT4G30560, AT1G01340, AT3G48010, and AT5G14870, respectively) obtained from the Arabidopsis eFP Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) [55] The expression of CNGC7 in dry seed was arbitrarily set to 1, and the rest of the data normalized accordingly. B) Relative expression levels of pollen expressed CNGCs at different stages of pollen development obtained from The Pollen Transcriptome Navigator (http://pollen.umd.edu/), which uses data from Honys and Twell, 2004 [25] (left half) and Qin et al., 2009 [26] (right half). Developmental stages are denoted as MS: microspore; BC: bicellular; TC: tricellular; MP: mature pollen; 0.5 h: pollen tube germinated in vitro for 30 minutes. 4 h: pollen tube germinated in vitro for 4 hours, and SIV: pollen tubes after semi-in vivo growth through a stigma. For each data set, the expression of CNGC7 in microspore and dry pollen were arbitrarily set to 1 and rest of the data normalized accordingly.
Figure 2. Diagram of the genomic structure of CNGC7 and 8 and related T-DNA insertions.
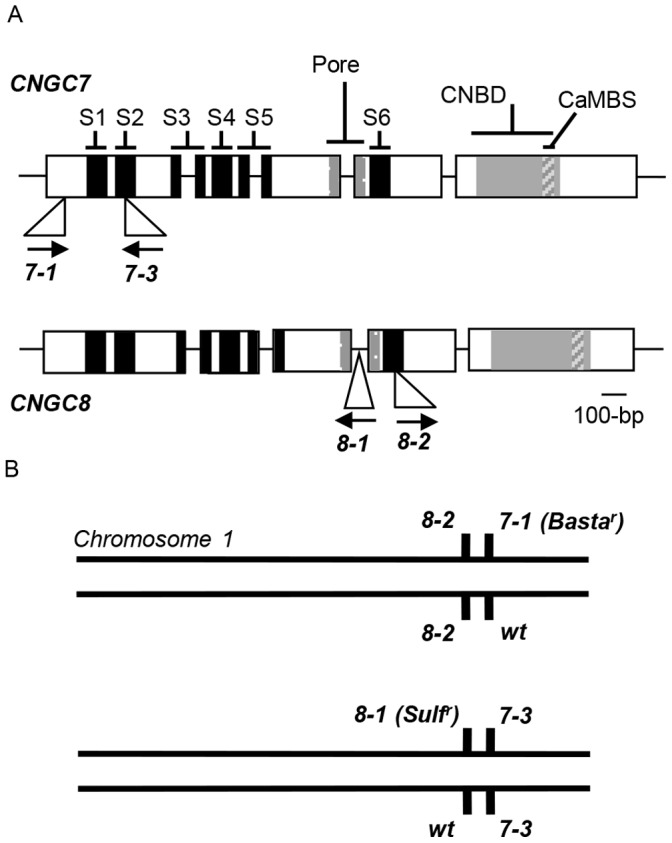
A) Locations of T-DNA insertions are shown for cngc7-1, cngc7-3, cngc8-1 and cngc8-2. Arrows indicate the direction of the T-DNA left border. Coding sequences are highlighted with expanded rectangles; lines indicate introns and flanking DNA sequences. CNGC7 and CNGC8 each have five exons, and encode proteins with six transmembrane spanning domains (S1-6; highlighted in black); a pore between S5 and S6 (shaded), a cyclic nucleotide binding domain (CNBD; shaded), and a calmodulin (CaM) binding site (CaMBS; shaded with gray lines) (after Köhler et al., 1999 [55] ). B) A diagram of chromosome 1 showing the arrangement of 2 different combinations of cngc7 and 8 alleles in which one of the two alleles is segregating with either a Bastar or Sulfr marker.
Table1. Segregation analysis showing a pollen transmission defect associated with a double knockout of cngc7/8.
| Cross | F1 | Segregation of +/− T-DNA | ||
| Female X Male | Total | Expect%a | Observed% | p- valueb |
| Crosses with single mutants | ||||
| cngc7-1+/−; SELFED | 559 | 75 | 74.1f, e | 0.99 |
| cngc7-3+/−; SELFED | 178 | 75 | 74.7c | 0.99 |
| cngc8-1+/−; SELFED | 1347 | 75 | 74.5d | 0.97 |
| cngc8-2+/−; SELFED | 1409 | 75 | 77.4d | 0.6 |
| WT X cngc7-1+/− | 37 | 50 | 54c | 0.95 |
| WT X cngc7-3+/− | 71 | 50 | 51c | 0.99 |
| WT X cngc8-2+/− | 206 | 50 | 50d | 1 |
| Crosses with double mutants (one gene −/−, second gene +/−) | ||||
| cngc7-1−/−, 8-2+/−; SELFED | 637 | 75 | 50d | <0.0001 |
| cngc7-1+/−, 8-2−/−;SELFED | 599 | 75 | 47.7g,h | <0.0001 |
| cngc7-3+/−, 8-1−/−; SELFED | 76 | 75 | 51c,h | ≤0.06 |
| cngc7-1−/−, 8-2+/− X WT | 143 | 50 | 55d | 0.7 |
| cngc7-3−/−, 8-1+/− X WT | 308 | 50 | 55d | 0.5 |
| WT X cngc7-1+/−, 8-2−/− | 727 | 50 | 0e | <0.0001 |
| WT X cngc7-1−/−, 8-2+/− | 756 | 50 | 0d | <0.0001 |
| WT X cngc7-3−/−, 8-1+/− | 5283 | 50 | 0d | <0.0001 |
Expected percentages based on Mendelian segregation.
Significance determined by the Pearson’s Chi-Squared test with two degrees of freedom.
PCR genotyping.
Mutant allele scored by Sulf r marker.
Mutant allele scored by Bastar maker.
117 by PCR genotyping.
313 by PCR genotyping.
no homozygous double knockout found.
