Abstract
The NF-κB pathway plays a critical role in regulating cellular processes such as immune responses, stress responses, apoptosis, proliferation and differentiation, whereas dysfunction of this pathway has been associated with numerous cancer and immune disorders. We have applied our Random Activation of Gene Expression technology to an NF-κB reporter cell line to facilitate the discovery of positive regulators of NF-κB activation. A small protein expression library, corresponding to ≈0.1× genome coverage, was generated and screened for clones exhibiting constitutive activation of NF-κB. After isolation of cellular clones displaying the relevant phenotypes, we identified two known components of the NF-κB pathway and a hypothetical gene that we have designated the human ortholog of Xenopus TAK1-binding protein 3 (TAB3). Overexpression of human TAB3 was found to activate both NF-κB and AP-1 transcription factors. Furthermore, the activation of NF-κB by TAB3 was blocked by the NF-κB inhibitor, SN50, and by expression of dominant-negative forms of tumor necrosis factor α-associated factor 6 and transforming growth factor β-activated kinase. Taken together, these data demonstrate that TAB3 transforming growth factor is a constituent of the NF-κB pathway functioning upstream of tumor necrosis factor α-associated factor 6/transforming growth factor β-activated kinase. Interestingly, increased expression of TAB3 was found in some cancer tissues, and its overexpression in NIH 3T3 cells resulted in cellular transformation, thus establishing a causative link between elevated TAB3 expression, constitutive NF-κB activation, and oncogenesis.
The NF-κB family of transcription factors comprises five structurally related proteins, RelA/p65, cRel, RelB, p50/p105, and p52/p100 (1, 2), in varied combinations to form hetero- and homodimeric complexes. A menagerie of NF-κB isoforms may exist even within a single cell; however, the p65/p50 heterodimer is predominant in many cell types (3, 4). In most normal cells, latent NF-κB is sequestered in an inactive form through tight association with a member of the IκB family of inhibitor proteins. Whereas the NFκB:IκB complex is capable of shuttling between the cytoplasm and nuclear compartment, it is predominantly resident in the cytoplasm (5). A variety of extracellular signals trigger signal transduction pathways that converge on a crucial regulatory junction: the phosphorylation, ubiquitination, and subsequent proteasome-dependent degradation of IκB to liberate active NF-κB (6). Once free of IκB, NF-κB rapidly translocates into the nucleus and binds to the κB motif within the regulatory regions of NF-κB-responsive genes.
NF-κB is well established as an activator of genes that regulate innate and acquired immune responses, as well as maintaining the fine balance between pro- and anti apoptotic cellular pathways (7). Currently, >100 human genes have been identified to be responsive to NF-κB (8, 9). The classes of gene products regulated through NF-κB activation pathways are diverse, including cytokines, growth effectors, receptors, transcription factors, enzymes, and matrix and adhesion proteins. Whereas normal cells tightly regulate the many pathways that lead to NF-κB activation, dysfunction in any of the pathways typically results in the onset of severe diseases (10, 11). A variety of cancers, rheumatoid arthritis, atherosclerosis, asthma, cachexia, diabetes, inflammatory bowel syndrome, and septic shock are all associated with the inappropriate regulation of NF-κB (10, 12, 13). Therefore, regulators of NF-κB activation command high interest as potential drug targets. The discovery of new NF-κB regulatory proteins continues to be made, and it is likely that many of these will prove to be important targets for the development of effective therapeutics (14, 15).
We have devised an efficient functional genomics strategy to facilitate the discovery and characterization of human genes encoding regulators of NF-κB. The random activation of gene expression (RAGE) technology (16) was used to generate a limited protein expression library in a transgenic NF-κB reporter cell line, and this library was selected for those cells that constitutively activated a positive regulator of NF-κB. Herein, we report the discovery and characterization of human transforming growth factor β-activated kinase (TAK1)-binding protein 3 (TAB3), a regulator of NF-κB and AP-1.
Materials and Methods
The 293TH8 Cell Line, RAGE Library Construction, and Selection. NF-κB-responsive cell line 293TH8 is derived from 293-EBNA cells (Invitrogen) engineered to contain 2.1 kb of human genomic DNA encoding the IL-8 promote, exon 1, and intron 1 (relative to ATG, sequence spanning nucleotides –1244 to +788 of the IL-8 gene) positioned to express a functional chimeric transcript encoding the HSV thymidine kinase ORF (1,120 nucleotides), 450 nt of human serum albumin intron 5, the hygromycin (Hyg)–resistance ORF (1,038 nucleotides), and the SV40 polyadenylation sequence. The 293TH8 cells were expanded in ganciclovir-containing medium to eliminate cells with unregulated expression of IL8-thymidine kinase/Hyg (TkHyg). The 293TH8 cells were electroporated with RAGE vector pFG114 to generate a protein expression library. pFG114 is similar to the RAGE vector described by Harrington et al. (16), except that the resistance gene is positioned in the opposite orientation to the RAGE activation exon. Electroporated cells were diluted into DMEM-C and were seeded at 5 × 106 cells per 15-cm culture dish. The culture medium was replaced with DMEM-C and 750 μg of Hyg per ml at 48 h postelectroporation to select for cells that converted to a phenotype of constitutive NF-κB activation. Selective media was exchanged every 3 days for 10 days, medium-supplemented with 750 μg of Hyg per ml, and 3 μg of puromycin per ml was added. Isolated colonies were expanded and 24-h culture supernatants were collected and assayed by IL-8 ELISA (R & D Systems). Library clone 495.74 was transfected with a Cre expression vector and cultured in DMEM-C and 10 μg of Gcv per ml for 14 days to select for phenotype reverted cells. GcvR 495.74:Cre cells were further assayed for IL-8 expression, as described above.
