Abstract
Induction of a family of phase 2 genes encoding for proteins that protect against the damage of electrophiles and reactive oxygen intermediates is potentially a major strategy for reducing the risk of cancer and chronic degenerative diseases. Many phase 2 genes are regulated by upstream antioxidant response elements (ARE) that are targets of the leucine zipper transcription factor Nrf2. Under basal conditions, Nrf2 resides mainly in the cytoplasm bound to its cysteine-rich, Kelch domain-containing partner Keap1, which is itself anchored to the actin cytoskeleton and represses Nrf2 activity. Inducers disrupt the Keap1-Nrf2 complex by modifying two (C273 and C288) of the 25 cysteine residues of Keap1. The critical role of C273 and C288 was established by (i) their high reactivity when purified recombinant Keap1 was treated with dexamethasone mesylate and the dexamethasone-modified tryptic peptides were analyzed by mass spectrometry, and (ii) transfection of keap1 and nrf2 gene-deficient mouse embryonic fibroblasts with constructs expressing cysteine to alanine mutants of Keap1, and measurement of the ability of cotransfected Nrf2 to repress an ARE-luciferase reporter. Reaction of Keap1 with inducers results in formation of intermolecular disulfide bridges, probably between C273 of one Keap1 molecule and C288 of a second. Evidence for formation of such dimers was obtained by 2D PAGE of extracts of cells treated with inducers, and by the demonstration that whereas C273A and C288A mutants of Keap1 alone could not repress Nrf2 activation of the ARE-luciferase reporter, an equal mixture of these mutant constructs restored repressor activity.
This paper describes the molecular mechanisms that control expression of phase 2 genes, which play central roles in protecting aerobic life against the relentless stresses imposed by electrophiles and reactive oxygen intermediates: the principal causes of many chronic diseases, including cancer. Many phase 2 proteins are enzymes that are highly inducible by transcriptional activation, and exert versatile, long-acting, and often catalytic protection against electrophile and oxidative damage. Phase 2 proteins comprise not only the “classical” phase 2 xenobiotic-metabolizing enzymes such as glutathione transferases and UDP-glucuronosyltransferases, which conjugate xenobiotics with endogenous ligands, but include also NAD-(P)H:quinone reductase (NQO1) (EC 1.6.99.2), epoxide hydrolase, heme oxygenase 1, ferritin, γ-glutamylcysteine ligase, glutathione reductase, aldehyde dehydrogenase, dihydrodiol dehydrogenase, leukotriene B4 dehydrogenase, and glutathione S-conjugate efflux pumps (reviewed in refs. 1 and 2).
Much evidence supports the notion that induction of the phase 2 response is an efficient strategy for reducing the risk of a variety of diseases (for reviews, see refs. 2–4). For example: (i) mice in which the phase 2 response has been silenced (nrf2 gene knockouts) have low and uninducible phase 2 enzymes, are much more susceptible to carcinogens and the toxicity of oxygen and electrophiles and, unlike cognate wild-type mice, cannot be protected by inducers (2, 5–7). (ii) Targeted disruption of two phase 2 enzymes in mice (glutathione transferase π and NQO1) increased incidence of skin tumors evoked by polycyclic hydrocarbons (8, 9). (iii) Bioassays of phase 2 inducer potency, based on quantifying NQO1 activity in murine hepatoma cells, resulted in the isolation of several potent anticarcinogenic inducers, including sulforaphane from broccoli, and have also guided the synthesis of potent inducers (for review, see ref. 2). (iv) Human populations polymorphic for certain phase 2 enzymes are more susceptible to toxicity and carcinogenesis (10–13). (ν) Administration of the phase 2 inducer oltipraz to individuals at very high risk of developing primary hepatocellular carcinoma, because of heavy dietary intakes of aflatoxin B1, substantially increased excretion of phase 2 metabolites of aflatoxin, a biomarker for carcinogen load (14).
Inducers of phase 2 genes belong to nine structurally highly diversified chemical classes (15, 16), and share only a few common properties (17): (i) all are chemically reactive; (ii) nearly all are electrophiles; (iii) most are substrates for glutathione transferases; and (iv) all can modify sulfhydryl groups by alkylation, oxidation, or reduction. Recognition of these properties suggested that cells contain primary sensor(s) equipped with highly reactive cysteine residues that are recognized and chemically modified by inducers, thereby initiating the enhanced transcription of phase 2 genes. We have recently obtained evidence that Kelch-like ECH-associated protein 1 (Keap1) is probably this regulatory sensor (18), a conclusion that is strongly supported by the accompanying paper (19), and other very recent studies (20, 21).
Phase 2 genes are regulated by 5′ upstream regulatory sequences which have been designated as antioxidant response elements (ARE) (22, 23). Nrf2, a member of the NF-E2 family of nuclear basic leucine zipper transcription factors, binds to the ARE, and accelerates transcription of the cognate genes (24–26). Under basal conditions, Keap1, a recently identified protein associated with the actin cytoskeleton, binds very tightly to Nrf2, anchors this transcription factor in the cytoplasm, and targets it for ubiquitination and proteasome degradation, thereby repressing the ability of Nrf2 to induce phase 2 genes (27–32). Inducers disrupt the Keap1-Nrf2 complex, allowing Nrf2 to translocate to the nucleus where, in heterodimeric combinations with other basic leucine zipper proteins, it binds to AREs of phase 2 genes and accelerates their transcription.