The creation of plants harboring independent sets of double knockouts required the identification of cross-over recombination events between different pairs of cngc7 and 8 T-DNA insertions, since CNGC7 and 8 are closely linked on chromosome 1 (Figure 1). Plant lines with different sets of alleles were allowed to self-fertilize and plant lines with the following 4 genotypes were identified in which only one of the two alleles was segregating: cngc7-3 (−/−)/8-1 (+/− Sulf r), cngc7-3 (+/−)/8-1 (−/−), cngc7-1 (−/−)/8-2 (+/− Sulf r), and cngc7-1 (+/− Bastar)/8-2 (−/−). For three of these genotype combinations, the segregating allele is linked to a unique selectable marker-gene associated with the T-DNA insertion, either providing resistance to glufosinate ammonium (Bastar) or sulfadiazine (Sulfr).
To try and identify a homozygous cngc7/8 double knockout, cngc7/8 combinations segregating only one of the mutant alleles were allowed to self-fertilize, and the progeny was genotyped by PCR assays. In more than 389 progeny analyzed, no plants were found harboring a double homozygous mutation (Table 1). This segregation distortion was corroborated by analyzing the transmission frequencies of the Bastar or Sulfr markers associated with two different cngc7/8 knockout combinations (∼ 49% marker transmission observed versus 75% expected, n = 1236).
To determine if the inability to segregate a homozygous cngc7/8 mutant was due to a male or female defect, reciprocal crosses were conducted with three of the different allele combinations. For transmission of the cngc7/8 double mutation through the female, we observed the expected 50% transmission frequency (n = 451, Table 1). In contrast, no male transmission events were ever detected in more than 6766 progeny analyzed, indicating that pollen transmission was reduced by more than 3000-fold.
To corroborate that the cngc7/8 mutations used here represent loss of function null alleles (i.e., knockout), we tested whether the pollen transmission phenotype could be rescued by pollen expression of a transgene encoding either CNGC7 or 8. The N-terminal ends of CNGC7 and 8 were engineered with either GFP or a FLAG-tag, and the transgenes were expressed under the control of either a strong or weak pollen promoter (derived from the regulatory regions upstream of the pollen-expressed Ca2+-pump ACA9 [19] or CNGC18 [13], [14], respectively). Outcrosses to a female cngc7-3 (−/−) were done using pollen from plants that were cngc7-3 (−/−)/8-1 (+/− Sulfr) and hemizygous for a transgene encoding either a GFP- or FLAG-tagged CNGC7 or 8. In this situation, meiosis produces pollen with the following 4 genotypes: cngc7/8 (+/− the transgene) and cngc7/CNGC8 (+/− the transgene). Since a cngc7/8 pollen without a transgene fails to show any transmission (see Table 1), only 3 of the 4 meiotic products have the potential for transmission. Thus, a transgene providing a perfect rescue of cngc7/8 pollen would result in 33% of the progeny showing the transmission of the cngc7/8 double knockout, as scored by the segregation of the Sulfr marker associated with the cngc8-1 allele. While all transgene variations tested were able to rescue the cngc7/8 pollen transmission defect to some extent, the best transmission frequencies (23 to 27%) were observed for pollen harboring a FLAG-CNGC7 transgene expressed under the control of the relatively weak CNGC18 promoter (Figure 3). These results indicate that the cngc7/8 mutations studied here result in loss of function phenotypes that can rescued by a transgene encoding either a CNGC7 for CNGC8.
Figure 3. The cngc7/8 pollen transmission defect can be rescued by CNGC7 and 8 transgenes.
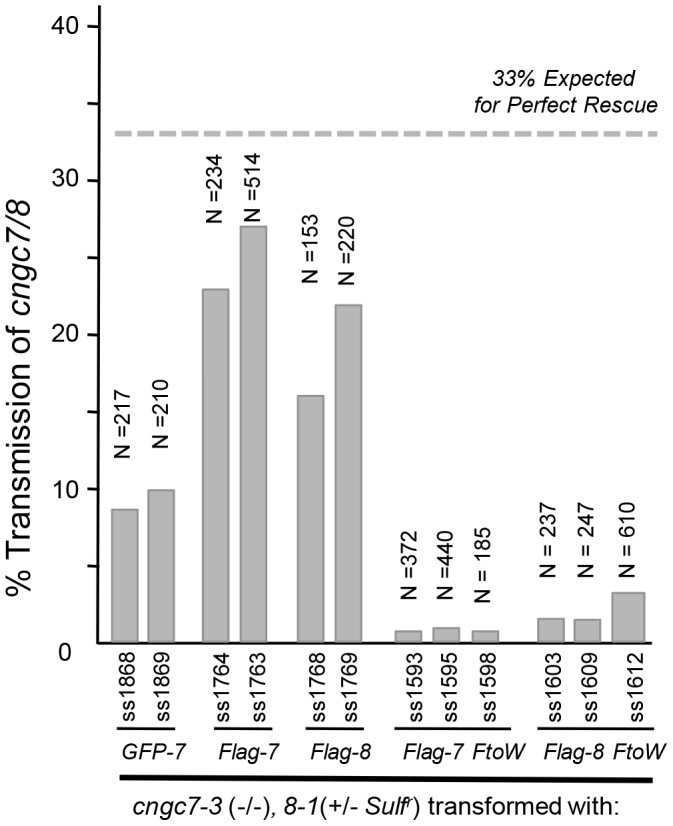
Pollen transmission efficiencies for cngc7/8 are shown, as scored by the transmission of the Sulfr marker to F1 progeny. The Sulf r marker was associated with the cngc8-1 allele that was segregating in the parental line. The cngc7-3 allele was homozygous. Pollen outcrosses were made by manually pollinating females that were wild type or cngc7-3(−/−) with equivalent results. All pollen outcrosses were done using male parents that were verified by reciprocal crosses to be hemizygous for the transgene. In these pollen outcross assays, a perfect rescue would result in 33% of the progeny carrying the Sulfr marker; because only 3 of the 4 meiotic products have the potential to show transmission to F1 progeny (see text). Numbers preceded by ss (seed stock) under each bar represent independent transgenic lines used for outcrossing. Homozygous double knockout seed stocks, created by selfing or out-crossing, are identified in Figure S4. Lines shown displayed typical rescue efficiencies mediated by transgene constructs for GFP-7 (9p-i-GFP-CNGC7), FLAG-7 or 8 (18p-i-FLAG-CNGC7 or 8), FLAG-7 or 8 F to W (18p-i-FLAG-CNGC7-F589W and 18p-i-FLAG-CNGC8-F624W). Three additional homozyogus rescued lines were obtained (not shown) using a transgene construct ps1687 18-i-GFP-CNGC8 (ss1402, ss1404, ss1405).
