Abstract
Mutations in the human DDB2 gene give rise to xeroderma pigmentosum group E, a disease characterized by increased skin tumorigenesis in response to UV-irradiation. Cell strains derived from xeroderma pigmentosum group E individuals also have enhanced resistance to UV-irradiation due to decreased p53-mediated apoptosis. To further address the precise function(s) of DDB2 and the consequence of non-naturally occurring DDB2 mutations, we generated mice with a disruption of the gene. The mice exhibited significantly enhanced skin carcinogenesis in response to UV-irradiation, and cells from the DDB2–/– mice were abnormally resistant to killing by the radiation and had diminished UV-induced, p53-mediated apoptosis. Notably, the cancer-prone phenotype and the resistance to cellular killing were not observed after exposure to the chemical carcinogen, 7,12-dimethylbenz[a]anthracene (DMBA), to which mice carrying defective nucleotide excision repair genes respond with enhanced tumors and cell killing. Although cells from heterozygous DDB2+/– mice appeared normal, these mice had enhanced skin carcinogenesis after UV-irradiation, so that XP-E heterozygotes might be at risk for carcinogenesis. In sum, these results demonstrate that DDB2 is well conserved between humans and mice and functions as a tumor suppressor, at least in part, by controlling p53-mediated apoptosis after UV-irradiation.
The disease xeroderma pigmentosum (XP) group E is one of eight subgroups of the cancer-prone syndrome, XP, and it has been associated with mutations in the DDB2 gene (1–4) that codes for the smaller subunit of the heterodimeric damage-specific DNA binding protein, DDB (5, 6). DDB strongly binds to DNA damages caused by UV light, including (6-4) photoproducts and trans,syn-cyclobutane pyrimidine dimers, yet it does not have a strong affinity for cis,syn-cyclobutane pyrimidine dimers, the most abundant UV photoproduct (5, 7–11).
The binding to DNA damages might suggest a direct role for DDB in nucleotide excision repair (NER) and DNA repair assays with rodent Chinese hamster ovary cell lines that lack DDB2 expression combined with ectopic overexpression of DDB2 cDNA led to the conclusion that DDB2 is a repair factor involved in global genomic repair, a subpathway of NER. However, the removal from DNA of many bulky lesions, including (6-4) photoproducts, occurs in the absence of DDB (8, 12, 13), and DDB2 is induced by UV-irradiation at a time by which NER has been completed. Moreover, the DDB2 promoter resembles that of a cell cycle-regulated gene (1, 14), and DDB2 is part of the COP9 signalosome complex (15, 16). Instead, DDB2 has been proposed to be a transcription transactivator through its stimulation of E2F1 (17), and we recently observed that DDB2 controls p53-mediated apoptosis after UV-irradiation of human diploid fibroblasts and that XP group E (XP-E) primary skin fibroblasts are abnormally resistant to killing by UV-irradiation (18). In sum, the functions of DDB are apparently varied and remain undefined.
The absence of DDB2 from Chinese hamster ovary cell lines led to the proposition that mouse models would not be appropriate for studying XP-E and related carcinogenesis (19, 20), although DDB activity for which both DDB1 and DDB2 proteins are required had been observed in mouse plasmacytoma cells (21). To elucidate any differences between mouse and human DDB2 function and to gain further insight into the role(s) of DDB2 and into the XP-E phenotype, we constructed mice with a disrupted DDB2 gene. Studies of these mice and their primary cells demonstrated that the mouse DDB2 gene is required for p53-mediated apoptosis and the suppression of skin carcinogenesis on exposure to UV-irradiation but, surprisingly, not to a chemical carcinogen.