Cloning of RAGE-Activated Genes. mRNA was purified from clone 495.74 cells by using Qiagen oligotex reagent and protocol. Reverse transcription was performed by using primer dTZ(Sfi) [5′-TCAGACTTTAGATGGCCAACGAGGCCT6ZT6ZT5ZT5ZT5ZT8; Z denotes 3-nitropyrrole (17)] with SuperScript II reagents and protocol (Invitrogen). Standard 3′ RACE reactions were performed by using a forward primer (5′-ACCTACTTAATACGACTCACTATAGGGCGA) complementary to sequence within the SfiI containing RAGE activation exon, paired with reverse primer dTZ(Sfi). Amplification products were SfiI-digested and cloned into an expression vector that contained SfiI sites identical to those of 3′ RACE products derived from RAGE transcripts.
Transcript Analyses. Quantitative PCR was performed by using mRNA from peripheral blood lymphocytes and clone 495.74 cells, or from human kidney, brain, and prostate tissues (BD Biosciences Clontech, Palo Alto, CA). The ratio of expressed TAB3a and TAB3b transcripts was determined by using an exon 12 reverse primer (5′-AGCCCCTTCGTAGTCTTCATCTC) paired with an exon 11 forward primer (5′-GCAGACATCCATGACACCCA) to determine total TAB3 transcripts. The same reverse primer was paired with an exon 10 forward primer (5′-AGTTGTACCACCCAAGCCATCTA) to determine the proportion of TAB3a transcripts. Quantitative PCR was performed by using SYBR green PCR Master Mix (Applied Biosystems) and standard quantitative PCR cycling conditions. β-glucuronidase provided an internal standard for normalization by using forward (5′-TTGTCGGCTGGGTGTGGTAC) and reverse (5-′AGCGTGTCGACCCCATTC) gene-specific primers.
Functional Analyses. NF-κB inhibitor studies were conducted by using SN50 and SN50M (Calbiochem) at culture concentrations of 100 μg/ml. Dominant-negative (DN) expression assays were performed by combining 2.5 μg of vectors expressing DN-TAK1 or DN-tumor necrosis factor α-associated factor 6 (TRAF6) with 0.5 μg of vectors expressing TAB3a or TAB3b, cotransfecting 293IL8-luc cells, and quantifying 48-h luciferase activities by using Promega kit reagents and protocol.
Electrophoretic mobility-shift assays (EMSAs) were conducted by using nuclear protein extracts prepared as described (18). Protein–DNA-binding reactions were performed by mixing 5 μg of nuclear protein, 1 μg of poly deoxyinosine-deoxycytosine in 10 mM Tris·HCl, pH 7.5/0.5 mM EDTA/0.5 mM DTT/50 mM NaCl/1 mM MgCl2/4% glycerol in a total volume of 10 μl. Nonlabeled competitor NF-κB oligo, AP-1 oligo, and mutant NF-κB oligo (Promega) were added in 50-fold excess to their respective binding mixes. Supershift assays were performed by using anti-p65 and anti-c-Jun antibodies (Santa Cruz Biotechnology). Antibody (0.2 μg) was added to the binding reaction and incubated at room temperature for 30 min, then 32P-labeled oligo was added and incubated for an additional 30 min. Bound probes were resolved by means of electrophoresis through nondenaturing 4% polyacrylamide gels in 0.5× TBE buffer.
Disease Association. The analyses of TAB3 expression in normal, autoimmune disease and cancer tissues was determined by using 32P-labeled cDNA as probe and hybridizing to the autoimmune disease profiling array and cancer profiling array II (BD Biosciences Clontech).
Transformation assays were performed by using NIH 3T3 cells stably transfected with either TAB3a or TAB3b expression vectors. For contact inhibition assays, 1 × 106 transgenic NIH 3T3 were seeded in 10-cm culture plates. For anchorage-independent growth assays, cells were seeded onto soft agar (DMEM-C and 1% agarose) surfaces. Contact-inhibited growth and colony formation were assessed 2 weeks after seeding.
Results
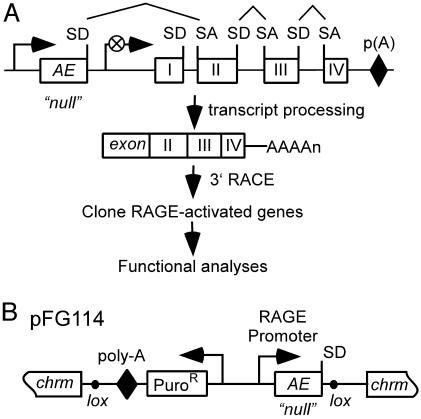
Functional Screens for Activators of NF-κB. In this study, we developed a function-to-gene strategy to discover regulators of the NF-κB pathway. This approach was based on the use of RAGE protein expression libraries in combination with transgenic NF-κB reporter cells. Compared with conventional cDNA-based protein expression libraries, RAGE libraries are relatively normalized and express a large percentage of the genes in the human genome, possibly allowing us to identify NF-κB pathway genes missed by previous approaches. The basic principle of the RAGE expression mechanism has been described (16) and is summarized in Fig. 1A.
