Abstract
Endothelin 1 (ET-1), a potent vasoconstrictor peptide expressed by endothelium, is also produced in the heart in response to a variety of stresses. It induces hypertrophy in cultured cardiac myocytes but only at concentrations far greater than those found in plasma. We tested whether ET-1 generated by cardiac myocytes in vivo is a local signal for cardiac hypertrophy. To avoid the perinatal lethality seen in systemic ET-1-null mice, we used the Cre/loxP system to generate mice with cardiac myocyte-specific disruption of the ET-1 gene. We used the α-myosin heavy chain promoter to drive expression of Cre and were able to obtain 75% reduction in ET-1 mRNA in cardiac myocytes isolated from these mice at baseline and after stimulation, in vivo, for 24 h with tri-iodothyronine (T3). Necropsy measurements of cardiac mass indexed for body weight showed a 57% reduction in cardiac hypertrophy in response to 16 days of exogenous T3 in mice homozygous for the disrupted ET-1 allele compared to siblings with an intact ET-1 gene. Moreover, in vivo MRI showed only a 3% increase in left ventricular mass indexed for body weight in mice with the disrupted allele after 3 weeks of T3 treatment versus a 27% increase in mice with an intact ET-1 gene. A reduced hypertrophic response was confirmed by planimetry of cardiac myocytes. We conclude that ET-1, produced locally by cardiac myocytes, and acting in a paracrine/autocrine manner, is an important signal for myocardial hypertrophy that facilitates the response to thyroid hormone.
Cardiac hypertrophy is characterized by an increase in myocardiocyte size and is accompanied by qualitative and quantitative changes in gene expression, protein synthesis, and physiological performance (1). Cardiac hypertrophy helps to maintain cardiac output in the presence of increased demand or afterload, yet it is one of the most important predisposing risk factors for sudden death and the development of heart failure in human populations. A greater understanding of hypertrophy, and the capacity to regulate it in human disease, could have profound clinical implications.
Many ligand/receptor systems, with their specific and interwoven downstream signaling pathways, have been implicated in cardiomyocyte hypertrophy in both cell culture and intact animals (reviewed in ref. 2). These include the peptide endothelin 1 (ET-1) and its receptors ETA and ETB on the cell surface of cardiomyocytes. Several lines of evidence suggest that ET-1 functions in a paracrine/autocrine manner in cardiac hypertrophy. In cell cultures of cardiomyocytes, ET-1 induces hypertrophy as assessed by cell size, increased myofibrillogenesis, and transcriptional changes associated with cardiac hypertrophy such as expression of atrial natriuretic factor (3, 4). Of note, in these studies the concentrations of ET-1 required to produce hypertrophy were 10-7 to 10-9 M, far greater than those encountered in plasma (10-12 to 10-13 M) (5). Although cardiac synthesis of ET-1 is negligible under normal circumstances, conditions that stimulate hypertrophy markedly increase expression of ET-1 mRNA in cardiac cells (6). Antagonists of the ETA receptor reduce hypertrophic responses to other stimuli, including angiotensin II (7) and aortic banding (8). Such inhibition reduces remodeling of the left ventricle under circumstances that otherwise elicit a hypertrophic response and improves survival (9). Thus a significant role for cardiomyocyte ET-1 functioning in a paracrine fashion is suggested by (i) plasma levels that are much lower than effective hypertrophic concentrations, (ii) the induction of cardiomyocyte ET-1 expression during hypertrophy, and (iii) the effect of ETA antagonism on other hypertrophic stimuli. If ET-1 does play an autocrine role, augmenting and modifying other stimuli, it becomes a particularly attractive candidate for pharmacological manipulation to ameliorate cardiac hypertrophy.
The hypothesis tested in this study, derived from this evidence of a paracrine/autocrine role for ET-1 in cardiac hypertrophy, is that disruption of ET-1 in cardiac myocytes would result in an altered cardiac response to hypertrophic stress. Accordingly, we generated mice in which ET-1 can be disrupted in a tissue-specific manner. By crossing these animals with α-myosin heavy chain (α-MHC)-Cre transgenic mice, we excised exon 2 of the ET-1 gene specifically in cardiomyocytes. We then challenged these mice as young adults with exogenous tri-iodothyronine (T3) and analyzed their capacity to develop cardiac hypertrophy.
Materials and Methods
Lox P Bracketed ET-1 Gene Transgenic Mice. A detailed description of the creation of the lox targeted ET-1 allele will be published elsewhere (Y.Y.K. and M.Y., unpublished work). Construction of targeting vectors and homologous recombination in embryonic stem (ES) cells was performed as described (10). Briefly, we constructed a targeting vector with loxP sites flanking both exon 2 of the ET-1 gene (which encodes mature ET-1 peptide) and the neo-thymidine kinase selection cassette. Successfully transfected ES cells were selected by neomycin resistance, then transiently transfected with a Cre expression plasmid and screened for excision of the selection cassette. Clones showing appropriate excision were used to generate chimeric mice by blastocyst injection.