The 624 amino acids of murine Keap1 include 25 cysteines and comprise five domains (Fig. 1). Because all cysteine residues are conserved, identification of those critical for induction presented special problems. Consequently, we resorted to the strategy of first identifying the most reactive cysteines by chemical modification with dexamethasone 21-mesylate (Dex-mes), an inducer that reacts irreversibly with thiols, and second, examining whether mutations of these residues alter the ability of Keap1 to repress Nrf2 function. Four cysteines [C257, C273, C288, and C297, all located in the intervening region (IVR) domain] of purified recombinant Keap1 were most reactive with Dex-mes (18), suggesting strongly that Keap1 is the molecular sensor for inducers. We now establish the critical functional role of these reactive cysteine thiols of Keap1 in signaling induction, by further analysis of the Dex-mes-modified tryptic peptides of Keap1, and by introducing systematic mutations of these cysteine residues.
Fig. 1.
Amino acid sequence of the five domains of Keap1: (i) N-terminal region (NTR, amino acids 1–60: two cysteines, blue). (ii) BTB (Broad complex, Tramtrack, Bric-a-Brac; amino acids 61–179: three cysteines, pink), an evolutionarily conserved protein–protein interaction motif that often dimerizes with other BTB domains (33). (iii) IVR (amino acids 180–314: eight cysteines, yellow). (iv) Double glycine (DGR, amino acids 315–598: nine cysteines, gray) comprising six Kelch motifs (amino acids 315–359, 361–410, 412–457, 459–504, 506–551, and 553–598). Repeated Kelch motifs give rise to a β-propeller structure with multiple protein-binding sites (34). The DGR of Keap1 binds tightly to the Neh2 segment (the 100 N-terminal amino acids) of Nrf2 (24, 27), and is also the region involved in anchoring Keap1 to the actin cytoskeleton (19). (v) C-terminal region (CTR, amino acids 599–624: three cysteines, green). The 25 cysteine residues are highlighted in bright yellow, the 39 arginine residues are blue, and the 17 lysine residues are red. The tryptic peptides labeled with dexamethasone (T-22, T-26, T-28, T-29, and T-55) are designated. The matrix-assisted laser desorption ionization/time-of-flight mass spectral analyses of all 57 tryptic peptides, including the 19 peptides that contain cysteine residues are recorded in Table 1 (see Supporting Text).
Experimental Procedures
Constructs. Each Cys to Ala mutant of Keap1 was produced by PCR and standard recombination techniques. Construct authenticity was confirmed by sequencing. pcDNA3 or pET vectors were used for mammalian or bacterial cell expression, respectively, as described (18). All primers and PCR conditions used for genotyping and for the preparation of the constructs used in this study are listed in Supporting Text, which is published as supporting information on the PNAS web site.
Production of keap1(–/–) and Combined keap1(–/–)::nrf2(–/–) Mouse Embryonic Fibroblasts. Fibroblasts were isolated from 13.5-day-old embryos of keap1(–/–) and keap1(–/–)::nrf2(–/–) mice and used to establish stable lines by standard procedures (35). Cells were maintained in Iscove's modified Eagle's medium, supplemented with 10% heat-inactivated FBS, at 37°C and 5% CO2.
Functional Reporter Assay. Transient reporter assays were performed by standard transfection methods using 2 × 105 keap(–/–)::nrf2(–/–) mouse embryonic fibroblast (MEF) cells (K0N0) in 60-mm diameter dishes. Each construct of plasmid [2 μg of reporter gene pNQO1AREluc, 2 μg of pRLTK-normalizing vector, 8 μg of pCMVmNrf2, and various amounts (see Fig. 4) of each Keap1 expression vector, with total DNA adjusted to 2 μg with pcDNA3] was introduced into K0N0 MEF cells by the calcium phosphate co-precipitation method. Two micrograms of pRLTK plasmid bearing the Renilla luciferase gene under the control of the HSV-tk promoter/enhancer were included in each transfection and used for normalization. Cells were harvested 24 h after transfection and luciferase activities were measured according to the manufacturer's instructions (Promega). Relative luciferase activity was obtained from six independent transfection experiments and is shown ±SEM in each figure.
Fig. 4.
Repression of the intensity of luciferase luminescence of ARE-luciferase in K0N0 mouse embryonic fibroblasts by wild-type Keap1 and its cysteine mutants. All cells were transfected with the ARE-luciferase (2 μg) and the Nrf2 (8 μg) constructs. (a) Repression of luminescence as a function of amount of wild-type Keap1 (black bars) and its reversal by exposure to 2 μM sulforaphane (gray bar). (b) Repression of luminescence by wild-type (WT) Keap1 (2 μg) and equivalent quantities of the following mutants: NTR (C23A and C38A); CTR (C613A, C622A, and C624A); IVR C226A–C297A (C226A, C241A, C249A, C257A, C273A, C288A, and C297A); IVR C226A–C249A (C226A, C241A, and C249A); and IVR C257A–C297A (C257A, C273A, C288A, and C297A). (c) Repressive activity of single or multiple cysteine mutants of Keap1 with 100 ng of each expression vector. The structures of the mutants are designated: + for cysteine, – for alanine. The structure of each wild-type and mutant Keap1 is shown below the bar indicating its repressor activity. Repressor activity is abrogated if C273 or C288, or both, are mutated to alanine.