Fig. 1.
(A) Activated expression of an endogenous gene by using RAGE. A RAGE vector randomly integrates into the host genome and becomes operably linked to the nearest downstream gene. The RAGE promoter drives transcription through the RAGE activation exon (AE) and downstream gene to adopt its poly(A) signal. RNA splicing removes genomic sequence between the splice donor (SD) of the AE and the first splice acceptor (SA) site, typically found preceding exon 2 of the endogenous gene. RAGE-activated transcripts are tagged at their 5′ ends with the AE sequence, thereby facilitating their rapid isolation. (B) RAGE vector pFG114 depicted integrated into a chromosome (chrm). The AE contains an SfiI site and unpaired SD but lacks a translation start site (“null”). Lox sites enable Cre-mediated excision of critical RAGE elements. Puro, puromycin.
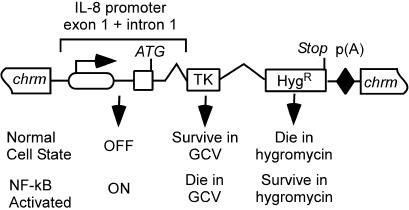
The identification of genes by using RAGE requires a cellular assay relevant to the biology of interest. To this end, we constructed a transgenic reporter cell line, 293TH8, containing the TkHyg gene downstream of the NF-κB-responsive IL-8 promoter (Fig. 2). In nonstimulated reporter cells, the NF-κB pathway is inactive, the IL-8 promoter is repressed (19), and the cells die under Hyg selection. However, after activation of the NF-κB pathway, the IL-8 promoter will become active, thereby driving expression of the Hyg-resistance gene and allowing the cells to survive under Hyg selection. The performance of this assay was tested by transfecting 293TH8 cells with a construct that expresses and secrets the mature-form IL-1 protein, a known activator of NF-κB. The demonstration of cells proliferating in Hyg-containing media, conditions that otherwise effectively kill these cells, provided validation of our NF-κB reporter cell line and assay (data not shown).
Fig. 2.
The 293TH8 NF-κB reporter cell line. The NF-κκ-responsive IL-8 promoter is operably linked to an ORF encoding Tk- and Hyg-resistance proteins. The selectable phenotypes of normal and NF-κB-activated cells are indicated. RAGE-activated expression of a gene that causes NF-κB activation will induce the expression of TkHyg, enabling cells to be selected in Hyg-containing medium.
To identify genes that, after overexpression, activate the NF-κB pathway, we created a RAGE library by using the activation vector pFG114 (Fig. 1B). In this study, we elected to use a single RAGE vector in which the activation exon lacked a translation start codon. This strategy allowed us to keep the library size at a minimum while maximizing genome coverage. Based on our library size of 2.5 × 105 clones, the estimated percentage of genes lacking a start codon in exon 1, and our empirical library coverage (16), we estimated that ≈5–10% of human genes would be expressed in a functional form in this small library.
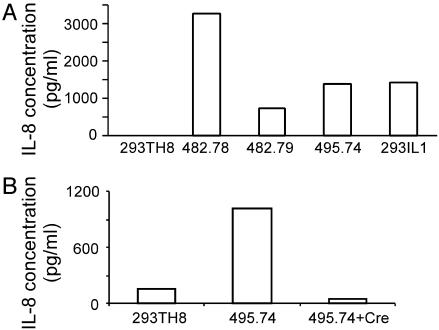
After creation of the RAGE library in 293TH8 cells, the library was subjected to Hyg selection to identify clones in which the transgene IL-8 promoter had become activated. Because activation of the NF-κB pathway should lead to activation of our transgene as well as endogenous IL-8 gene, we further analyzed HygR library clones for expression of IL-8. Three clones were identified that expressed IL-8 at levels comparable to our positive control IL-1-stimulated 293-EBNA cells (Fig. 3A), suggesting that these clones had converted to a state of constitutive NF-κB activation. Accordingly, the RAGE-activated gene from each of these three library clones was cloned by 3′ RACE and identified. Two of the RAGE-activated genes were found to encode ACT1 (20–22) and XEDAR (23), both known regulatory components of the NF-κB pathway, thus providing important validation for our approach. The RAGE-activated gene isolated from the third library clone, 495.74, was determined to encode a predicted gene, MGC45404 (gi:22749540), of unknown function. Consistent with MCG45404 being a bona fide gene, we found that MCG45404 transcript was expressed in the parental 293TH8 cells and that this putative gene was overexpressed by 50-fold in the RAGE-activated clone 495.74. No other RAGE-activated transcripts were detected in clone 495.74, suggesting that MCG45404 was responsible for the activation of the IL-8 gene.
Fig. 3.
(A) IL-8 ELISA analyses of clones selected from the 293TH8 RAGE library. Clones 482.78 and 482.79 were determined to express RAGE-activated Act1 and XEDAR, respectively. Clone 495.74 was found to express an unknown RAGE-activated gene, now designated TAB3. Cell line 293TH8 is the parental cell line. For a positive control, 293(EBNA)-IL1 cells were used. (B) IL-8 expression from clone 495.74 and 495.74 cells manipulated to express Crerecombinase (+Cre) and selected in Gcv-medium.