Materials and Methods
Production of DDB2–/– Mice. DDB2 mouse genomic DNA was isolated from a mouse 129 genomic DNA bacterial artificial chromosome library (Incyte Pharmaceuticals, Palo Alto, CA). The targeting vector was constructed by replacing exons 4–7 (and thus also introns 4–6) of the mouse DDB2 gene with the PGK-neor gene and was then linearized and electroporated into 129/SVE embryonic stem (ES) cells. About 120 G418-resistant ES clones were screened by Southern blotting with a probe that hybridizes to a 6.5-kb XbaI restriction fragment in wild-type cells and an 8.0-kb XbaI restriction fragment in homologous recombinants. Three independent ES clones with homologous integration at the targeting site were injected into C57BL/6 blastocysts to generate chimeric mice. These chimeras were subsequently crossed with C57BL/6 females, and heterozygous mice (F1) with successful germ line transmission of the targeted allele were used to generate DDB2–/– mice (F2). For PCR genotyping, the mutant allele produces a 478-bp product with primers Pneo (5′-GCCTGCTTTGCCGAATATCATGGTGGAAAAT) and P7 (5′-ATTCTGAGATTGTAGGCTGTGTATGTGACC) and the wild-type allele produces a 258-bp product with primers P6 (5′-TTTGTCAGCTTGTTTTAGCCCAGATGGAGC) and P7.
Biochemical Analyses. RT-PCR analysis was performed as described (3, 18). Primers were 5′-GTTTAACCATCTCAATACCA and 5′-GTGTGAGGTGCTGTAACCA for DDB2, or 5′-GACAGAGGCAACTGAGCACC and 5′-CAAACGTCAAGACGGCCGTGTG for tubulin. Electrophoretic mobility-shift assays and Western blot analysis were as described (1, 3, 18), using 50% ammonium sulfate precipitate fractions (5). Antibodies against DDB2 (3, 18), BAX, p21CDKN1A, actin (Santa Cruz Biotechnology), MDM2 (SMP14, Santa Cruz Biotechnology; Ab-2, Oncogene Research Products), DDB1 (Zymed Laboratories), and p53 (1C12, Cell Signaling Technology) were used.
Mouse Fibroblasts and UV-Sensitivity Assays. MEFs (mouse embryonic fibroblasts) and neonatal fibroblasts were obtained from e13.5 embryos and p0 neonates (the day of birth), respectively, by a standard procedure. Cell-killing as measured by dye exclusion after UV-irradiation (18) or 7,12-dimethylbenz[a]anthracene (DMBA) exposure (22–24) and caspase-3 assays (18) were as described.
Tumorigenesis Studies. For UV-B treatment, shaved dorsal skin of 8-week-old mice was irradiated for 5 days per week at a dose of 2,500 J·m–2 per day by using a narrow-band UV-B TL-01 lamp (311 nm; Phillips). Individual mice were irradiated until either skin tumors appeared or for a maximum of 20 weeks. For treatment with DMBA, 10 μg of DMBA in 0.1 ml of acetone was applied to shaved dorsal skin once per week for 20 weeks. Mice were killed if profoundly ill or if external tumors exceeded 2 cm in diameter and these mice were scored as a death in the tumor incidence Kaplan–Meier analyses. Statistical significance was measured by using the log-rank test. For histology, skin tumors were fixed with 10% neutral-buffered formalin and stained with hematoxylin/eosin by conventional methods.
Results
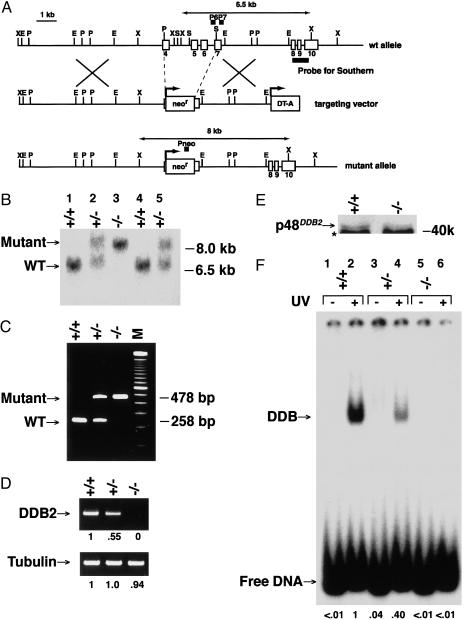
Consequences of Disruption of the Mouse DDB2 Gene. The mouse DDB2 gene was disrupted in ES cells by replacing exons 4–7 (and concomitantly introns 4–6) with the PGK-neor gene (Fig. 1A). Notably, exons 4–7 contain the known sites of mutations in human XP-E patients (1–4) and intron 4 contains a p53-mediated important regulatory element of human DDB2 (18). Transformed ES cells were screened for the replacement with Southern blot and PCR analyses, and the selected cells were then injected into BL/6 blastocysts to obtain chimeric mice. After crossing chimeras with BL/6 mice, progeny were screened by Southern blot (Fig. 1B) and PCR (Fig. 1C). The complete absence of DDB2 expression only from DDB2–/– mouse embryonic fibroblasts (MEFs) (e13.5), but not heterozygous DDB2+/– MEFs, was confirmed by RT-PCR (Fig. 1D), immunoblots (Fig. 1E), and activity assays (electrophoretic mobility-shift assays; Fig. 1F).