A Regulatory Site Mutation Impairs the Function of CNGC7 and 8
To generate a mutant plant with only a partial rescue of cngc7/8, rescue constructs were engineered to encode mutant versions of CNGC7 and 8 that contained an F589W or F624W substitution, respectively. These substitutions are positioned at a site conserved in plant CNGCs near the carboxyl end of the predicted cyclic nucleotide binding domain (CNBD) and the beginning of a potentially overlapping calmodulin binding-site (CaMBS) (Figure S1).
The respective rescue constructs harboring F to W substitutions were introduced into plants in which the cngc7 allele was homozygous and the 8 allele was segregating (i.e., cngc7-3 (−/−)/8-1 (+/− Sulfr)). Pollen was then outcrossed and the transmission frequency of a cngc7/8 double knockout scored in progeny by either PCR genotyping or the expression of a Sulfr phenotype. In contrast to a robust rescue using a wild type version of a FLAG-CNGC7 or 8, the incorporation of an F to W substitution (at amino acids 589 and 624, respectively) reduced the pollen transmission efficiency by 10 to 20-fold (Figure 3).
To evaluate whether the F to W substitutions would also compromise the seed set potential in a homozygous mutant, homozygous cngc7-3/8-1 lines rescued with a FLAG-CNGC7-F589W were identified by PCR genotyping. Although individual plants sometimes showed a reduction in seed set compared to wild type controls, this phenotype was not consistently observed. To understand the cause of this variation, three different plants displaying poor seed set were manually fertilized with the plant’s own pollen. In these cases, the manual self-fertilization was able to restore full seed set. This indicates that the variation in seed set is not a defect associated with the female gametophyte. Rather, the variation is either a result of less pollen being delivered to the stigma, and/or a further decrease in pollen fitness due to unknown variations in growth environments or plant health.
cncg7/8 Pollen Grains Burst as they Germinate
To determine why cngc7/8 mutant pollen are sterile, we first conducted a semi- in vivo pollen tube growth assay using pollen from a double knockout mutant segregating a GFP-CNGC7 rescue construct to 50% of the pollen grains. To set up these assays, receptive stigmas were manually pollinated and then cut and transferred to an agar surface for semi-in vivo growth. The only tubes observed to grow were those that showed GFP fluorescence, and therefore were rescued by a GFP-CNGC7 transgene (n = 27). The absence of any tubes without a GFP-CNGC7 suggested that non-rescued mutant tubes were defective at some early stage of pollen grain germination or tube growth.
In vitro pollen germination assays were then used to specifically evaluate potential defects at early stages of tip growth initiation. In these assays, we evaluated two different combinations of cngc7/8 alleles in which only one of the alleles was segregating. For both allele combinations, we observed a high frequency (50 to 60%) of pollen grains bursting (Figure 4). In contrast, wild type controls showed an average bursting frequency of less than 10%. For cngc7/8 mutants, the bursting events usually occurred before any tube growth could be detected (see Figure 5D for example). Similar bursting phenotypes and frequencies were observed using two different standard germination media.
Figure 4. Mutant cngc7/8 pollen grains burst during in vitro germination.
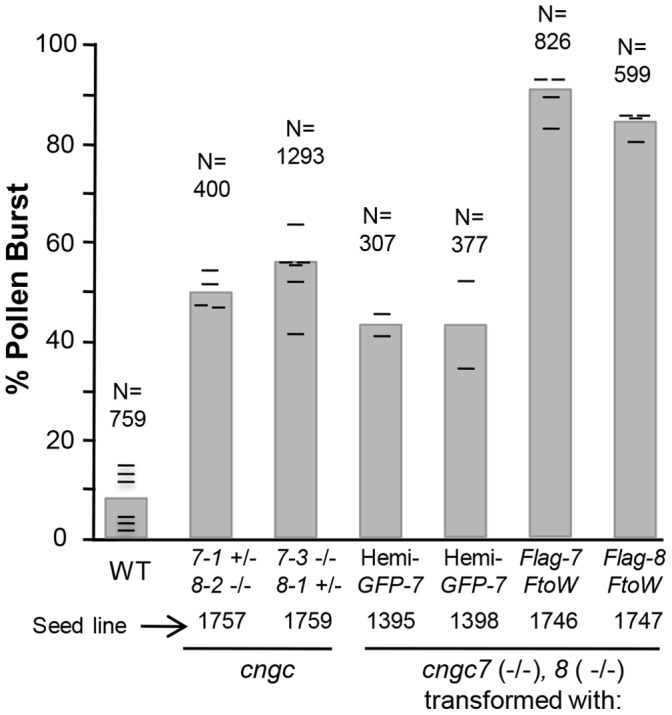
Pollen grains were germinated in vitro and scored for bursting at the time of germination. A ∼5-fold higher bursting frequency was observed for two different combinations of cngc7/8 alleles (cngc7-1+/−, 8-2−/− and cngc7-3−/−, 8-1+/−) in which only one of the alleles was segregating. An equivalent bursting frequency was observed for a cngc7-3/8-1 double knockout in which only 50% of the pollen harbored a rescue construct encoding GFP-CNGC7 (GFP-7) (denoted by “Hemi-”). A ∼90% bursting frequency was observed when a cngc7/8 double knockout was rescued with a FLAG-CNGC7 or 8 harboring an F to W substitution. In these “F to W” examples, the pollen came from a mixed pool of parental plants that were either hemizygous or homozygous for the transgene (i.e., at least 50% of the pollen had a “rescue” construct). Each hash mark indicates the % bursting for each of the six independent experiments. Results using two different media (standard, and 10% PEG media) showed equivalent results (n = 3 experiments each).