To confirm that the RAGE activation of MCG45404 was responsible for the observed cellular phenotype, we exploited the fact that the RAGE vector pFG114 was flanked by lox sites (Fig. 1B), thereby allowing it to be excised from the genome by site-specific recombination. Accordingly, clone 495.74 was transfected with a Cre recombinase expression vector, and clones that excised the RAGE vector were analyzed for phenotypic reversion. As expected, vector excised clones became Hyg-sensitive and no longer expressed endogenous IL-8 (Fig. 3B). Taken together, these data demonstrate that RAGE-activated MCG45404 was indeed responsible for activation of the IL-8 gene.
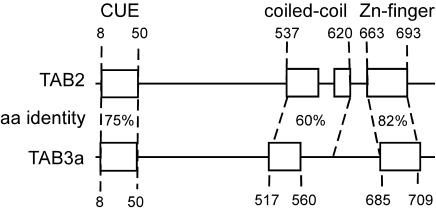
Bioinformatic and Expression Analyses. Having established a causal link between MCG45404 overexpression and IL-8 activation, we then carried out sequence analysis of the isolated RAGE-activated transcripts to gain additional insight into the structure and function of this gene. MCG45404 maps to human chromosome X and, consistent with a role in NF-κB signaling, was found to be 67% identical at the amino acid level to Xenopus laevis TAB3 (gi:27228108). Xenopus TAB3 has previously been shown to interact with TAK1 (24), suggesting the RAGE-activated protein may have a similar interaction. Furthermore, we have also identified three regions of MCG45404 that have coincident organization and high sequence identity with putative functional domains of TAK1-binding protein 2 (TAB2) (25), a known regulator of NF-κB activation (Fig. 4). Amino acids at the N terminus of MCG45404 and TAB2 share 75% identity and conform to a CUE domain, a sequence demonstrated to direct self-monoubiquitylation (26). Coiled-coil regions sharing 60% identity are located toward the C terminus of each protein, and sequences of 82% identity that conform to Zn-finger structures are positioned at the C termini of each protein. Coiled-coil structures facilitate protein–protein interactions and are required for the interaction of TAB2 with TAK1 (25). Based on these homologies and apparent common function, we have designated MCG45404 and the gene expressed in RAGE clone 495.74 as the human ortholog of TAB3.
Fig. 4.
Regions of high sequence identity between human proteins TAB3 and TAB2.
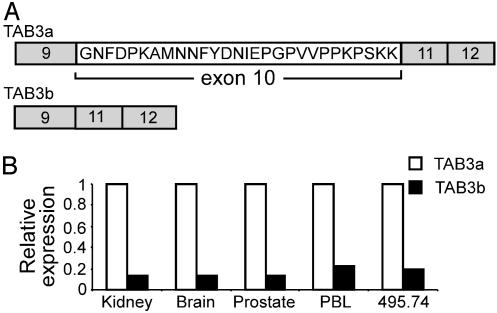
Further analyses of human TAB3 revealed that it is minimally comprised of 12 exons, with the 712-aa ORF beginning in what we have designated exon 6 and terminating in exon 12. Because RAGE vectors integrated upstream of endogenous genes typically produce a transcript in which the activation exon replaces the endogenous exon 1 through RNA splicing, an uncharacterized upstream exon may exist that contributes to the 5′ UTR of native TAB3 transcripts. For clarity in describing the results of this study, we refer to the first exon that the RAGE activation exon spliced onto as “exon 1.” Analyses of 3′ RACE sequences revealed TAB3 to express a splice-variant transcript that excludes exon 10 to eliminate 28 internal amino acids, but otherwise maintains the correct reading frame (Fig. 5A). PCR analyses confirmed that both the full-length and splice-variant transcript are naturally coexpressed, and that the short splice variant form comprises only 13–20% of the total TAB3 transcripts expressed in the cell types surveyed (Fig. 5B). The full-length 712-aa protein was designated TAB3a (GenBank accession no. AY331591), and the 684-aa protein encoded by the splice-variant transcript lacking exon 10 was designated TAB3b (GenBank accession no. AY331592). It is noteworthy that the 28 amino acids encoded within exon 10 of TAB3a reside between the coiled-coil and Zn-finger homology domains of TAB2 and TAB3 and does not alter the sequences of these conserved domains.
Fig. 5.
(A) RAGE clone 495.74 expresses splice-variant transcripts that encode 712-aa TAB3a and 684-aa TAB3b isoforms. (B) The expression level of splice-variant TAB3b transcripts normalized to the level of TAB3a transcripts expressed within each of four different tissues and RAGE clone 495.74. TAB3b is the minor form of TAB3 transcripts expressed in each cell type examined.