Fig. 1.
Targeted disruption of the mouse DDB2 gene. (A) Maps of the genomic locus, the targeting vector, and the targeted locus. (B) Southern blot analysis of XbaI-digested genomic MEF DNAs by using a genomic DNA probe from exons 8–10. (C) Multiplex PCR genotype analysis of MEF DNAs with a primer set for the mutant and wild-type alleles. (D) RT-PCR analysis of DDB2. PCR was performed for 40 (DDB2) or 27 (tubulin) cycles by using MEF RNAs. Relative band intensities are shown. (E) Western blot analysis for DDB2 of cell-free extracts (50% ammonium sulfate precipitates) from MEFs. Note that the p48DDB2 antibody made against human DDB2 only weakly detects mouse p48DDB2 (*, a nonspecific antigen). (F) Electrophoretic mobility-shift assays for DDB activity of MEF cell-free extracts (50% ammonium sulfate precipitates). Probe irradiated with 6,000 J·m–2 of UV-C or unirradiated probe were incubated with cell extracts and then analyzed. Relative band intensities are shown.
DDB2–/– mice were viable and fertile, and heterozygote crosses produced the expected Mendelian proportions among 488 offspring: 128 (26%) +/+, 236 (48%) +/–, and 125 (26%) –/–. Eight weeks after birth, DDB2–/– mice were somewhat smaller than their DDB2+/+ or DDB2+/– littermates {males, 24.49 ± 0.28 g (n = 51) [+/+], 24.13 ± 0.29 g (n = 82) [+/–], 22.28 ± 0.40 g (n = 49) [–/–]; females, 19.44 ± 0.26 g (n = 52) [+/+], 19.26 ± 0.24 g (n = 76) [+/–], 18.14 ± 0.24 g (n = 47) [–/–]}.
DDB2–/– MEFs Are Abnormally Resistant to Killing After UV-Irradiation but Not After DMBA Exposure. To determine the effect of the DDB2 disruption on susceptibility to killing by UV-irradiation or chemical carcinogens, MEFs (e13.5) were treated with UV-C (254 nm) or DMBA, a chemical carcinogen that forms bulky DNA adducts. As predicted from human XP-E diploid fibroblasts (18), MEFs from the DDB2–/– mice were more resistant than those from DDB2+/+ or DDB2+/– mice to killing by UV-irradiation as measured by dye exclusion (Fig. 2 A and B). By contrast, all three cell types showed equal killing by the bulky chemical carcinogen (Fig. 2 C and D). Colony-forming assays could not be used for the MEF studies because of the extremely low plating efficiency and clonogenic ability of MEFs (23–25). (In any case, colony-forming assays do not measure specifically early events in killing processes, notably apoptosis.)
Fig. 2.
Effects of UV-irradiation or DMBA treatment on cell viability and caspase-3 induction. (A and C) Relative viable cell number after 8 J·m–2 of UV-irradiation (A) or exposure to 20 μg/ml DMBA (C). Early-passage (1–3) MEFs were irradiated with UV-C (A) or pretreated with DMBA for3h(C), and then viable cell number was determined by dye exclusion in three independent experiments (n = 3). (B and D) Relative viable cell number after various doses of UV-irradiation (B) or DMBA (D). MEFs were irradiated as indicated with UV-C (B) or pretreated as indicated with DMBA (D), and then viable cell numbers were determined by dye exclusion 5 days after treatment (n = 3). (E) Caspase-3 activity after 8 J·m–2 of UV-irradiation (n = 2). The ordinate represents units of activity per 2 × 106 cells.