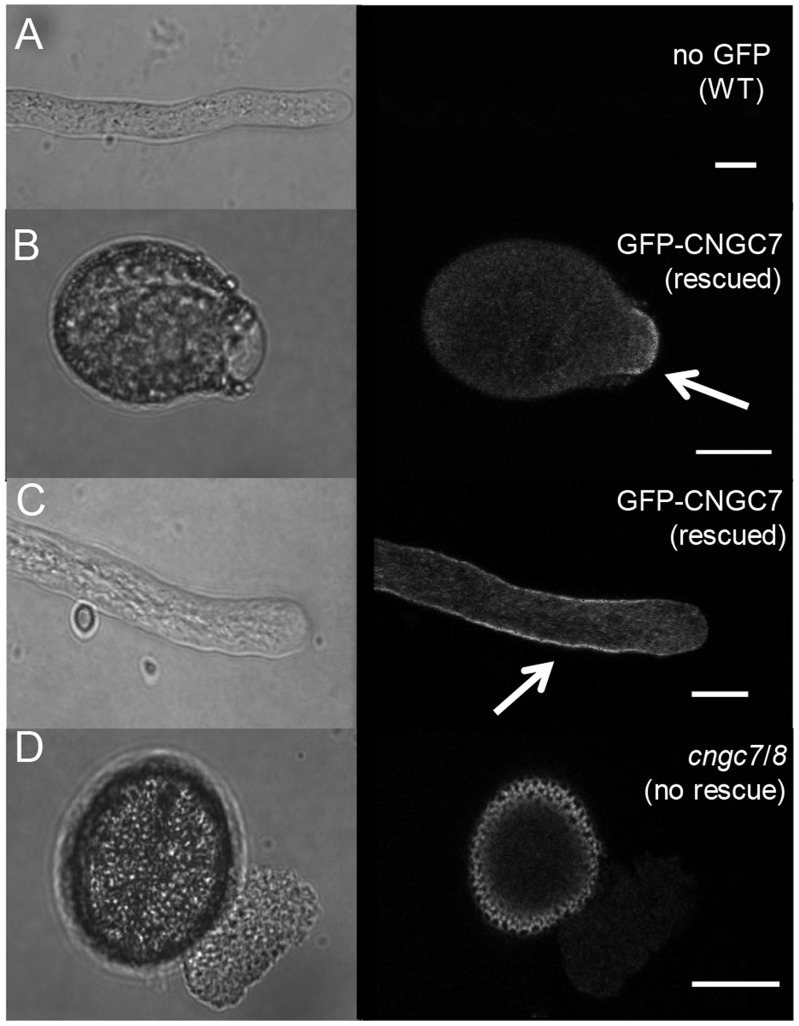
Figure 5. Confocal microscopy showing PM localization for GFP-CNGC7, and the cngc7/8 bursting defect.
Pollen were germinated in vitro and imaged. DIC images are shown to the left, and corresponding confocal fluorescence micrographs to the right. A) A negative control showing a wild type pollen tube without any GFP. B and C), GFP-CNGC7 in cngc7-3−/−, 8-1−/− showing a tip focused PM (plasma membrane) localization at the emerging tube (B) and the tip shank during tube extension (C). D) A non-rescued pollen from cngc7-3−/−, 8-1−/− (segregating a GFP-CNGC7) showing a typical bursting event at germination. Scale bar = 10 µm.
A cngc7/8 -dependent bursting phenotype was confirmed in two ways (Figure 4). First, in vitro germination assays were done with homozygous cngc7/8 mutants in which 50% of the pollen were expressing a rescue construct encoding GFP-CNGC7 (i.e., parent plants were hemizygous for the transgene). Plants segregating 50% of their pollen with a rescue construct were identified by imaging pollen from each plant for the expression of GFP. Using pollen from these plants, the bursting frequency was near 50% (n = 684). This is consistent with the expectation that 50% of the mutant pollen would be rescued from bursting through the expression of a GFP-CNGC7. This was corroborated by confocal fluorescence microscopy, which revealed that the only tubes to grow beyond the budding stage were those that showed GFP fluorescence (n >50). A second approach was to examine the frequency of bursting in mutant pollen grains from cngc7/8 plants that harbor transgenes that conferred only a partial rescue. For these pollen expressing either CNGC7-F589W or CNGC8-F624W, the bursting frequency was around 90%, which was about 10-fold higher than wild type controls.