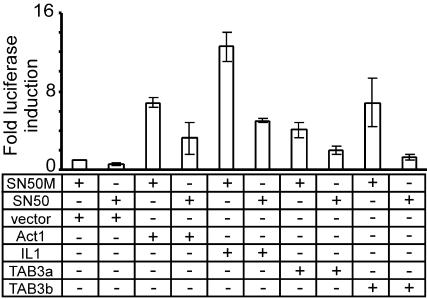
Transgene Expression and EMSA Analyses. The homology between human TAB3 and TAB2, as well as the observed ability of TAB3 to activate expression of an NF-κB-inducible IL8 gene, suggests that TAB3 is a constituent of the NF-κB pathway. To investigate this hypothesis, TAB3a and TAB3b cDNA expression vectors were introduced into HEK293 cells containing the IL-8 promoter linked to the luciferase gene, and were tested for their ability to activate IL8 promoter activity in the presence or absence of SN50, a specific inhibitor of NF-κB nuclear translocation (27). As expected, TAB3a and TAB3b induced a robust up-regulation of IL8 promoter activity, and in both cases, this effect was blunted in the presence of SN50 (Fig. 6). Thus, TAB3 isoforms function to induce the IL-8 promoter through a cellular pathway involving NF-κB activation.
Fig. 6.
The effect of NF-κB inhibitor SN50 on induced IL-8 promoter activity. TAB3a, TAB3b, Act1, and IL-1α were expressed in an IL8 luciferase cell line. Positive controls are Act1- and IL-1α-expressing cells. SN50M is an inactive control peptide.
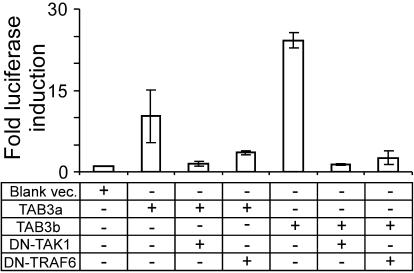
Based on the structural similarities between TAB3 and TAB2, as well as the known association between Xenopus TAB3 and TAK1 (25), we hypothesize that human TAB3 isoforms interact with TAK1. Results from dominant-negative coexpression studies were consistent with this hypothesis (Fig. 7). The coexpression of TAB3a and TAB3b with DN-TAK1 or DN-TRAF6 (28, 29) demonstrated that each of these DN proteins effectively blocked TAB3-induced expression of the IL-8 promoter. Thus, human TAB3 interacts with either TAK1 or TRAF6, or at some upstream regulatory point within the signal transduction pathway.
Fig. 7.
The effect on IL-8 promoter activity of coexpressing TAB3a or TAB3b with DN forms of TAK1 or TRAF6. Cotransfected cDNAs were expressed in IL8 luciferase cells.
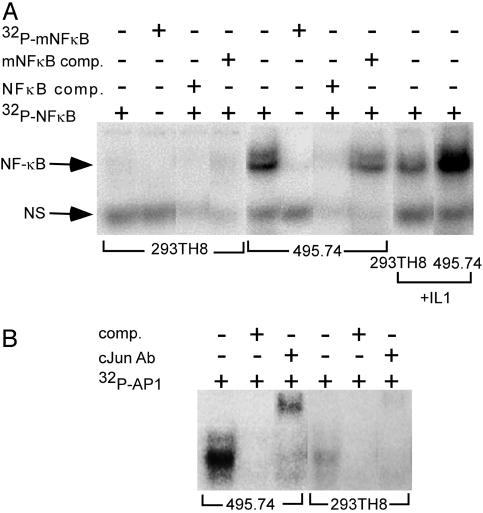
Transgene expression data provide strong evidence that human TAB3 isoforms function as regulators of NF-κB activation. If true, constitutive RAGE-activated expression of TAB3 in library clone 495.74 would be expected to confer persistent cellular activation of NF-κB. Nuclear proteins prepared from clone 495.74 cells were tested by EMSA assay and found to exhibit specific, high-affinity NF-κB probe-binding activity, whereas nuclear proteins isolated from the host cell line failed to show this activity (Fig. 8A). Treating the binding reactions with anti-p65 antibody produced prominent supershifted bands (data not shown), confirming that the bound nuclear proteins were NF-κB transcription factors. Nuclear extracts prepared from IL-1α-treated 495.74 cells showed enhanced binding activity to the NF-κB probe compared with 495.74 cells alone or IL-1α-treated 293TH8 cells, indicting an additive effect of TAB3 with at least one other NF-κB activator. Because some cellular triggers cause coordinated activation of both NF-κB and AP-1 (30), EMSAs were conducted to determine whether RAGE-activated TAB3 also caused activation of the JNK signal transduction pathway (Fig. 8B). EMSA demonstrated that the overexpression of human TAB3 does, indeed, activate AP-1 transcription factors. Like NF-κB, AP-1 transcription factors regulate the expression of a wide variety of genes involved in mediating cell differentiation, proliferation, and death (31). Therefore, proteins that are positive regulators of both NF-κB and AP-1 activation may be valuable targets for drug discovery.
Fig. 8.
EMSA analyses. (A) Nuclear extracts were prepared from 293TH8 and clone 495.74 cells with or without IL-1α treatment. Nuclear proteins were assayed for binding specificity to [32P]NF-κB consensus oligo with or without nonlabeled NF-κB competitor oligo, and with or without 32P-labeled and nonlabeled mutated (m)NF-κB oligo. (B) Nuclear extracts of 293TH8 cells or 495.74 cells were assayed for activity in binding [32P]AP-1 consensus oligo with or without anti-cJun antibody.