Nonreducing-Reducing 2D SDS/PAGE. HEK293 cells (5 × 105 per 60-mm dish) were transfected with 20 μg of pEFmKeap1. After transfection (48 h), the medium was exchanged with serum-free medium containing 250 μM DTT, and cells were incubated for 3 h. After extensive washing with PBS (5×), cells were exposed to an inducer or vehicle and incubated in medium at 37°C, 5% CO2 for 6 h, and finally harvested with 150 μl of RIPA buffer (10 mM Tris·HCl, pH 7.5/1% Nonidet P-40/0.1% Nadeoxycholate/0.1% SDS/150 mM NaCl/1 mM EDTA). Samples were boiled with DTT-free SDS buffer, and 25 μg of protein were subjected to nonreducing disk-gel (2-mm diameter disk) SDS/PAGE. Gels were ejected and reduced with 1× SDS buffer including 5% 2-mercaptoethanol at room temperature for 1 h, with one buffer change. Disk gels were inserted into the well of a second SDS/PAGE and subjected to electrophoresis.
Immunoblotting. Keap1 was detected by using rabbit anti-mKeap1 polyclonal antibody (25, 28). For normalizing cell number, nuclear Lamin B was detected from the same extracts with goat anti-Lamin B antibody (Santa Cruz Biotechnology). For normalizing transfection efficiency, Bsd-GFP protein, encoded from the same plasmid as Keap1, was detected by goat anti-GFP antibody (Santa Cruz Biotechnology). Membranes were blocked and treated with primary antibody followed by reaction with the appropriate secondary antibodies conjugated to horseradish peroxidase (Zymed, Bio-Rad). Immune complexes were visualized with enhanced chemiluminescence (Amersham Pharmacia).
Results and Discussion
Identification of the Most Reactive Cysteine Residues of Keap1 and Their Importance for Inducer and Nrf2-Binding Activity. In further validation that Keap1 contains highly reactive cysteine thiols that sense inducers (18), we carried out additional experiments with Dex-mes, an inducer which alkylates thiols irreversibly, and increases their masses substantially (374.19 atomic mass units). Homogeneous recombinant Keap1 (600 pmol) was incubated with a modest excess of Dex-mes (6 nmol) at 25°C and pH 8.0, for 2 h. Excess Dex-mes was removed by gel filtration, and the denatured protein was treated with N-ethylmaleimide to alkylate unreacted thiols. Tryptic peptides were separated by reversedphase HPLC, fractions were analyzed by matrix-assisted laser desorption ionization/time-of-flight (MALDI-TOF) MS. The 57 tryptic peptides were designated T-1 to T-57. All of the predicted 19 cysteine-containing tryptic peptides were recovered in the fractions (either as single peptides or as incompletely cleaved di- or tripeptides). Their masses agreed within 0.5 atomic mass units with calculated values (see Table 1, which is published as supporting information on the PNAS web site). Peptides containing C257 (T-22), C273 (T-25 to T-28), C288 (T-28), C297 (T-29), and C613 (T-55) (see Fig. 1) were found to have mass increases corresponding to the covalent addition of the steroid. All other cysteine-containing peptides showed mass increases of 125.05 atomic mass units (for each cysteine residue), indicating alkylation by N-ethylmaleimide. Thus, in agreement with our earlier findings, C257, C273, C288, and C297, all located in the IVR domain, as well as C613, are the most reactive cysteine residues of native Keap1 in vitro (18).
The reproducible labeling of 4 specific cysteines of Keap1 prompted us to examine the properties of recombinant Keap1 in which these cysteine residues were replaced by alanine. The mutant protein (C257A–C297A) was overexpressed in E. coli and purified to homogeneity by our procedure used to purify wild-type Keap1 (18). Both wild-type and mutant proteins migrated identically on native and SDS/PAGE (molecular weight ≈ 65,000).
Comparison of rates of binding of [3H]Dex-mes and sulforaphane to purified wild-type and mutant (C257A–C297A) Keap1, measured by tritium incorporation and dithiocarbamate formation, respectively, revealed that the reaction rates of the mutant were ≈50% slower, confirming that the modified cysteines were indeed the most reactive in the native protein.
We next examined whether the four most reactive cysteine residues of Keap1 influenced binding to Nrf2. Incubation of Keap1 with the Neh2 (Keap1 binding) domain of Nrf2 under reducing conditions led to formation of a complex that can be detected by native PAGE (Fig. 2a). At a constant Keap1 concentration, the intensity of the band corresponding to the complex increases with increasing amounts of Neh2, reaching saturation at a ratio of Keap1 to Neh2 of ≈2:1. Both wild-type and mutant Keap1 can form complexes with Neh2, but the mutant protein has ≈50% lower affinity for Neh2 based on band density (Fig. 2b).
Fig. 2.
Comparison of binding of wild-type and mutant (C257A–C297A) Keap1 to the Neh2 domain of Nrf2. (a) Native gel electrophoresis showing complex between Keap1 (100 pmol) and the Neh2 domain of Nrf2 (10, 20, 40, and 80 pmol, lanes 4–7 and 8–11, respectively). Lane 1, Neh2; lane 2, wild-type Keap1; lane 3, mutant Keap1. Lanes 4–7 show progressively increasing quantities of complex between wild-type Keap1 and Neh2; lanes 8–11 show lower quantities of complex formation between mutant Keap1 and Neh2. Arrowhead, arrow, and asterisk show Keap1, Neh2, and Neh2-Keap1 or mutant Keap1 complex bands, respectively. (b) Densitometric quantification of the intensities of bands of complexes between Neh2 and wild-type Keap1 (black bars) and mutant Keap1 (hatched bars).