CNGC7 is Localized to the Plasma Membrane of Pollen Tubes
To provide evidence for the subcellular location of CNGC7, fluorescence confocal microscopy was used to image GFP-CNGC7 in pollen. All imaging was done with homozygous cngc7/8 mutants that had been rescued by pollen expression of a GFP-CNGC7. Two different promoters were used to drive GFP-CNGC7 expression. We failed to see detectable levels of GFP using a weak promoter from CNGC18, although this promoter was capable of providing low levels of expression sufficient for functional rescues (see Figure 3). Therefore, to obtain high enough expression levels for imaging, we employed a stronger promoter from ACA9 [19], which resulted in a range of expression levels, from high to barely detectable. Figure 5 shows representative images of cells that have relatively weak but detectable levels of expression. Pollen with very high levels of expression always showed strong fluorescence throughout the cell, including endomembranes (as also observed in transient expression by [13], [14]). However, since functional rescues were observed with very low expression levels (e.g., provided by the CNGC18 promoter), we posit that images corresponding to low expression levels are more likely to reflect a normal distribution for a CNGC7, and less likely to be an artifact of over-expression [35]. With the imaging parameters used here, autofluorescence was occasionally seen associated with the cell wall (for example, Figure 5D). However, no other significant background fluorescence was detected within cells. In comparison, pollen expressing relatively low levels of GFP-CNGC7 showed strong fluorescent signals predominately associated with the PM at the bud site (Figure 5B), and in growing tubes, predominately at a region flanking the growing tip (Figure 5C).
Discussion
Genetic evidence presented here indicates that CNGC7 and 8 function together to provide at least one redundant activity that is essential for pollen fertility in Arabidopsis thaliana. Pollen harboring a cngc7/8 double knockout failed to show any transmission events in pollen outcrosses yielding more than 6000 progeny (expected frequency = 50%, Table 1).
Three lines of evidence suggest that the primary defect in cngc7/8 pollen occurs at the initiation of pollen tube tip growth, as shown with in vitro pollen growth assays (Figure 4). First, pollen grain bursting was observed for approximately 50% of the pollen assayed from mutant plants segregating 50% of their pollen as a cngc7/8 double knockout. Second, an equivalent bursting frequency was observed for pollen from a plant homozygous for cngc7/8 in which only half of the pollen harbored a GFP-tagged CNGC7 rescue construct. Third, a higher bursting frequency near 90% was observed for cngc7/8 pollen partially rescued by a transgene encoding a CNGC7 or 8 that was functionally compromised by an F to W substitution near the end of the proposed cyclic nucleotide binding domain (F589W or F624W, respectively). These in vitro results are consistent with the failure to observe tube growth for a cngc7/8 mutant in a semi-in vivo growth assay in which pollen was allowed to germinate on a stigma surface.
Of more than 50 mutations identified with defects associated with pollen germination, only two others are well characterized with an increased bursting frequency, vgd1 and anx1/anx2 [6], [7], [21]. AtAnx1 and 2 encode receptor-like kinases preferentially expressed in pollen, and are proposed to function redundantly in a signaling pathway that controls the timing of pollen tip bursting and sperm discharge when pollen tubes reach the synergid [7]. AtVGD1 (Vanguard1) encodes a pectin methyltransferase that is important for modifying the pollen cell wall to increase its rigidity [6]. In the absence of a rigid wall, pollen tubes, which have turgor pressure, are more likely to burst.
A CNGC18-7/8 Regulatory Node for Pollen Tube Tip Growth?
The cngc7/8 bursting defect also has similarities to the phenotype observed for cngc18 null mutants [13], [14]. In the case of cngc18, mutant pollen produced short kinky tubes that would often terminate by bursting. While some of the cngc7/8 pollen also germinated with similar projections, the dominant phenotype appeared to be a bursting projection directly from the pollen grain (Figure 5D). Given the similarities in phenotypes, further research is warranted to determine if CNGC18 might form multimeric complexs with CNGC7 and/or 8. In both plants and animals, CNGCs are thought to function as hetero-multimers [36]–[39], Assuming that hetero-multimers do form between CNGC18 and either CNGC7 or 8, a mutation that disrupts one of the subunits (e.g., CNGC18) might create a dysfunctional or destabilized complex.
Models for CNGC Regulation of Tip Growth
There are at least two reasonable models, not mutually exclusive, to explain the bursting phenotype associated with a dysfunctional CNGC7/8 multimeric complex. First, the channel complex might be essential for an ion homeostasis mechanism that regulates turgor. When the channel complex is dysfunctional, turgor pressure might increase to a bursting point, as the pollen grain cell wall begins to weaken during germination [6], [40], [41]. Since CNGCs are also permeable to K+, they might directly contribute a K+ transport involved in turgor regulation. Regulation of K+ transport has been proposed as a key feature in the mechanism of tube bursting at the time of sperm discharge [5].
In a second model, the CNGC complex might provide a signaling function that helps coordinate growth cycles at the pollen tube tip. For example, a cyclic nucleotide triggered Ca2+ signal might function as a “stop signal” to terminate a growth cycle and restrict growth to a manageable rate. In the absence of such a signal, growth processes might become uncoordinated and thereby make pollen tubes or buds highly susceptible to bursting. This speculation is consistent with a model in which Ca2+ signals can block signaling pathways, for example, ROP GTPases, which are implicated in promoting tip growth in pollen tubes and root hairs [42]–[45]. Alternatively, uncoordinated growth cycles might disrupt proper cell wall assembly at the growing tip, and give rise to a structurally weak wall, with a bursting phenotype analogous to that seen with the vanguard mutant [6].
While additional insights will be required to distinguish between these models, evidence here supports a model in which CNGCs 7 and 8 have redundant functions that are essential for the initiation or maintenance of pollen tube tip growth. It remains to be determined as to whether CNGC7 and 8 can form functional interactions with the other four pollen-expressed CNGCs in A. thaliana. Regardless, loss of function mutations for CNGC18 and 7/8 identify at least one CNGC activity that has evolved to be essential to the life cycle of a flowering plant.
Materials and Methods
Metadata for CNGC7 and CNGC8 can be found at TAIR, The Arabidopsis Information Resource (http://www.arabidopsis.org/), under the following accession numbers: At1g15990 and At1g19780, respectively.