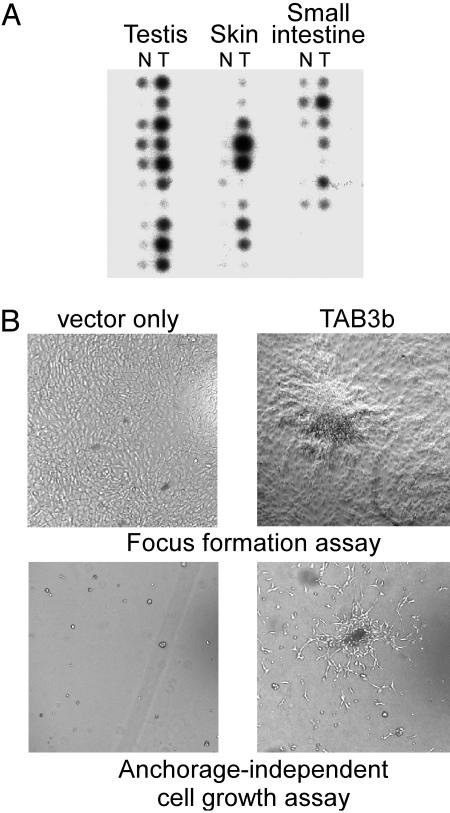
Disease Correlation. Dysfunctional regulation of NF-κB-responsive genes is involved in two broad areas of disease: those leading to aberrant immune system function, and those leading to cancer (10, 32–35). Therefore, to determine whether there is a link between either of these disease areas and the expression of human TAB3, dot-blot hybridization analyses were performed by hybridizing TAB3 cDNA probe against human autoimmune disease and cancer expression profiling arrays. The autoimmune array revealed that TAB3 expression was uniformly low, and did not exhibit significant differences between normal and diseased tissue samples (data not shown). Furthermore, 293TH8, 293-EBNA, and peripheral blood lymphocyte cells were treated with tumor necrosis factor α, IL-1, LPS, double-stranded RNA, and transforming growth factor β, and their RNAs were analyzed. None of these cell treatments altered TAB3 expression (data not shown). Thus, no association was found between altered TAB3 expression and autoimmune disease or cells treated with immune response triggers. In contrast, however, cancer expression profile analyses demonstrated that TAB3 is markedly elevated in a variety of cancer types (Fig. 9A). TAB3 is expressed at high levels in various skin, testis, and small intestine cancers, as compared with its expression in their respective normal tissues. Numerous studies establish a correlation between cancer phenotypes and the aberrant constitutive activation of NF-κB (36, 37), suggesting these findings have physiological significance.
Fig. 9.
(A)[32P]TAB3 probe hybridization of dot-blotted cDNAs derived from normal (N) and tumor (T) cells of testis, skin, and small intestine. Positive control hybridization by using [32P]ubiquitin probe confirmed uniform hybridization signal between all samples (data not shown). (B) Solid surface and soft agar culture assays of TAB3b-expressing NIH 3T3 cells.
To determine whether elevated human TAB3 expression is sufficient to induce cellular transformation, NIH 3T3 cells were transfected to stably express TAB3a and TAB3b, and were then evaluated for acquired anchorage-independent growth. Cells transfected with the blank vector continued to exhibit contact-inhibited growth; however, NIH 3T3 cells expressing TAB3a or TAB3b demonstrated the ability to form discreet, dense foci (Fig. 9B). In a similar assay, NIH 3T3 cells expressing TAB3b were found to flourish when cultured on soft agar medium, whereas NIH 3T3 cells expressing TAB3a (or the blank expression vector) consistently died in soft agar culture (Fig. 9B). This finding raises the interesting possibility that TAB3a and 3b have different activities or potencies in regulating NF-κB activation. Of possible significance, the assay methods used in this study consistently found TAB3b to provide 2-fold greater activity in activating NF-κB as compared with TAB3a (e.g., Figs. 6 and 7). Further investigations are needed to fully characterize any activity differences, and their significance, between TAB3 isoforms. Nonetheless, our finding that TAB3 is constitutively overexpressed in certain tumor tissues, and that TAB3b overexpression is sufficient to transform NIH 3T3 cells, compel us to hypothesize that the overexpression of human TAB3b may cause it to function as an oncogenic factor.
Discussion
Proteins encoded by NF-κB-dependent genes function in the cellular processes of immune responses, stress responses, apoptosis, and proliferation and/or differentiation. Consequently, stringent regulation of these genes is paramount to maintaining normal cellular function, whereas dysfunctional regulation of NF-κB often leads to disease. Because of this result, proteins that function as regulators of NF-κB activation may provide premier targets for the development of drugs that provide therapeutic intervention in cancer and immune-related diseases. To facilitate the discovery of NF-κB regulators, we have devised a function-to-gene discovery strategy that combines the RAGE technology with a robust NF-κB reporter cell line. From our initial attempt of this approach, we isolated two RAGE clones expressing known activators of NF-κB, and a third clone expressing an uncharacterized putative regulator of NF-κB. The protein encoded by the RAGE-activated gene shares 67% amino acid identity with Xenopus TAB3, a protein that binds TAK1 (25). Because of their highly conserved sequences and respective associations with signal transduction pathways involving NF-κB, we have designated this RAGE-activated protein as the human ortholog of TAB3. It should be noted that, during the final review of this manuscript, Ishitani et al. (38) reported the characterization of this same human gene, and similarly designated it TAB3. In this report, complementary experimental evidence is presented that further establishes the function of TAB3 as an activator of NF-κB.