Two separate lines of evidence, rates of reaction with inducers and binding to the Neh2 domain, indicate that the mutant Keap1 in which four specific cysteine residues in the IVR (C257A–C297A) were mutated not only reacted markedly more slowly with inducers, but also displayed a parallel decrease in affinity for the Neh2 domain of Nrf2. This is powerful evidence for the importance of these cysteines of Keap1 for repression of Nrf2.
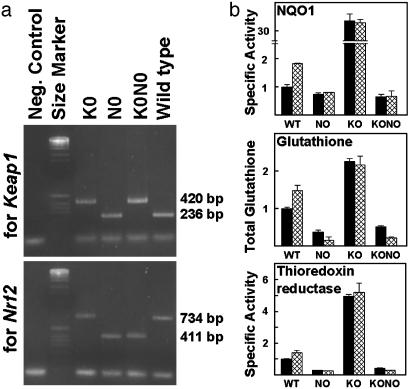
Preparation and Properties of Mouse Embryonic Fibroblasts in Which keap1, nrf2, or both Genes Have Been Disrupted. Further understanding of the regulatory functions of Keap1 and its mutants by transfection analyses of whole cells required cell lines in which interference from endogenous Keap1 and Nrf2 was eliminated. Consequently, we established cell lines from primary cultures of embryo fibroblasts of wild-type, nrf2-knockout (N0), keap1-knockout (K0), and K0N0 mice (36) (Fig. 3a). Wild-type cells have readily detectable basal levels of NQO1, glutathione, and thioredoxin reductase, which were elevated upon exposure to 1.5 μM sulforaphane (Fig. 3b). N0 and K0N0 cells have lower and essentially uninducible levels of these components. Moreover, the already lower levels of glutathione were further reduced by >50% upon treatment with sulforaphane, most likely because of direct conjugation with reduced glutathione (37). In sharp contrast, K0 cells have much higher basal levels compared to their wild-type counterparts, and essentially no induction was observed upon exposure to sulforaphane. These findings confirm the critical role of the Keap1/Nrf2 system for both basal and inducible expression of these phase 2 responses, and are consistent with the proposed repression of Nrf2 by Keap1.
Fig. 3.
Genotyping and levels of expression of phase 2 gene products in established lines of embryonic fibroblasts obtained from mice in which the keap1 (K0), nrf 2 (N0), or both genes (K0N0) were disrupted. (a) Electrophoresis of the PCR products derived from keap1 wild-type (236 bp) and mutant (420 bp) alleles, and nrf2 wild-type (734 bp) and mutant (411 bp) alleles. (b) Comparison (normalized to wild-type controls) of the specific activities of NQO1 and thioredoxin reductase, and concentration of glutathione in cell-free extracts of the three mutant and wild-type mouse embryonic fibroblasts. Black bars, cells untreated with inducers; hatched bars, cells exposed for 24 h to 1.5 μM sulforaphane.
Repression by Keap1 of ARE- and Nrf2-Dependent Gene Expression: Mutations of Cysteine Residues of Keap1. Effects of specific cysteine to alanine mutations on the ability of Keap1 to repress Nrf2 in vivo were examined by transient transfection experiments in mouse embryonic fibroblasts from keap1/nrf2 double knockout animals (K0N0). K0N0 cells were transiently transfected with controlled quantities of three vectors: (i) an Nrf2 expression vector (pCMVmNrf2); (ii) a vector that expresses either wild-type or mutants of Keap1; and (iii) a luciferase reporter vector controlled by the ARE of NQO1 (pNQO1AREluc) (38). The effect of mutating cysteine residues of Keap1 on the luciferase activity provided a measure of how such mutations affect repressor function.
Because Keap1 contains 25 cysteine residues, mutation of all possible combinations of cysteine residues was impractical, and we therefore mutated cysteines in selected domains. The BTB and DGR domains of Keap1 (see Fig. 1) are involved in dimerization (33) and Nrf-2/actin binding (19, 27), respectively; consequently, we did not mutate cysteine residues in these regions. The following groups of cysteine to alanine mutations were generated in Keap1: (i) N-terminal region (NTR), i.e., C23A and C38A, (ii) C-terminal region (CTR), i.e., C613A, C622A, and C624A, (iii) seven cysteines in the IVR, and (iv) 10 other combinations of mutations of these residues. Control cells were “mock” transfected with all constructs including the Keap1 expression vector, which lacked the cDNA sequence for Keap1.
In the absence of Keap1, the luciferase reporter of transfected cells showed high levels of luminescence (Fig. 4a). In agreement with previous studies (27), expression of increasing amounts of wild-type Keap1 in the presence of a constant amount of Nrf2 repressed the ARE-driven reporter luciferase activity in a concentration-dependent manner, and this repression was reversed by sulforaphane (Fig. 4a). Mutations of all cysteines in the N-terminal (NTR) or C-terminal (CTR) domains had no effect on the repressor activity of Keap1 (Fig. 4b). Thus, although C613 was labeled by Dex-mes, it is apparently not critical for the binding of Keap1 to Nrf2. In sharp contrast, mutants of Keap1 lacking either seven cysteine residues of the IVR (C226A–C297A), or four of these cysteines (C257A–C297A) cannot repress the expression of the luciferase reporter (Fig. 4b). This finding strongly suggests that the cysteine residues of Keap1 that are most reactive with an inducer are also those that influence binding and repression of Nrf2.
