Asymmetric cell division is a conserved mechanism to generate progeny with divergent fates. In the context of somatic stem cells, it provides a mode of self-renewal that retains the stem cell identity of one daughter cell while producing another daughter cell that is committed to differentiation. This allows for a balance between the requirement for committed progenitors during regeneration and the maintenance of tissue stem cells. Recently, skeletal muscle stem cells have stepped into the spotlight as a tractable paradigm to study the molecular determinants regulating asymmetric stem cell divisions. By examining this system, a recent study has yielded important insight into the molecular mechanisms that lead to the distinct identities of stem cell progeny.
Muscle regeneration depends on the participation of a heterogeneous population of stem cells and progenitors known as satellite cells. The satellite stem cell population, which is marked by the absence of expression of the myogenic regulatory factor (MRF) Myf5, is able to resist differentiation and maintain the satellite cell compartment after regeneration; on the other hand, committed satellite myogenic cells, which express the MRFs Myf5 and MyoD, are the predominant cell type that differentiates and fuses into myofibers.1 While Myf5- satellite stem cells can asymmetrically divide to give rise to Myf5+ satellite myogenic cells, other modes of asymmetric divisions have been observed in myoblasts, which give rise to either MyoD- reserve cells or Pax7-/MyoG+ differentiating myocytes.1,2 Although these modes of divisions are distinct from asymmetric satellite stem cell divisions, there are likely overlapping fate determinants, such as paracrine signaling and cell polarity pathways, governing these processes during regeneration.
The mode of satellite stem cell division is extrinsically pre-determined in response to physiological demand. The symmetric vs. asymmetric outcome of satellite stem cell division is regulated by cumulative signals from a variety of cell types in the local milieu. For example, enhanced Wnt7a signaling during regeneration promotes symmetric expansion of satellite stem cells by stimulating the Frizzled7 planar-cell-polarity (PCP) pathway.3 By polarizing the PCP effector Vangl2 to opposing poles of the satellite stem cell parallel to its host myofiber, Wnt7a facilitates a planar division, thus giving rise to identical stem cell progeny. Additionally, feedback from proliferating Myf5+ satellite cells can modulate the effects of Wnt7a by contributing high levels of Fibronectin into the satellite stem cell niche.4 This allows for a multi-faceted regulation of stem cell numbers through the progression of regeneration.
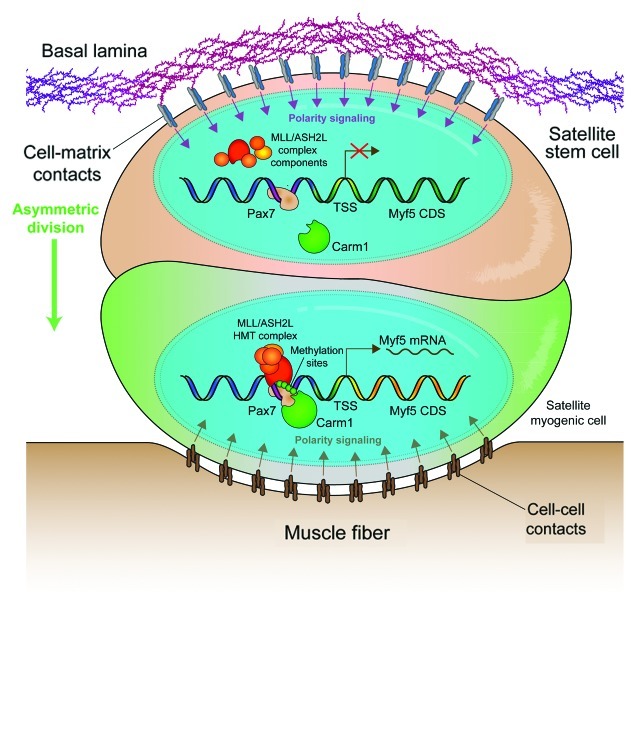
The satellite cell niche plays an important role in the subsequent processes leading up to an asymmetric division. The basal surface of satellite cells adheres to the basal lamina surrounding individual myofibers; whereas, the apical surface of satellite cells maintains cell-cell contact with its host myofibers (Fig. 1). This polarized environment serves as an intrinsic cue for mitotic spindle orientation and leads to the asymmetric segregation of fate determinants. In an analogous mechanism to asymmetric Drosophila neuroblast divisions, segregation of Par-3 proteins and the Notch-inhibitor Numb to opposing poles create asymmetry in dividing satellite cells.5,6 Interestingly, Par-3 associates with active p38α/β and leads to differentiation,6 whereas Numb segregation co-localizes with the asymmetric retention of the template DNA strand.5 Moreover, asymmetric segregation of template DNA correlates to a subpopulation of differentiation resilient Pax7High satellite cells.7 Taken together, polar segregation of cellular components intrinsically determine divergent cell fates.