Nontransformed fibroblasts from XP-E patients are resistant to UV-irradiation because they have an abnormally low or no apoptotic response to the radiation (18). To clarify the effect of the mouse DDB2 gene disruption on p53-mediated apoptosis, we examined in MEFs the activity of caspase-3, an effector of the apoptosis pathway. Indeed, whereas the basal levels of caspase-3 were not significantly different between the DDB2–/– and DDB2+/+ MEFs, the induction of caspase-3 activity by UV-irradiation was greatly impaired in the DDB2–/– cells (Fig. 2E).
The amounts of several other proteins involved in p53-mediated responses to UV-irradiation were also examined in extracts from neonatal cells from the mouse strains (Fig. 3A). As determined by immunoblots, the basal and induced amounts of p53 and its downstream proteins, MDM2, BAX, and p21CDKN1A, were generally reduced in the DDB2–/– cells (Fig. 3A). (The BAX induction was underestimated in the DDB2+/+ cells, but not the DDB2–/– cells because of the absence of the lysed apoptotic cells from the DDB2+/+ sample.) Therefore, diminished p53-mediated apoptosis could explain the resistance of the mouse DDB2–/– cells to killing by UV-irradiation.
Fig. 3.
Effects of UV-irradiation on amounts of several p53-mediated UV response proteins and DDB activity. (A) Protein levels were estimated by immunoblotting after 8 J·m–2 of UV-irradiation of early-passage (1-5) p0 neonatal cells as described in Materials and Methods.(B) DDB activity after 8 J·m–2 of UV-irradiation of p0 neonatal cells. The same cell-free extracts were used in the experiments. Relative band intensities are shown.
As seen in Fig. 1E, mouse DDB2 protein is difficult to detect, because available antibodies to DDB2 are each directed against human DDB2 and poorly cross-react with mouse DDB2. Therefore, levels of DDB2 protein in the mouse cells were estimated from damaged DNA binding assays (Fig. 3B). [In human diploid fibroblasts, levels of DDB2 protein are limiting for the DDB heterodimer and paralleled damaged DNA binding activity (1, 3).] Consistent with the observations in normal human cells (1, 18), DDB activity decreased on irradiation and then reappeared after DNA repair was largely completed in the mouse DDB2+/+ cells (Fig. 3B). However, no DDB activity was detectable in DDB2–/– cells before or after UV-irradiation (Fig. 3B). As with human cells (18), mouse DDB1 protein amounts were not affected by UV-irradiation (Fig. 3A).
DDB2–/– and DDB2+/– Mice Have Dramatically Enhanced Skin Tumorigenesis After Exposure to UV Light but Not After Exposure to DMBA. To establish whether DDB2–/– mice are susceptible to developing skin cancers, mice with shaved back skin were treated with UV-B (311 nm). No significant changes in the skin (e.g., sunburn) were found in DDB2+/+, DDB2+/–, or DDB2–/– mice after 4 days of exposure to 2,500 J·m–2 per day. Mice were also irradiated with 2,500 J·m–2 of UV-B five times per week for 20 weeks or until tumors developed. DDB2–/– mice started to develop skin tumors 13 weeks after the start of treatment, whereas both DDB2+/+ and DDB2+/– mice first showed tumors after 19 weeks (Fig. 4A). Once started, however, tumor development proceeded at a much greater rate in the DDB2+/– mice than in the DDB2+/+ mice. There were also significant differences in the number of UV-induced tumors among the three genotypes (Fig. 4B). For example, there were 0.08, 0.3, or 13 tumors per mouse for DDB2+/+, DDB2+/–, or DDB2–/– mice, respectively, 25 weeks after the start of treatment.
Fig. 4.
Tumorigenesis analysis after chronic treatment with UV light or DMBA. (A and C) Kaplan–Meier curves of skin tumor-free mice after chronic treatment. (B and D) Tumor number per mouse after chronic treatment. (A–D) Solid bars indicate the exposure period for all animals; dotted bars indicate the exposure period for tumor-free animals only. (E and F) Histopathological examination of UV-B-induced skin tumors of DDB2–/– mice. (Original magnification: E, ×40; F, ×400.) The figure shows a typical example of a squamous cell carcinoma predominantly found in these mice. Arrows indicate a nest of atypical squamous cells and inflammatory cells including neutrophils (E) or mitotic figures (F). (G) Histopathological examination of DMBA-induced skin tumors of DDB2–/– mice. (Original magnification: ×20.) The figure shows a typical example of a papilloma predominantly found in this experiment.