Other Transgenic Mice. Cardiac α-MHC-Cre transgenic mice were the generous gift of Peter Frenkel and Michael Schneider (Baylor College of Medicine, Houston) (11). CAG-CAT-Z transgenic mice, carrying a silent lacZ reporter under the control of the CAG promoter, were a generous gift from Jun-ichi Miyazaki (Osaka University, Osaka) (12).
Genotyping and Analysis of Cre-Mediated Recombination. Genomic DNA was prepared from tail biopsies from postnatal day 21 mice or tissues harvested from mice at the age of 8-12 weeks. For genotyping, this DNA was subjected to PCR and/or Southern blot analysis. Oligonucleotide primers used for the detection of the WT ET-1 allele were 5′-GCTGCCCAAAGAT TCTGAATTCTG-3′ (forward) and 5′-GATGATGTCCAGGTGGCAGAAG-3′ (reverse). A different forward primer was used for the detection of the floxed ET-1 allele, 5′-CTGCCCAAAGATTCTGAATTGAT-3′. Cre-mediated recombination that produced a deleted ET-1 allele (dlox) was detected by using the latter forward primer with the reverse primer, 5′-GAG TGACCCTGTGACAGGTTTGAATAAG-3′. These primers generated an ≈800-bp fragment for both the WT and floxed-allele of ET-1 and a 1.4-kb fragment for the dlox allele. Primers for detection of the α-MHC-Cre transgene were 5′-TTAGCAAACCTCAGGCACCCTTAC-3′ (forward) and 5′-CGCATAACCAGTGAAAC AGCATTGC-3′ (reverse). Primers for the detection of CAG-CAT-Z transgene were 5′-GACACCAGACCAACTGGTAATGG-3′ (forward) and 5′-GCATCGAGCTGGGTAATAAGCG-3′ (reverse). These primers amplified an ≈750-bp product for the α-MHC-Cre transgene and a 825-bp product for the CAG-CAT-Z transgene. For Southern blot analysis, 10 μg of genomic DNA was digested with EcoRI and EcoRV and hybridized with a radiolabeled 3′ external probe. Signal was quantitated by using a PhosphorImager (Molecular Dynamics).
Isolation of Cardiac Myocytes. Six-week-old sibling male mice with appropriate genotypes were treated with T3 (Sigma) (1 mg/kg by i.p. injection) or saline control and killed after 24 h. Cardiac myocytes were isolated by a modification of previously published methods (13). After anesthesia with i.p. injection of avertin (Sigma 1.25%, 0.02 ml/g of body weight), beating hearts were quickly removed from the chests. The aorta was cannulated with a 22-gauge catheter, flushed with Ca2+ free buffer (MEM, Joklik-modified, pH 7.2; GIBCO/BRL), and mounted onto a Langendorf perfusion apparatus. All perfusates were maintained at 37°C and continuously bubbled with 95% O2/5%CO2. The hearts were perfused with constant pressure (100 cm H2O) at 2.2 ml/min initially with calcium-free Joklik buffer (4 min), followed by Joklik buffer supplemented with 0.25 mM Ca2+, 75 units/ml collagenase I and II (Worthington), 1% BSA (Sigma), and 2% calf serum (pH 7.2). When swollen and soft (12-15 min), the heart was removed from the apparatus and the ventricles were minced into Joklik buffer supplemented with 0.25 mM Ca2+ and 2% calf serum. Aliquots of these cells showed 80-85% rod cells (corresponding to cardiac myocytes) consistent with previous reports. The isolated cells were washed in a physiological buffer (132 mM NaCl/4.8 mM KCl/1.2 mM MgCl2·6H2O/5 mM glucose/10 mM Hepes, pH 7.2) and frozen for RNA isolation.