We next refined the mutation strategy to focus on the effects of mutating various combinations of the aforementioned seven cysteine residues of the IVR. Again mutation of both C226A–C257A and C297A did not disturb repressor activity, nor did mutation of C257A alone. The critical finding (Fig. 4c) is that absence of either C273 or C288 (or both) abrogates the repressor activity. Both of these cysteines react avidly with Dex-mes. C273 and C288 are flanked by basic amino acids (RC273H, and KC288E, respectively; see Fig. 1) which lower their pKa values markedly, and thereby increase their reactivity (39). These results are pleasingly consistent with two very recent studies: Zhang and Hannink (21) showed that C273 or C288 are required for Keap1-dependent ubiquitination of Nrf2, and Levonen et al. (20) demonstrated that reactive thiols of Keap1 are targets of electrophilic lipid oxidation products, and mutation of C273 or C288 to serine renders Keap1 unable to prevent Nrf2 nuclear translocation.
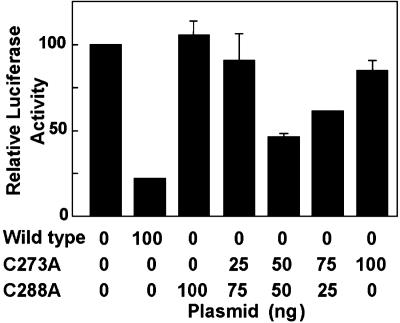
The question of how mutation of either C273 or C288 could lead to loss of repressor function was further illuminated by experiments in which mixtures of vectors expressing each of these mutants were transfected simultaneously into the reporter system (Fig. 5). The striking finding was that, whereas each Keap1 mutant alone lacked repressor activity, transfection of a mixture of equal quantities of both of these Keap1 mutant expression vectors led to substantial restoration of repressor activity. This observation supports the view that Keap1 acts as a dimer and suggests that the simultaneous presence of C273 on one monomer and C288 on the other is compatible with repressor activity.
Fig. 5.
Repression of the intensity of luciferase luminescence of ARE-luciferase in K0N0 mouse embryo fibroblasts by wild-type Keap1 and its C273A and C288A mutants. All cells were transfected with the ARE-luciferase (2 μg) and the Nrf2 (8 μg) constructs. The luminescence observed in the absence of Keap1 is expressed as 100 units. Addition of 100 ng of wild-type Keap1 plasmid reduced the luminescence to ≈20%. The plasmids (100 ng) coding for C273A or C288A showed little, if any, repression of the fluorescence, whereas mixtures of these plasmids restored the repressor activity. A mixture of 50 ng of each plasmid repressed ≈40% of the luminescence of the system.
Formation of Intermolecular Disulfide Bonds by Binding of Phase 2 Enzyme Inducers to Keap1 in Cells. Our previous observation that reaction of Keap1 with stoichiometric equivalents of dipyridyl disulfide led to formation of two equivalents of pyridine thione (18) strongly suggested that the initially formed mixed disulfide between reagent and protein was rapidly reduced by another highly reactive cysteine thiol of Keap1 to form a disulfide linkage. To determine the oligomerization state of Keap1 after inducer treatment, we first transfected human embryonic kidney (HEK) 293 cells with the Keap1 vector and then exposed the cells to inducers for 6 h in serum-free medium. Inducers of three different types were used, i.e., the Michael reaction acceptor bis(2-hydroxybenzylidene)acetone, the isothiocyanate sulforaphane, and 1,2-dithiole-3-thione. At the end of the exposure period, cell-free extracts were subjected to 2D SDS/PAGE (40) under nonreducing conditions in the first dimension and under reducing conditions (2-mercaptoethanol) in the second dimension. Western blot analysis was then used to localize Keap1 (Fig. 6). Only a single immunoreactive product corresponding to monomeric Keap1 was detected in uninduced cells. In contrast, the anti-Keap1 antibody recognized two products in extracts of cells treated with inducers, corresponding to a monomer and an intermolecular disulfide-linked dimer of Keap1 (Fig. 6c). This observation provides strong, independent evidence that reaction with inducers leads to the formation of intermolecular dimers between Keap1 subunits.
Fig. 6.
Exposure to inducers causes formation of a disulfide-linked dimer of Keap1 in HEK 293 cells transfected with a construct encoding for Keap1 and GFP (normalization control). (a) Coomassie brilliant blue (CBB) staining of 2D SDS/PAGE of cell-free extracts. (b) Immunoblots of SDS/PAGE of control (lanes 1 and 2) and inducer-treated (lanes 3 and 4) cells showing (Top) reduced binding of the anti-Keap1 antibody for Keap1 in inducer-treated cells compared with control cells, (Middle) equal expression of GFP, and (Bottom) equal cell numbers as judged by the expression of Lamin B. (c) Immunoblots for Keap1 of 2D SDS/PAGE of extracts of control cells and cells exposed to inducers of three different chemical types. SF, sulforaphane; D3T, 1,2-dithiole-3-thione; 2-HBA, bis(2-hydroxybenzylidene)acetone.