Plant Growth Conditions
Arabidopsis thaliana ecotype Columbia (wild type Col-0 and transgenic plants) were germinated on half-strength MS medium (Murashige and Skoog, 1962) with 0.05% (w/v) MES, 0.5% (w/v) sucrose, pH 5.7, and 1% (w/v) agar, under a 24-h light regime, at 21°C. MS medium was supplemented, when necessary, with the appropriate selection marker. Concentrations were as follows: 25 µg/ml hygromycin; 10 µg/ml basta (glufosinate ammonium); 50 µg/ml kanamycin; and 75 µg/ml sulfadiazine.
10-days old seedlings were transplanted to Metro-Mix 200 Series soil (Hummert), fertilized with Triple Ten 10-10-10 containing 40% slow release nitrogen (Growth Products) and grown under a 16-h light/8-h dark regime, at 21°C. All experiments were conducted by comparison of wild type and mutant plants grown side-by-side.
Isolation of cngc7 and cngc8 T-DNA Insertions
T-DNA insertions were identified using the SIGnAL “T-DNA Express” Arabidopsis Gene Mapping Tool (http://signal.salk.edu/cgi-bin/tdnaexpress). CNGC7 T-DNA insertion lines were obtained from Syngenta Arabidopsis Insertion Library collection (cngc7-1, SAIL_59_F03, harboring a glufosinate-resistance gene, Basta r; [32]), and SALK collection (cngc7-3, Salk_060871 [33]). T-DNA insertion lines for cngc8 were obtained from GABI-Kat collection (cngc8-1, GABI_101C03; cngc8-2, GABI_462B04; [34]), all harboring a sulfadiazine-resistance (Sulfr) marker within the T-DNA insertion. The glufosinate-ammonium used for Basta r selection and sulfadiazine used for Sulfr are obtained from Sigma-Aldrich (St. Louis,).
The genotypes of all plant lines were confirmed by PCR analysis of genomic DNA using gene-specific and T-DNA left border primers. The presence of a wild type CNGC7 was diagnosed using gene specific primers 1345a and 1345b (Figure S2). The cngc7-1 and 7-3 insertion alleles were diagnosed using primers 1345br and 638, and 1345a and 792, respectively. CNGC8 was diagnosed using gene specific primers 960a and 960b. The cngc8-1 and 8-2 insertion alleles were diagnosed using primers 960a and 958, and 960b and 958, respectively. T-DNA border fragments were amplified and sequenced for each line to verify the site of T-DNA insertion.
Plasmid Constructs Encoding CNGC7 and CNGC8
Plant expression constructs were made in a modified pGreenII vector system [46], with a kanamycin selection marker for bacteria, and a hygromycin marker for plants. The DNA sequence of each construct is provided as a supplemental file (Figure S3). The 9p promoter corresponds to the upstream regulatory region for calcium pump ACA9 [19]. The 18p promoter corresponds to the upstream regulatory region of CNGC18 [13], [14]. In each construct, the 5′ UTR contains an intron corresponding to a 5′UTR intron from AHA3 [13], [14], [47]. All CNGC7 constructs contain a genomic sequence for CNGC7, which was PCR amplified from Col-0 genomic DNA using primers 1147a and 1147br (Figure S2). All CNGC8 constructs were made with a CNGC8 cDNA, which was amplified from a Col-0 pSPORT cDNA library (Invitrogen) using primers 1148a and 1148br (Figure S2). F to W substitutions were engineered by a two-step PCR [48]. All sequences derived from PCR reactions were verified by DNA sequencing.
9p-i-GFP-CNGC7 (ps1300) encodes a GFP-tagged CNGC7, expressed under the control of a 9p promoter. 18p-i-FLAG-CNGC 7 (ps1692) encodes a FLAG epitope [49], [50] tagged CNGC7, expressed under the control of the CNGC18 promoter. 18p-i-FLAG-CNGC7(F589W) (ps1650) is the same as ps1692, but encodes a CNGC7 with an F589W substitution. 18p-i-FLAG-CNGC8 (ps1687) encodes a FLAG epitope tagged CNGC8, expressed under the control of the CNGC18 promoter. 18p-i-FLAG-CNGC8(F624W) (ps1685) is the same as ps1687, but encodes a CNGC8 with an F624W substitution. Representative transgenic plants with these constructs are listed in Figure S4 seed stock table.
Plant Transformation
Transgenic Arabidopsis thaliana plants were generated by floral dipping with Agrobacterium tumefaciens strain GV3101 [51]. Transgenic plants were selected on MS medium containing hygromycin.
Pollen Germination
Pollen from open flowers was germinated on standard medium containing 1% low-melting agarose with 0.01% H3BO3,1 mM CaCl2, 5 mM KCl, 10% sucrose, pH 7.5, as modified from [52]. An alternative medium with 10% PEG (polyethylene glycol 4000) was modified from [53], [54] and contained, 0.01% H3BO3, 3 mM Ca(NO3)2, 1 mM MgSO4, 1 mM KNO3, 10% (w/v) PEG, 10% (w/v) sucrose, pH 7.5 with KOH. To make solid medium with PEG, the liquid medium minus PEG was first solidified with 1% low-melting agarose, and then equilibrated with liquid medium including 10% PEG. To enhance the germination rate, one pistil was placed in proximity of the pollen on the germination medium.
Image Acquisition
Images of GFP fluorescence were collected on a Olympus confocal system (FluoView FV10-ASW 1.5; Olympus) attached to an Olympus microscope (Inverted IX81) using a 60X objective (N.A. = 1.39) and an argon gas laser for generating a 488-nm excitation line. Emission was detected with band pass between 510 and 530 nm. Differential interference contrast (DIC) images were collected on the same system by using a single transmitted light detector. Images were processed by using FluoView software.
Supporting Information
Sequence alignment of cyclic nucleotide binding domains (CNBDs) from CNGCs.
(TIF)
Primers used in this study.
(TIF)
DNA sequences of plasmid constructs used in this study.
(PDF)
Seed stocks used in this study.
(TIF)
Acknowledgments
We thank Norman Groves, Jason Stubich, Miguel F. Carvalho and Kelly Zinn for advice and technical assistances.