Determination of Cardiac Myocyte ET-1 mRNA. Endothelin mRNA was quantitated by both real-time RT-PCR and Southern blotting of PCR products. Total RNA was isolated from isolated cardiomyocytes by TRIzol reagent (GIBCO/BRL) according to the manufacturer's instruction. Triplicate real-time RT-PCRs were performed with SYBR green PCR master mix on an Applied Biosystem's GeneAmp 5700 System according to the manufacturer's protocol. One microgram of total RNA was reverse-transcribed (Roche Applied Science, AMV reverse transcriptase), and product was amplified from cDNA representing 5 ng of total RNA. Expression level was interpolated from a standard curve generated from a series of dilutions at a minimal cycle number where threshold intensity was clearly exceeded. GAPDH message was used as an internal control. The primers were 5′-TGCACCACCAACTGCTTAG-3′ (GAPDH, forward), 5′-GGATGCAGGGATGATGTTC-3′ (GAPDH, reverse), 5′-GTGTCTACTTCTGCCACCTGGACAT-3′ (ET-1 forward), and 5′-GGGCTCGCACTATATAAGGGATGAC-3′ (ET-1 reverse). Standard two-step reaction conditions were used (95°C 15 sec, 60°C 1 min). Conventional RT-PCR was also performed on these samples. Aliquots of PCR products were obtained during amplification and subjected to agarose gel electrophoresis. After these gels were transferred to nitrocellulose membranes, Southern hybridizations were performed with 32P-labeled probes (random primer labeling system, Roche Applied Science). The amount of PCR products from samples in the linear amplification range were quantitated on a PhosphorImager (Molecular Dynamics). GAPDH was used as a reference control.
For determination of ET-1 peptide, tissues were homogenized by using a Polytron (20,000 rpm) in 1 M acetic acid containing 0.01 mM pepstatin A for 1 min and were immediately placed in boiling water for 10 min. After centrifugation at 25,000 × g, the supernatant was concentrated with Sep-Pak C18 columns (Waters) as described (14). Concentration of mature ET-1 peptide was determined by enzyme immunoassay as described (15).
Induction and Analysis of Cardiac Hypertrophy. Age-matched (12-16 weeks old) male mice with appropriate genotypes were treated with T3 (Sigma) for 3 weeks (1 mg/kg daily by i.p. injection). Left ventricular mass and function before and after treatment was quantified by MRI analysis performed under Avertin anesthesia. Electrocardiographically gated, spin-echo images at multiple stages of the cardiac cycle were obtained by using a clinical 1.5 Tesla system (Philips, Eindhoven, The Netherlands) as described (16). Short axis slices were obtained spanning the left ventricle. Endocardial and epicardial contours were drawn for each slice, and myocardial wall volume was calculated by using Simpson's rule. The wall volume was multiplied by the specific gravity of myocardium (1.055 mg/mm3) to calculate the estimated left ventricular mass. Similar calculations of minimal and maximal chamber volume corresponding to end-systole and end-diastole were used to determine ejection fractions as an assessment of ventricular function. In a separate set of experiments, hearts were obtained from similarly matched and T3-treated animals, and the entire hearts were excised, dissected free of noncardiac tissue, rinsed with saline, carefully dabbed dry, and weighed for determination of total cardiac mass.
We obtained quantitative histological assessment of hypertrophy from another similar group of animals. After treatment for 1 or 3 weeks with T3, hearts were rapidly excised and, while still beating, incubated in Krebs-Henseleit solution (112 mM NaCl/4.7 mM KCl/1.2 mM KH2PO4/25 mM NaHCO3/0.1 mM glucose/1.2 mM MgSO4) for 5 min to relax the muscle, and then fixed in 4% paraformaldehyde overnight. After embedding in paraffin, images of hematoxylin and eosin-stained 5-μ sections were analyzed with planimetry software (IMAGE 1.62, Scion, Frederick, MD) by an observer blinded to genotype. A grid was applied to ×40 fields obtained from six levels of the papillary muscle and the area of the cell closest to 11 dispersed registration marks was obtained. In this manner 66 cells were sampled from each of three mice in each experimental group.
Statistical Analysis. Comparison of hypertrophy between groups was performed by using a two-tailed, unpaired Student's t test with an assumption of homoskedasticity. Peptide concentrations were compared by a two-tailed t test. Mean values are reported ±1 SD for cardiac mass or with the SEM for morphometry, and P < 0.05 was considered significant.
Results
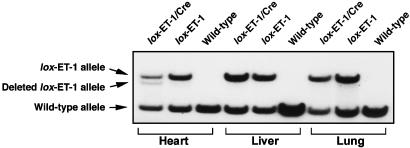
Generation of Cardiomyocyte-Specific ET-1 Knockout Mice. Mice heterozygous for the floxed ET-1 allele were bred to homozygosity. ET-1flox/flox mice have no detectable abnormalities. These animals were then bred with the α-MHC-Cre transgenic mice to generate mice homozygous for the floxed ET-1 allele carrying the hemizygous α-MHC-Cre transgene (ET-1flox/flox;Cre+). These animals were also born with no defects and were apparently healthy into adulthood. To demonstrate that the floxed ET-1 exon 2 is disrupted by Cre-mediated recombination, we harvested tissues from both heterozygous ET-1flox/+;Cre+ and ET-1flox/+;Cre-sibling mice, isolated genomic DNA, and performed Southern blot analysis using a probe that distinguished the intact and deleted floxed allele of ET-1 (Fig. 1). The band representing the deleted allele has 17% of the total counts detected in the two bands. As this Southern blot is performed on mice heterozygous for the floxed allele, we estimate that 34% of the cellular nuclei of the heart would exhibit the deletion in a homozygous animal. Because the ratio of interstitial cell nuclei to myocyte nuclei is estimated to be between 2:1 and 3:1 (17) we conclude that 70-90% of cardiac myocytes showed Cre-mediated deletion of the ET-1 gene. We further confirmed the functional expression of the α-MHC-Cre transgene in vivo by demonstrating cardiac-specific, cre-dependent activation of a β-galactosidase transgene in 83% of cardiac myocytes (18).