Comparison of the Keap1/Nrf2 System with Other Genes Regulated by Oxidative Stress and Sensed by Cysteine Thiol Groups. Although the behavior of the Keap1-Nrf2 system is in many ways unique, it resembles regulation of other proteins in which cysteine residues are modified. The prokaryotic transcription factors OxyR and SoxR are activated by oxidation, resulting in formation of intramolecular disulfide bonds in response to hydrogen peroxide and superoxide, respectively (41–44). In response to oxidants, two intramolecular disulfide bonds are formed and Zn is released from the redox-sensitive chaperone Hsp33, leading to large conformational changes which increase its affinity for protein folding intermediates, thus protecting them from oxidative damage (45, 46). Regulation of protein function can also occur by disulfide bond reduction as exemplified by activation of integrins that control cell adhesion and migration (47).
To our knowledge, the Keap1-Nrf2 system may be unique in that it depends on chemical modification of specific cysteine thiols and that the modifying agents then appear to be displaced by intermolecular sulfhydryl disulfide interchange to lead to a covalent disulfide dimer of Keap1. This eukaryotic system resembles the RsrA-SigR system from Streptomyces coelicolor (48–50). Both Keap1-Nrf2 and RsrA-SigR are essential components of signal transduction pathways involved in protection against oxidative stress and electrophiles by up-regulating systems that detoxify damaging agents, the Phase 2 gene products in mammals and the thioredoxin operon in S. coelicolor. Under basal conditions, both Keap1 and RsrA are negative regulators of the transcription factors Nrf2 and SigR, respectively. Oxidative stress or exposure to inducers disrupts the complexes, allowing the transcription factors to activate gene expression via their respective enhancer elements. Deletion of the genes encoding for Keap1 or RsrA leads to constitutive activation of the transcription factors and overexpression of the genes that are under their control, even in the absence of any stimulus. Interestingly, the IVR of Keap1 is similar in size and cysteine content to the RsrA. Individual mutations of critical cysteines of Keap1 and RsrA render them unable to repress their partner transcription factors. In both cases, this finding was completely unexpected, because the wild-type repressors are active in the reduced state and the inability to form disulfide bond(s) by mutating the participating cysteine residues is expected to lock the repressor in its constitutively active form.
These two systems present a similar paradox: (i) the repressors are sensors of the signal and are therefore indispensable components in the signal transduction pathway leading to induction, but if they are absent (i.e., knocked out), the same inducible genes, instead of being uninducible, are constitutively upregulated; (ii) specific cysteines must be able to form disulfide bonds for repressor activity, but disulfide formation leads to release of repression, pointing out that these cysteines are not only important for inducer sensing, but also for direct interaction between the two partner (repressor/transcription factor) molecules. Their modification, for example, by disulfide bond formation, can lead to conformational changes that do not allow binding to occur. In RsrA, the critical cysteines are involved in Zn coordination that controls the thiol-disulfide reactivity of RsrA, the Zn is expelled during oxidation with concomitant disulfide bond formation that then causes major conformational changes and does not allow binding to SigR. It is conceivable that C273 and C288 from each monomer are involved in metal coordination that, in addition to the proximity of these cysteines to basic amino acid residues, can stabilize the negative charges on the thiolate anions, lower their pKa values, and explain their unusually high reactivity.
Summary. These results are consistent with the model shown in Fig. 7. Under basal (reducing) conditions, Keap1 exists as a dimer in which two monomers are bound to each other, possibly by hydrophobic interactions via their BTB domains. The cysteines C273 and C288 of the intervening region are in the reduced state. In this conformation Keap1 sequesters one molecule of Nrf2 between two DGR domains in the cytoplasm and ensures its rapid turnover by targeting to the proteasome. Upon exposure to inducers, the reactive C273 and C288 residues form intermolecular disulfide bonds, thus covalently linking two monomers of Keap1. The resulting conformation separates the DGR domains, liberates Nrf2, and allows its translocation to the nucleus and enhanced expression of phase 2 genes.
Fig. 7.
Mechanism of regulation of the phase 2 response. Nrf2 (black) is retained in the cytoplasm by interaction with two molecules of Keap1, which are dimerized through their BTB domains (pink) and anchored to the actin cytoskeleton via the Kelch or DGR region (gray propeller). Inducers of the phase 2 response interact with cysteine thiol groups in the intervening region (IVR, yellow) of Keap1, causing the formation of disulfide bonds (most likely between C273 of one monomer and C288 of the other). This results in conformational change that renders Keap1 unable to bind to Nrf2, which then translocates to the nucleus. The Nrf2 in heterodimeric combination with other transcription factors such as small Maf binds to the ARE regulatory region of phase 2 genes and enhances their transcription.
Supplementary Material
Acknowledgments
We thank Philip A. Cole, Jed W. Fahey, and James T. Stivers for much perceptive advice. Robert N. Cole provided consultation and advice on the matrix-assisted laser desorption ionization/time-of-flight mass spectrometry, Osamu Ohneda helped in establishing the cell lines from the knockout mice, and Pamela Talalay provided valuable editorial consultation. These studies were supported by grants from the National Cancer Institute, Department of Health and Human Services (Grant CA 94076), the American Institute for Cancer Research (Washington, DC), and the Brassica Foundation for Cancer Chemoprotection Research (Baltimore). The AB-Mass Spectrometry Facility at the Johns Hopkins School of Medicine is funded by National Center for Research Resources Shared-Instrumentation Grant 1S10-RR14702. These studies were also supported by generous gifts from the Lewis B. and Dorothy Cullman Foundation, the Barbara Lubin Goldsmith Foundation, and the McMullan Family Fund.