Funding Statement
This work was supported by grants to JFH from National Institutes of Health (1RO1 GM070813-01) for genetic analyses into the formation of calcium signals in pollen, and Department of Energy (DE-FG03-94ER20152) for subcellular localization studies on membrane proteins and regulation of plant cell growth. Funding for operation of the confocal microscope core came from COBRE NIH grant GM103554. This work was also supported by a grant to RM from Fundação Ciência e Tecnologia (PTDC/AGR-GPL/108156/2008; FEDER). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kessler SA, Grossniklaus U (2011) She’s the boss: signaling in pollen tube reception. Current Opinion in Plant Biology 14: 622–627. [DOI] [PubMed] [Google Scholar]
- 2. Chae K, Lord EM (2011) Pollen tube growth and guidance: roles of small, secreted proteins. Annals of Botany 108: 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takeuchi H, Higashiyama T (2011) Attraction of tip-growing pollen tubes by the female gametophyte. Current Opinion in Plant Biology 14: 614–621. [DOI] [PubMed] [Google Scholar]
- 4. Higashiyama T, Kuroiwa H, Kawano S, Kuroiwa T (2000) Explosive discharge of pollen tube contents in Torenia fournieri. Plant Physiology 122: 11–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amien S, Kliwer I, Márton ML, Debener T, Geiger D, et al. (2010) Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biology 8: e1000388 doi:10.1371/journal.pbio.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jiang L, Yang S, Xie L, Puah CS, Zhang X, et al. (2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell Reports 17: 584–596 doi:10.1105/tpc.104.027631.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boisson-Dernier A, Roy S, Kritsas K, Grobei M a, Jaciubek M, et al. (2009) Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development (Cambridge, England) 136: 3279–3288 doi:10.1242/dev.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qin Y, Yang Z (2011) Rapid tip growth: insights from pollen tubes. Seminars in Cell & Developmental Biology 22: 816–824 doi:10.1016/j.semcdb.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spalding EP, Harper JF (2011) The ins and outs of cellular Ca(2+) transport. Current Opinion in Plant Biology 14: 715–720 doi:10.1016/j.pbi.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hepler PK, Kunkel JG, Rounds CM, Winship LJ (2012) Calcium entry into pollen tubes. Trends in Plant Science 17: 32–38 doi:10.1016/j.tplants.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 11. Iwano M, Entani T, Shiba H, Kakita M, Nagai T, et al. (2009) Fine-tuning of the cytoplasmic Ca2+ concentration is essential for pollen tube growth. Plant Physiology 150: 1322–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou L, Fu Y, Yang Z (2009) A genome-wide functional characterization of Arabidopsis regulatory calcium sensors in pollen tubes. Journal of Integrative Plant Biology 51: 751–761 doi:10.1111/j.1744-7909.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- 13. Chang F, Yan A, Zhao L-N, Wu W-H, Yang Z (2007) A Putative Calcium-Permeable Cyclic Nucleotide-Gated Channel, CNGC18, Regulates Polarized Pollen Tube Growth. Journal of Integrative Plant Biology 49: 1261–1270 doi:10.1111/j.1672–9072.2007.00524.x. [Google Scholar]
- 14. Frietsch S, Wang Y-F, Sladek C, Poulsen LR, Romanowsky SM, et al. (2007) A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proceedings of the National Academy of Sciences of the United States of America 104: 14531–14536 doi:10.1073/pnas.0701781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao F, Han X, Wu J, Zheng S, Shang Z, et al.. (2012) A heat-activated calcium-permeable channel - Arabidopsis cyclic nucleotide-gated ion channel 6 - is involved in heat shock responses. The Plant Journal. doi:10.1111/j.1365-313X.2012.04969.x. [DOI] [PubMed]
- 16. Konrad KR, Wudick MM, Feijó JA (2011) Calcium regulation of tip growth: new genes for old mechanisms. Current Opinion in Plant Biology 14: 721–730 doi:10.1016/j.pbi.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 17. Michard E, Lima PT, Borges F, Silva AC, Portes MT, et al. (2011) Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science (New York, NY) 332: 434–437. [DOI] [PubMed] [Google Scholar]
- 18. Chaiwongsar S, Strohm AK, Roe JR, Godiwalla RY, Chan CWM (2009) A cyclic nucleotide-gated channel is necessary for optimum fertility in high-calcium environments. New Phytologist 183: 76–87. [DOI] [PubMed] [Google Scholar]
- 19. Schiøtt M, Romanowsky SM, Baekgaard L, Jakobsen MK, Palmgren MG, et al. (2004) A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proceedings of the National Academy of Sciences of the United States of America 101: 9502–9507 doi:10.1073/pnas.0401542101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Myers C, Romanowsky SM, Barron YD, Garg S, Azuse CL, et al. (2009) Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. The Plant Journal 59: 528–539. [DOI] [PubMed] [Google Scholar]
- 21. Cheung AY, Wu H-M (2008) Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annual Review of Plant Biology 59: 547–572 doi:10.1146/annurev.arplant.59.032607.092921. [DOI] [PubMed] [Google Scholar]
- 22. Cárdenas L, Lovy-Wheeler A, Kunkel JG, Hepler PK (2008) Pollen tube growth oscillations and intracellular calcium levels are reversibly modulated by actin polymerization. Plant Physiology 146: 1611–1621 doi:10.1104/pp.107.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potocký M, Pejchar P, Gutkowska M, Jiménez-Quesada MJ, Potocká A, et al.. (2012) NADPH oxidase activity in pollen tubes is affected by calcium ions, signaling phospholipids and Rac/Rop GTPases. Journal of Plant Physiology. [DOI] [PubMed]