When the shaved backs were exposed to two DMBA treatments of 10 μg per week, no significant changes in the skin were noted. After treatment of 10 μg of DMBA per week for 20 weeks, however, tumor development started after 19, 16, or 17 weeks for DDB2+/+, DDB2+/–, or DDB2–/– mice, respectively (Fig. 4C). Moreover, the pattern of the development was not significantly different among the three genotypes (Fig. 4C) and the DMBA treatment resulted in no significant difference in the number of tumors among the three genotypes (Fig. 4D).
All tumors appeared only on UV-B or DMBA exposed skin areas and none appeared internally. The tumors were predominantly squamous cell carcinomas after UV exposure (Fig. 4 E and F) or papillomas after DMBA exposure (Fig. 4G) among all three genotypes. [The papillomas would histopathologically be classified in humans as trichilemmal tumors (proliferating trichilemmal cysts or trichilemmal horns) rather than papillomas.]
Discussion
The near-normal development and fertility of DDB2–/– mice demonstrate that DDB2 does not normally have a major function in these processes. This is distinct from the major disruption of chromosome segregation in a DDB1 knockout strain of Schizosaccharomyces pombe (26) or of development in a DDB1 knockdown situation in Drosophila melanogaster (27). Although a DDB1 mutant is lacking for vertebrates, these distinctions support previous suggestions for individual roles for DDB1 and DDB2 aside from the heterodimeric damage binding (1, 18).
The apparent conservation of DDB2 in mice and the development of UV-induced tumors in DDB2–/– animals strongly support the validity of mouse models for studying XP-E and DDB2 function. Importantly, the DDB2–/– (XPE) mice did not exhibit any abnormality in DMBA-induced carcinogenesis, whereas mouse models that are defective in NER genes such as XPA (23, 28), XPD (29), or CSB (24) show abnormal skin cancer predisposition after either UV-irradiation or exposure to DMBA, presumably because these mice are defective in proteins directly involved in NER and cannot normally repair bulky DNA adducts in general. By contrast, TP53–/– mice do not increase initiation or promotion of DMBA-induced skin tumors (30), strongly supporting the proposal that DDB2 is involved in controlling p53 levels and/or its activity after UV-irradiation. We are currently crossing p53–/– and DDB2–/– mice to further study the DDB2–p53 interactions.
Does DDB2 affect responses to DNA damages brought about by agents other than UV-irradiation? The mismatch repair proteins hMLH1, hMSH2, hMSH6, and hPMS2 have been implicated in p53-dependent and p53-independent apoptosis induced by alkylating agents, suggesting that these have dual roles of DNA repair and apoptotic signaling (refs. 31–36; reviewed in ref. 37). Conversely, DDB2 mRNA appears to be up-regulated in response to γ-radiation (38). How mismatch repair proteins, DDB2, or other proteins might each regulate p53 in response to DNA damage are clearly questions that warrant intense study.
As a final note, the enhanced tumorigenesis after UV-irradiation of the DDB2+/– mice even at the subsunburn doses used also emphasizes not only the utility of the DDB2 mouse model but also the need to carefully assess susceptibilities to cancer of individuals heterozygous for XP-E or other genes involved in signaling or repair in response to DNA damages. Moreover, the apparent normality of DDB2–/– mice in development observed here and the late onset of tumors in XP-E individuals (2, 3) could indicate that XP-E is often not diagnosed and might be more common in the human population then has been appreciated. In this regard, we look forward to data from the Environmental Genome Project of the National Institute of Environmental Health Sciences (www.genome.utah.edu/genesnps) in which correlations of single-nucleotide polymorphisms of some 200 human genes including DDB2 are being matched against susceptibility to environmental diseases.