Abbreviations: ARE, antioxidant response element; NQO1, nicotinamide quinone oxidoreductase 1, NAD(P)H:quinone acceptor oxidoreductase 1; K0N0, nrf2/keap1 double knockout; Dex-mes, dexamethasone 21-mesylate; Keap1, Kelch-like ECH-associated protein 1; IVR, intervening region of Keap1; C257A, C257 replaced by alanine; C257A–C297A, all cysteine residues from C257 to C297 replaced by alanine.
References
- 1.Hayes, J. D. & McLellan, L. I. (1999) Free Radical Res. 31, 273–300. [DOI] [PubMed] [Google Scholar]
- 2.Talalay, P., Dinkova-Kostova, A. T. & Holtzclaw, W. D. (2003) Adv. Enzyme Regul. 43, 121–134. [DOI] [PubMed] [Google Scholar]
- 3.Talalay, P. (1999) Proc. Am. Philos. Soc. 143, 52–72. [Google Scholar]
- 4.Talalay, P. (2000) Biofactors 12, 5–11. [DOI] [PubMed] [Google Scholar]
- 5.Ramos-Gomez, M., Kwak, M. K., Dolan, P. M., Itoh, K., Yamamoto, M., Talalay, P. & Kensler, T. W. (2001) Proc. Natl. Acad. Sci. USA 98, 3410–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fahey, J. W., Haristoy, X., Dolan, P. M., Kensler, T. W., Scholtus, I., Stephenson, K. K., Talalay, P. & Lozniewski, A. (2002) Proc. Natl. Acad. Sci. USA 99, 7610–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, H. Y., Jedlicka, A. E., Reddy, S. P., Kensler, T. W., Yamamoto, M., Zhang, L. Y. & Kleeberger, S. R. (2002) Am. J. Respir. Cell Mol. Biol. 26, 175–182. [DOI] [PubMed] [Google Scholar]
- 8.Henderson, C. J., Smith, A. G., Ure, J., Brown, K., Bacon, E. J. & Wolf, C. R. (1998) Proc. Natl. Acad. Sci. USA 95, 5275–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long, D. J., Jr., Waikel, R. L., Wang, X. J., Perlaky, L., Roop, D. R. & Jaiswal, A. K. (2000) Cancer Res. 60, 5913–5915. [PubMed] [Google Scholar]
- 10.Clairmont, A., Sies, H., Ramachandran, S., Lear, J. T., Smith, A. G., Bowers, B., Jones, P. W., Fryer, A. A. & Strange, R. C. (1999) Carcinogenesis 20, 1235–1240. [DOI] [PubMed] [Google Scholar]
- 11.Lafuente, M. J., Casterad, X., Trias, M., Ascaso, C., Molina, R., Ballesta, A., Zheng, S., Wiencke, J. K. & Lafuente, A. (2000) Carcinogenesis 21, 1813–1819. [DOI] [PubMed] [Google Scholar]
- 12.Smith, M. T., Wang, Y., Skibola, C. F., Slater, D. J., Lo Nigro, L., Nowell, P. C., Lange, B. J. & Felix, C. A. (2002) Blood 100, 4590–4593. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, J., Schulz, W. A., Li, Y., Wang, R., Zotz, R., Wen, D., Siegel, D., Ross, D., Gabbert, H. E. & Sarbia, M. (2003) Carcinogenesis 24, 905–909. [DOI] [PubMed] [Google Scholar]
- 14.Kensler, T. W., Qian, G.-S., Chen, J.-G. & Groopman, J. D. (2003) Nat. Rev. Cancer 3, 321–329. [DOI] [PubMed] [Google Scholar]
- 15.Talalay, P., De Long, M. J. & Prochaska, H. J. (1988) Proc. Natl. Acad. Sci. USA 85, 8261–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prestera, T., Zhang, Y., Spencer, S. R., Wilczak, C. A. & Talalay, P. (1993) Adv. Enzyme Regul. 33, 281–296. [DOI] [PubMed] [Google Scholar]
- 17.Dinkova-Kostova, A. T., Massiah, M. A., Bozak, R. E., Hicks, R. J. & Talalay, P. (2001) Proc. Natl. Acad. Sci. USA 98, 3404–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinkova-Kostova, A. T., Holtzclaw, W. D., Cole, R. N., Itoh, K., Wakabayashi, N., Katoh, Y., Yamamoto, M. & Talalay, P. (2002) Proc. Natl. Acad. Sci. USA 99, 11908–11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang, M.-I., Kobayashi, A., Wakabayashi, N., Kim, S.-G. & Yamamoto, M. (2004) Proc. Natl. Acad. Sci. USA 101, 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levonen, A.-L., Landar, A., Ramachandran, A., Ceasar, E. K., Dickinson, D. A., Zanoni, G., Morrow, J. D. & Darley-Usmar, V. M. (2003) Biochem. J., in press. [DOI] [PMC free article] [PubMed]