- 24. Yan A, Xu G, Yang Z-B (2009) Calcium participates in feedback regulation of the oscillating ROP1 Rho GTPase in pollen tubes. Proceedings of the National Academy of Sciences of the United States of America 106: 22002–22007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biology 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, et al. (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genetics 5: e1000621 doi:10.1371/journal.pgen.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rato C, Monteiro D, Hepler PK, Malhó R (2004) Calmodulin activity and cAMP signalling modulate growth and apical secretion in pollen tubes. The Plant Journal 38: 887–897. [DOI] [PubMed] [Google Scholar]
- 28. Moutinho A, Hussey PJ, Trewavas AJ, Malhó R (2001) cAMP acts as a second messenger in pollen tube growth and reorientation. Proceedings of the National Academy of Sciences of the United States of America 98: 10481–10486 doi:10.1073/pnas.171104598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu J, Qu H, Jin C, Shang Z, Wu J, et al. (2011) cAMP activates hyperpolarization-activated Ca2+ channels in the pollen of Pyrus pyrifolia. Plant Cell Reports 30: 1193–1200. [DOI] [PubMed] [Google Scholar]
- 30. Christopher DA, Borsics T, Yuen CYL, Ullmer W, Andème-Ondzighi C, et al. (2007) The cyclic nucleotide gated cation channel AtCNGC10 traffics from the ER via Golgi vesicles to the plasma membrane of Arabidopsis root and leaf cells. BMC Plant Biology 7: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hua B-G, Mercier RW, Leng Q, Berkowitz GA (2003) Plants do it differently. A new basis for potassium/sodium selectivity in the pore of an ion channel. Plant Physiology 132: 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sessions A, Burke E, Presting G, Aux G, McElver J, et al. (2002) A high-throughput Arabidopsis reverse genetics system. The Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science (New York, NY) 301: 653–657 doi:10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 34. Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, et al. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology 53: 247–259. [DOI] [PubMed] [Google Scholar]
- 35. Moore I, Murphy A (2009) Validating the location of fluorescent protein fusions in the endomembrane system. The Plant Cell 21: 1632–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaupp UB, Seifert R (2002) Cyclic nucleotide-gated ion channels. Physiological Reviews 82: 769–824. [DOI] [PubMed] [Google Scholar]
- 37. Chen TY, Peng YW, Dhallan RS, Ahamed B, Reed RR, et al. (1993) A new subunit of the cyclic nucleotide-gated cation channel in retinal rods. Nature 362: 764–767. [DOI] [PubMed] [Google Scholar]
- 38. Chen TY, Illing M, Molday LL, Hsu YT, Yau KW, et al. (1994) Subunit 2 (or beta) of retinal rod cGMP-gated cation channel is a component of the 240-kDa channel-associated protein and mediates Ca(2+)-calmodulin modulation. Proceedings of the National Academy of Sciences of the United States of America 91: 11757–11761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Körschen HG, Illing M, Seifert R, Sesti F, Williams A, et al. (1995) A 240 kDa protein represents the complete beta subunit of the cyclic nucleotide-gated channel from rod photoreceptor. Neuron 15: 627–636. [DOI] [PubMed] [Google Scholar]
- 40. Ma LG, Fan QS, Yu ZQ, Zhou HL, Zhang FS, et al. (2000) Does aluminum inhibit pollen germination via extracellular calmodulin? Plant & Cell Physiology 41: 372–376. [DOI] [PubMed] [Google Scholar]
- 41. Hill AE, Shachar-Hill B, Skepper JN, Powell J, Shachar-Hill Y (2012) An osmotic model of the growing pollen tube. PloS One 7: e36585 doi:10.1371/journal.pone.0036585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin Y, Yang Z (1997) Inhibition of Pollen Tube Elongation by Microinjected Anti-Rop1Ps Antibodies Suggests a Crucial Role for Rho-Type GTPases in the Control of Tip Growth. The Plant Cell 9: 1647–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H, Lin Y, Heath RM, Zhu MX, Yang Z (1999) Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. The Plant Cell 11: 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li H, Shen JJ, Zheng ZL, Lin Y, Yang Z (2001) The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiology 126: 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jones MA, Shen J-J, Fu Y, Li H, Yang Z, et al. (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. The Plant Cell 14: 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Molecular Biology 42: 819–832. [DOI] [PubMed] [Google Scholar]
- 47. Robertson WR, Clark K, Young JC, Sussman MR (2004) An Arabidopsis thaliana plasma membrane proton pump is essential for pollen development. Genetics 168: 1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kammann M, Laufs J, Schell J, Gronenborn B (1989) Rapid insertional mutagenesis of DNA by polymerase chain reaction (PCR). Nucleic Acids Research 17: 5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Einhauer A, Jungbauer A (2001) The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. Journal of Biochemical and Biophysical Methods 49: 455–465. [DOI] [PubMed] [Google Scholar]
- 50. Chiang CM, Roeder RG (1993) Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Peptide Research 6: 62–64. [PubMed] [Google Scholar]
- 51. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 52. Boavida LC, McCormick S (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. The Plant Journal 52: 570–582 doi:10.1111/j.1365-313X.2007.03248.x. [DOI] [PubMed] [Google Scholar]
- 53. Palanivelu R, Brass L, Edlund AF, Preuss D (2003) Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114: 47–59. [DOI] [PubMed] [Google Scholar]
- 54. Jayaprakash P, Sarla N (2001) Development of an improved medium for germination of Cajanus cajan (L.) Millsp. pollen in vitro. Journal of Experimental Botany 52: 851–855. [DOI] [PubMed] [Google Scholar]
- 55. Köhler C, Merkle T, Neuhaus G (1999) Characterisation of a novel gene family of putative cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana. The Plant Journal 18: 97–104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of cyclic nucleotide binding domains (CNBDs) from CNGCs.
(TIF)
Primers used in this study.
(TIF)
DNA sequences of plasmid constructs used in this study.
(PDF)
Seed stocks used in this study.
(TIF)