Figure 1. Extrinsic and intrinsic determinants of cell fate in muscle stem cells. The satellite cell niche is highly asymmetric. Satellite cells can be in contact with the muscle fiber on their apical pole or with the extracellular matrix of the basal lamina at the basal pole. Cell-cell contacts and cell-matrix receptors induce polarity signals, allowing for positioning of the mitotic spindle apparatus and the induction of fate determinants after division. Myf5- satellite stem cells (ochre) express high levels of Pax7. However, transcription of the Myf5 locus is repressed. Upon asymmetric division, Carm1 methylates Pax7 and allows for the recruitment of the MLL/ASH2L HMT complex, which leads to the remodeling of chromatin at the Myf5 locus followed by transcription of Myf5 mRNA and myogenic commitment of the apical daughter cell (green).
Although polarity is the most puzzling aspect of asymmetric cell division, one of the least understood mechanisms is the post-mitotic establishment of divergent fates. Certainly, the asymmetric segregation of factors such as Notch, active p38α/β or even newly synthesized DNA strands may induce differentiation. However, the processes leading to the transcription of fate determination factors are largely unknown. Myf5 transcription in asymmetric satellite stem cell division clearly marks the myogenic commitment of the apical daughter cell.1 In a recent paper by Kawabe et al. published in Cell Stem Cell, it was demonstrated that expression of Myf5 in the apical daughter cell requires the post-translational activation of Pax78 (Fig. 1).
Pax7 directly regulates Myf5 mRNA expression by recruiting the MLL/ASH2L histone methyltransferase (HMT) complex to enhancers upstream of the Myf5 coding region.9 However, the substantial Pax7 expression in Myf5- satellite stem cells suggests that this mechanism is subject to post-translational regulation. Kawabe et al. identified Carm1 as a novel Pax7 binding protein and subsequently demonstrated its ability to methylate multiple arginine residues in the N terminus of Pax7.8 The methylation status of Pax7 affects its ability to recruit the MLL/ASH2L HMT complex and, thus, its ability to activate Myf5 expression. Importantly, knockdown of Carm1 expression by siRNA transfection resulted in a loss of Myf5 activation in asymmetric divisions, which is congruent to the phenotype observed with the knockdown of Pax7. This result suggests that the expression of Pax7 and Carm1 primes satellite stem cells for the myogenic fate.
Pax7-Carm1 interactions were only observed in the nuclei of committed Myf5+ satellite cells. Therefore, there remains an additional asymmetrically activated determinant that regulates the arginine methylation of Pax7. Both active Carm1 and Par-3 localize in the committed daughter cells of asymmetric divisions and are possibly connected, since they have been implicated in the establishment of polarity in blastomeres.10 However, whether Par-3 regulates Carm1 activity will have to be resolved by future studies.
These recent studies shed light on the post-mitotic mechanism leading to asymmetric commitment, but there is still much to discover. The mechanisms regulating satellite stem cell self-renewal could well apply to other stem cell populations and allow for the identification of therapeutic targets.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/23068
References
- 1.Kuang S, et al. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu W, et al. Development. 2012;139:2857–65. doi: 10.1242/dev.079665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Grand F, et al. Cell Stem Cell. 2009;4:535–47. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentzinger C. F., et al. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2012.09.015. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shinin V, et al. Nat Cell Biol. 2006;8:677–87. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 6.Troy A, et al. Cell Stem Cell. 2012;11:541–53. doi: 10.1016/j.stem.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rocheteau P, et al. Cell. 2012;148:112–25. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 8.Kawabe Y, et al. Cell Stem Cell. 2012;11:333–45. doi: 10.1016/j.stem.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKinnell IW, et al. Nat Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parfitt D-E, et al. Mol Biol Cell. 2010;21:2649–60. doi: 10.1091/mbc.E10-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]