Acknowledgments
We thank S. Aizawa, M. Yasunami, and A. Winoto for technical advice and reagents, H. Asahara and H. Kishi for assistance in mouse genotyping, and Y. Itoh for encouragement. This work was supported in part by the Nakatomi Foundation, the Ministry of Education, Science, Sports, and Culture of Japan, and U.S. Public Health Service Grants P30ES08196 and R01GM59424.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DMBA, 7,12-dimethylbenz[a]anthracene; ES, embryonic stem; MEF, mouse embryonic fibroblast; NER, nucleotide excision repair; XP, xeroderma pigmentosum; XP-E, XP group E.
References
- 1.Nichols, A. F., Itoh, T., Graham, J. A., Liu, W., Yamaizumi, M. & Linn, S. (2000) J. Biol. Chem. 272, 21422–21428. [DOI] [PubMed] [Google Scholar]
- 2.Itoh, T., Linn, S., Ono, T. & Yamaizumi, M. (2000) J. Invest. Dermatol. 114, 1022–1029. [DOI] [PubMed] [Google Scholar]
- 3.Itoh, T., Nichols, A. & Linn, S. (2001) Oncogene 20, 7041–7050. [DOI] [PubMed] [Google Scholar]
- 4.Rapic-Otrin, V., Navazza, V., Nardo, T., Botta, E., McLenigan, M., Bisi, D. C., Levine, A. S. & Stefanini, M. (2003) Hum. Mol. Genet. 12, 1507–1522. [DOI] [PubMed] [Google Scholar]
- 5.Keeney, S., Chang, G. J. & Linn, S. (1993) J. Biol. Chem. 268, 21293–21300. [PubMed] [Google Scholar]
- 6.Dualan, R., Brody, T., Keeney, S., Nichols, A. F., Admon, A. & Linn, S. (1995) Genomics 29, 62–69. [DOI] [PubMed] [Google Scholar]
- 7.Reardon, J. T., Nichols, A. F., Keeney, S., Smith, C. A., Taylor, J.-S., Linn, S. & Sancar, A. (1993) J. Biol. Chem. 268, 21301–21308. [PubMed] [Google Scholar]
- 8.Reardon, J. T. & Sancar, A. (2003) Genes Dev. 17, 2539–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiwara, Y., Masutani, C., Mizukoshi, T., Kondo, J., Hanaoka, F. & Iwai, S. (1999) J. Biol. Chem. 274, 20027–20033. [DOI] [PubMed] [Google Scholar]
- 10.Wakasugi, M., Shimizu, M., Morioka, H., Linn, S., Nikaido, O. & Matsunaga, T. (2001) J. Biol. Chem. 276, 15434–15440. [DOI] [PubMed] [Google Scholar]
- 11.Sugasawa, K., Shimizu, Y., Iwai, S. & Hanaoka, F. (2002) DNA Repair 1, 95–107. [DOI] [PubMed] [Google Scholar]
- 12.Mu, D., Hsu, D. S. & Sancar, A. (1996) J. Biol. Chem. 271, 8285–8294. [DOI] [PubMed] [Google Scholar]
- 13.Rapic-Otrin, V., Kuraoka, I., Nardo, T., McLenigan, M., Eker, A. P. M., Stefanini, M., Leveine, A. S. & Wood, R. D. (1998) Mol. Cell. Biol. 18, 3182–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols, A. F., Itoh, T., Zolezzi, F., Hutsell, S. & Linn, S. (2003) Nucleic Acids Res. 31, 562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nag, A., Bondar, T., Shiv, S. & Raychaudhuri, P. (2001) Mol. Cell. Biol. 21, 6738–6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groisman, R., Polanowska, J., Kuraoka, I., Sawada, J., Saijo, M., Drapkin, R., Kisselev, A. F., Tanaka, K. & Nakatani, Y. (2003) Cell 113, 357–367. [DOI] [PubMed] [Google Scholar]