- 21.Zhang, D. D. & Hannik, M. (2003) Mol. Cell. Biol. 23, 8137–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes, J. D. & McMahon, M. (2001) Cancer Lett. 174, 103–113. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen, T., Sherratt, P. J. & Pickett, C. B. (2003) Annu. Rev. Pharmocol. Toxicol. 43, 233–260. [DOI] [PubMed] [Google Scholar]
- 24.Chui, D. H., Tang, W. & Orkin, S. H. (1995) Biochem. Biophys. Res. Commun. 209, 40–46. [DOI] [PubMed] [Google Scholar]
- 25.Itoh, K., Chiba, T., Takahashi, S., Ishii, T., Igarashi, K., Katoh, Y., Oyake, T., Hayashi, N., Satoh, K., Hatayama, I., et al. (1997) Biochem. Biophys. Res. Commun. 236, 313–322. [DOI] [PubMed] [Google Scholar]
- 26.Venugopal, R. & Jaiswal, A. K. (1996) Proc. Natl. Acad. Sci. USA 93, 14960–14965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itoh, K., Wakabayashi, N., Katoh, Y., Ishii, T., Igarashi, K., Engel, J. D. & Yamamoto, M. (1999) Genes Dev. 13, 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itoh, K., Wakabayashi, N., Katoh, Y., Ishii, T., O'Connor, T. & Yamamoto, M. (2003) Genes Cells. 8, 379–391. [DOI] [PubMed] [Google Scholar]
- 29.McMahon, M., Itoh, K., Yamamoto, M. & Hayes, J. D. (2003) J. Biol. Chem. 278, 21592–21600. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen, T., Sherratt, P. J., Huang, H. C., Yang, C. S. & Pickett, C. B. (2003) J. Biol. Chem. 278, 4536–4541. [DOI] [PubMed] [Google Scholar]
- 31.Sekhar, K. R., Yan, X. X. & Freeman, M. L. (2002) Oncogene 21, 6829–6834. [DOI] [PubMed] [Google Scholar]
- 32.Stewart, D., Killeen, E., Naquin, R., Alam, S. & Alam, J. (2003) J. Biol. Chem. 278, 2396–2402. [DOI] [PubMed] [Google Scholar]
- 33.Zipper, L. M. & Mulcahy, R. T. (2002) J. Biol. Chem. 277, 36544–36552. [DOI] [PubMed] [Google Scholar]
- 34.Prag, S. & Adams, J. C. (2003) BMC Bioinformatics 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hogan, B., Constantini, F. & Lacy, Y. (1986) Manupulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 36.Wakabayashi, N., Itoh, K., Wakabayashi, J., Motohashi, H., Noda, S., Takahashi, S., Imakado, S., Kotsuji, T., Otsuka, F., Roop, D. R., et al. (2003) Nat. Genet. 35, 238–245. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, Y. & Callaway, E. C. (2002) Biochem J. 364, 301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Favreau, L. V.& Pickett, C. B. (1995) J. Biol. Chem. 270, 24468–24474. [DOI] [PubMed] [Google Scholar]
- 39.Snyder, G. H., Cennerazzo, M. J., Karalis, A. J. & Field, D. (1981) Biochemistry 20, 6509–6519. [DOI] [PubMed] [Google Scholar]
- 40.Hynes, R. O. & Destree, A. (1977) Proc. Natl. Acad. Sci. USA 74, 2855–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng, M., Aslund, F. & Storz, G. (1988) Science 279, 1718–1721. [DOI] [PubMed] [Google Scholar]
- 42.Zheng, M. & Storz, G. (2000) Biochem. Pharmacol. 59, 1–6. [DOI] [PubMed] [Google Scholar]
- 43.Sun, Y. & Oberley, L. W. (1996) Free Radical Biol. Med. 21, 335–348. [DOI] [PubMed] [Google Scholar]
- 44.Georgiou, G. (2002) Cell 111, 607–610. [DOI] [PubMed] [Google Scholar]
- 45.Graumann, J., Lilie, H., Tang, X., Tucker, K. A., Hoffmann, J. H., Vijayalakshmi, J., Saper, M., Bardwell, J. C. & Jakob, U. (2001) Structure (Cambridge, Mass.) 9, 377–387. [DOI] [PubMed] [Google Scholar]
- 46.Linke, K. & Jakob, U. (2003) Antioxid. Redox Signal. 5, 425–434. [DOI] [PubMed] [Google Scholar]
- 47.Yan, B. & Smith, S. W. (2001) Biochemistry 40, 8861–8867. [DOI] [PubMed] [Google Scholar]
- 48.Kang, J.-G., Paget, M. S. B., Seok, Y.-J., Hahn, M.-Y., Bae, J.-B., Hahn, J.-S., Kleanthous, C., Buttner, M. J. & Roe, J.-H. (1999) EMBO J. 18, 4292–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paget, M. S., Bae, J. B., Hahn, M. Y., Li, W., Kleanthous, C., Roe, J. H. & Buttner, M. J. (2001) Mol. Microbiol. 39, 1036–1047. [DOI] [PubMed] [Google Scholar]
- 50.Li, W., Bottrill, A. R., Bibb, M. J., Buttner, M. J., Paget, M. S. & Kleanthous, C. (2003) J. Mol. Biol. 333, 461–472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.