An attribute of the RAGE technology is the ability to express naturally occurring splice-variant transcripts. Consequently, several splice-variant forms of human TAB3 were expressed from the RAGE-activated gene, which were then cloned and characterized. Most notable is a transcript variant that excludes exon 10, and translates a protein isoform lacking 28 internal amino acids. The protein encoded by the full-length transcript has been designated by us as TAB3a, and is the same as recently described by Ishitani et al. (38). The smaller protein isoform has been designated TAB3b. TAB3b is found to be the minor transcript expressed in all of the cell lines and tissues types examined thus far, although the physiological significance of these findings remains to be determined.
The specificity of TAB3 in regulating NF-κB activation was demonstrated through transgene expression studies by using NF-κB responsive reporters in combination with NF-κB inhibitors and expressed DN mutants of TRAF6 and TAK1. These studies, in combination with EMSA analyses, definitively established that the overexpression of TAB3 is sufficient to induce and maintain constitutive cellular activation of NF-κB. However, additional studies are required to fully elucidate the molecular mechanism of TAB3 function. Nonetheless, our data provide hints as to a possible role for TAB3. Comparative bioinformatic analyses revealed that TAB3 is similar to TAB2 in that they both contain three putative functional domains with the same relative organization. In addition, the overexpression of TAB2 likewise causes activation of NF-κB and AP-1 transcription factors (39, 40). Such striking similarities lead us to hypothesize that TAB2 and TAB3 play similar roles in mediating signal transduction pathways involving NF-κB activation. TAB2 is a binding partner of TAK1, and is believed to function as an adapter molecule linking TAK1 and TRAF6 to mediate NF-κB and JNK activation in the IL-1 pathway (25), and it is reasonable that TAB3 may have similar binding partners and function.
Many studies establish a link between the dysfunctional regulation of NF-κB and cancer, and provide evidence that constitutive activation of NF-κB plays an important role in tumorigenesis (33, 41–43). Our study supports this hypothesis. We demonstrated that TAB3 overexpression causes constitutive activation of both NF-κB and JNK signal transduction pathways. Further, the expression of TAB3b in NIH 3T3 cells is sufficient to induce transformation and confer cellular proliferation on soft agar medium, indicating that its overexpression may increase the potential for metastases of transformed cells. Indeed, TAB3 expression is found to be significantly elevated in the tumors of skin, testis, and small intestine, but is expressed at very low levels in normal samples of these same tissues. Taken together, these results establish a strong correlation between elevated TAB3 expression and tumorigenesis. Further studies are required to fully characterize the interactions of TAB3 isoforms in regulating signal transduction pathways, to elucidate their function in disease processes, and to critically assess their potential as targets for drug development.
Acknowledgments
We thank Kurt Brunden and David Jackson for their valuable input.
Abbreviations: RAGE, random activation of gene expression; Hyg, hygromycin; EMSA, electrophoretic mobility-shift assay; TAK1, transforming growth factor β-activated kinase 1; TAB2, TAK1-binding protein 2; TRAF6, tumor necrosis factor α receptor-associated factor 6; TAB3, TAK1-binding protein 3; DN, dominant-negative.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY331591 and AY331592).
References
- 1.Baldwin, A. S., Jr. (1996) Annu. Rev. Immunol. 14, 649–683. [DOI] [PubMed] [Google Scholar]
- 2.Miyamoto, S. & Verma, I. M. (1995) Adv. Cancer Res. 66, 255–292. [PubMed] [Google Scholar]
- 3.Ghosh, S., May, M. J. & Kopp, E. B. (1998) Annu. Rev. Immunol. 16, 225–260. [DOI] [PubMed] [Google Scholar]
- 4.Chen, F. E. & Ghosh, G. (1999) Oncogene 18, 6845–6852. [DOI] [PubMed] [Google Scholar]
- 5.Huang, T. T., Kudo, N., Yoshida, M. & Miyamoto, S. (2000) Proc. Natl. Acad. Sci. USA 97, 1014–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixit, V. & Mak, T. W. (2002) Cell 111, 615–619. [DOI] [PubMed] [Google Scholar]
- 7.Karin, M. (1999) Oncogene 18, 6867–6874. [DOI] [PubMed] [Google Scholar]
- 8.Vincenti, M. P. & Brinckerhoff, C. E. (2001) Arthritis Res. 3, 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinz, M., Lemke, P., Anagnostopoulos, I., Hacker, C., Krappmann, D., Mathas, S., Dorken, B., Zenke, M., Stein, H. & Scheidereit, C. (2002) J. Exp. Med. 196, 605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldwin, A. S., Jr. (2001) J. Clin. Invest. 107, 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darnell, J. E., Jr. (2002) Nat. Rev. Cancer 2, 740–749. [DOI] [PubMed] [Google Scholar]