- 17.Shiyanov, P., Hayes, S. A., Donepudi, M., Nichols, A. F., Linn, S., Slagle, B. L. & Raychaudhuri, P. (1999) Mol. Cell. Biol. 19, 4835–4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh, T., O'Shea, C. & Linn, S. (2003) Mol. Cell. Biol. 23, 7540–7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang, Y. J., Hwang, B. J., Ford, J. M., Hanawalt, P. C. & Chu, G. (2000) Mol. Cell 5, 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan, T. & Chu, G. (2002) Mol. Cell. Biol. 22, 3247–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zolezzi, F. & Linn, S. (2000) Gene 245, 151–159. [DOI] [PubMed] [Google Scholar]
- 22.Miyata, M., Motoki, K., Tamura, E., Furukawa, M., Gonzalez, F. J. & Yamazoe, Y. (2002) Biochem. Pharmacol. 63, 1077–1084. [DOI] [PubMed] [Google Scholar]
- 23.de Vries, A., van Oostrom, C. T., Hofhuis, F. M., Dortant, P. M., Berg, R. J., de Gruijl, F. R., Wester, P. W., van Kreijl, C. F., Capel, P. J., van Steeg, H., et al. (1995) Nature 377, 169–173. [DOI] [PubMed] [Google Scholar]
- 24.van der Horst, G. T. J., van Steeg, H., Berg, R. J., van Gool, A. J., de Wit, J., Weeda, G., Morreau, H., Beems, R. B., van Kreijl, C. F., de Gruijl, F. R., et al. (1997) Cell 89, 425–435. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Cao, I., Garcia-Cao, M., Martin-Caballeo, J., Criado, L. M., Klatt, P., Flores, J. M., Weill, J.-C., Blasco, M. A. & Serrano, M. (2002) EMBO J. 21, 6225–6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zolezzi, F., Fuss, J., Uzawa, S. & Linn, S. (2002) J. Biol. Chem. 277, 41183–41191. [DOI] [PubMed] [Google Scholar]
- 27.Takata, K., Ishikawa, G., Hirose, F. & Sakaguchi, K. (2002) Nucleic Acids Res. 30, 3795–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakane, H., Takeuchi, S., Yuba, S., Saijo, M., Nakatsu, Y., Murai, H., Nakatsuru, Y, Ishikawa, T., Hirota, S., Kitamura, Y., et al. (1995) Nature 377, 165–168. [DOI] [PubMed] [Google Scholar]
- 29.de Boer, J., de Wit, J., van Steeg, H., Berg, R. J., Morreau, H., Visser, P., Lehmann, A. R., Duran, M., Hoeijmakers, J. H., Weeda, G., et al. (1998) Mol. Cell 1, 981–990. [DOI] [PubMed] [Google Scholar]
- 30.Kemp, C. J., Donehower, L. A., Bradley, A. & Balmain, A. (1993) Cell 74, 813–822. [DOI] [PubMed] [Google Scholar]
- 31.Agami, R., Blandino, G., Oren, M. & Shaul, Y. (1999) Nature 399, 809–813. [DOI] [PubMed] [Google Scholar]
- 32.Duckett, D. R., Bronstein, S. M., Taya, Y. & Modrich, P. (1999) Proc. Natl. Acad. Sci. USA 96, 12384–12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong, J. G., Costanzo, A., Yang, H. Q., Melino, G., Kaelin, W. G., Jr., Levrero, M. & Wang, J. Y. J. (1999) Nature 806–809. [DOI] [PubMed]
- 34.Wu, J., Gu, L., Wang, H., Geacintov, N. E. & Li, G. M. (1999) Mol. Cell. Biol. 19, 8292–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan, Z. M., Shioya, H., Ishiko, T., Sun, X., Gu, J., Huang, Y. Y., Lu, H., Kharbanda, S., Weichselbaum, R. & Kufe, D. (1999) Nature 399, 814–817. [DOI] [PubMed] [Google Scholar]
- 36.Shimodaira, H., Yoshioka-Yamashita, A., Kolodner, R. D. & Wang, J. Y. J. (2002) Proc. Natl. Acad. Sci. USA 100, 2420–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sancar, A., Lindsey-Boltz, L. A., Ünsal-Kaçmaz, K. & Linn, S. (2004) Annu. Rev. Biochem. 73, in press. [DOI] [PubMed]
- 38.Amundson, S. A., Do, K. T., Shahab, S., Bittner, M., Meltzer, P., Trent, J. & Fornace, A. J., Jr. (2000) Radiat. Res. 154, 342–346. [DOI] [PubMed] [Google Scholar]