- 12.Hinz, M., Loser, P., Mathas, S., Krappmann, D., Dorken, B. & Scheidereit, C. (2001) Blood 97, 2798–2807. [DOI] [PubMed] [Google Scholar]
- 13.Marok, R., Winyard, P. G., Coumbe, A., Kus, M. L., Gaffney, K., Blades, S., Mapp, P. I., Morris, C. J., Blake, D. R., Kaltschmidt, C. & Baeuerle, P. A. (1996) Arthritis Rheum. 39, 583–591. [DOI] [PubMed] [Google Scholar]
- 14.Bolotin, D. & Fuchs, E. (2003) Nature 421, 594–595. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh, S. & Karin, M. (2002) Cell 109, Suppl., S81–S96. [DOI] [PubMed] [Google Scholar]
- 16.Harrington, J. J., Sherf, B., Rundlett, S., Jackson, P. D., Perry, R., Cain, S., Leventhal, C., Thornton, M., Ramachandran, R., Whittington, J., et al. (2001) Nat. Biotechnol. 19, 440–445. [DOI] [PubMed] [Google Scholar]
- 17.Guo, Z., Liu, Q. & Smith, L. M. (1997) Nat. Biotechnol. 15, 331–335. [DOI] [PubMed] [Google Scholar]
- 18.Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. (1983) Nucleic Acids Res. 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nourbakhsh, M., Kalble, S., Dorrie, A., Hauser, H., Resch, K. & Kracht, M. (2001) J. Biol. Chem. 276, 4501–4508. [DOI] [PubMed] [Google Scholar]
- 20.Li, X., Commane, M., Nie, H., Hua, X., Chatterjee-Kishore, M., Wald, D., Haag, M. & Stark, G. R. (2000) Proc. Natl. Acad. Sci. USA 97, 10489–10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leonardi, A., Chariot, A., Claudio, E., Cunningham, K. & Siebenlist, U. (2000) Proc. Natl. Acad. Sci. USA 97, 10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia, Y. F., Li, Y. D., Li, X. & Geng, J. G. (2002) Biochem. Biophys. Res. Commun. 296, 406–412. [DOI] [PubMed] [Google Scholar]
- 23.Yan, M., Wang, L. C., Hymowitz, S. G., Schilbach, S., Lee, J., Goddard, A., de Vos, A. M., Gao, W. Q. & Dixit, V. M. (2000) Science 290, 523–527. [DOI] [PubMed] [Google Scholar]
- 24.Munoz-Sanjuan, I., Bell, E., Altmann, C. R., Vonica, A. & Brivanlou, A. H. (2002) Development (Cambridge, U.K.) 129, 5529–5540. [DOI] [PubMed] [Google Scholar]
- 25.Takaesu, G., Kishida, S., Hiyama, A., Yamaguchi, K., Shibuya, H., Irie, K., Ninomiya-Tsuji, J. & Matsumoto, K. (2000) Mol. Cell 5, 649–658. [DOI] [PubMed] [Google Scholar]
- 26.Shih, S. C., Prag, G., Francis, S. A., Sutanto, M. A., Hurley, J. H. & Hicke, L. (2003) EMBO J. 22, 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altmeyer, A., Klampfer, L., Goodman, A. R. & Vilcek, J. (1995) J. Biol. Chem. 270, 25584–25590. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, Z., Johnson, H. J., Nie, H., Qin, J., Bird, T. A. & Li, X. (2003) J. Biol. Chem. 278, 10952–10956. [DOI] [PubMed] [Google Scholar]
- 29.Wald, D., Commane, M., Stark, G. R. & Li, X. (2001) Eur. J. Immunol. 31, 3747–3754. [DOI] [PubMed] [Google Scholar]
- 30.Dinarello, C. A. (1996) Blood 87, 2095–2147. [PubMed] [Google Scholar]
- 31.Shaulian, E. & Karin, M. (2002) Nat. Cell Biol. 4, E131–E136. [DOI] [PubMed] [Google Scholar]
- 32.Chen, F., Demers, L. M., Vallyathan, V., Lu, Y., Castranova, V. & Shi, X. (2000) Am. J. Physiol. 279, C709–C716. [DOI] [PubMed] [Google Scholar]
- 33.Bharti, A. C. & Aggarwal, B. B. (2002) Biochem. Pharmacol. 64, 883–888. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, G. & Ghosh, S. (2001) J. Clin. Invest. 107, 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tak, P. P. & Firestein, G. S. (2001) J. Clin. Invest. 107, 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang, Y., Cui, L., Yie, T. A., Rom, W. N., Cheng, H. & Tchou-Wong, K. M. (2001) Oncogene 20, 2254–2263. [DOI] [PubMed] [Google Scholar]
- 37.Rong, R., He, Q., Liu, Y., Sheikh, M. S. & Huang, Y. (2002) Oncogene 21, 1062–1070. [DOI] [PubMed] [Google Scholar]
- 38.Ishitani, T., Takaesu, G., Ninomiya-Tsuji, J., Shibuya, H., Gaynor, R. B. & Matsumoto, K. (2003) EMBO J. 22, 6277–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang, Z., Ninomiya-Tsuji, J., Qian, Y., Matsumoto, K. & Li, X. (2002) Mol. Cell. Biol. 22, 7158–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian, Y., Commane, M., Ninomiya-Tsuji, J., Matsumoto, K. & Li, X. (2001) J. Biol. Chem. 276, 41661–41667. [DOI] [PubMed] [Google Scholar]
- 41.Rayet, B. & Gelinas, C. (1999) Oncogene 18, 6938–6947. [DOI] [PubMed] [Google Scholar]
- 42.Baldwin, A. S. (2001) J. Clin. Invest. 107, 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karin, M., Cao, Y., Greten, F. R. & Li, Z. W. (2002) Nat. Rev. Cancer 2, 301–310. [DOI] [PubMed] [Google Scholar]