Fig. 1.
Detection of in vivo Cre-mediated recombination of the floxed ET-1 allele in various tissues from mice with and without the α-MHC-Cre transgene. Tissue DNA from ET-1flox/+;Cre- (lox ET-1) and ET-1flox/+;Cre+ (lox ET-1/Cre) and WT mice was digested with EcoRI and EcoRV and hybridized with a probe that distinguishes the floxed and deleted alleles (Y.Y.K. and M.Y., unpublished work). Southern blot analysis shows detectable recombination only in the hearts from ET-1flox/+;Cre+ mice. There is no recombination seen in the other tissues.
We used an enzyme immunoassay to determine the level of mature ET-1 peptide in tissue homogenates from four mice of each genotype under normal conditions. The tissue level of ET-1 was not significantly different among these mice in any tissues tested, including the hearts (in pg/g for the hearts: ET-1flox/flox;Cre+ = 977 ± 236, ET-1flox/flox;Cre- = 624 ± 110, ET-1+/+;Cre+ = 980 ± 145, ET-1+/+;Cre- = 814 ± 147, P > 0.05 for all comparisons). The level of ET-1 mRNA, assessed by poly(A) Northern blots, was not different between ET-1flox/flox;Cre+ and ET-1flox/flox;Cre-mice even with the addition of 2 weeks of T3 stimulation (data not shown). These observations suggest that the floxed ET-1 allele functions normally and also that, even with T3 stimulation, cardiac myocytes produce only a small fraction of the ET-1 synthesized in the heart (compared to other cell types in the heart).
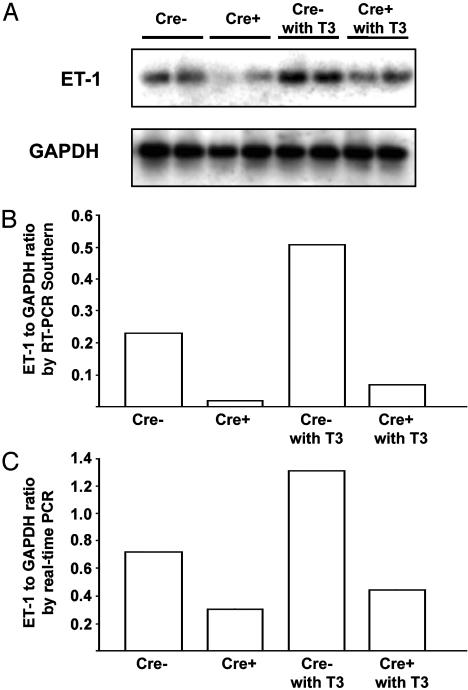
To assess the effect of Cre-directed excision of the floxed allele we isolated cardiac myocytes from the hearts of ET-1flox/flox mice, both Cre+ and Cre-, with and without treatment with T3 for 24 h. We determined mRNA abundance with amplification-based methods (real-time RT-PCR and Southern blotting of PCR products at a low number of amplification cycles). At baseline the Cre+ mice had substantially lower ET-1 expression by both methods and this difference was maintained at the higher levels of ET-1 message detected after T3 treatment (Fig. 2). The persistence of low levels of ET-1 message in the Cre+ mice could be caused either by contamination of the cardiomyocyte preparation with other cell types or mosaicism of expression of Cre in cardiomyocytes. This latter possibility is suggested by the less than completely uniform expression of β-galactosidase staining described above (18). We also examined the expression of the mRNAs for B-type natriuretic peptide (BNP) and atrial natriuretic peptide (ANP) in whole heart RNA, as a molecular correlate of hypertrophy. There was a modest increase in both the ANP/GAPDH and BNP/GAPDH ratios with 16 days of thyroid treatment and no statistically significant difference in the response of Cre- and Cre+ animals (data not shown), consistent with the previous finding that hyperthyroidism produces negligible elevation of ANP in the rat heart, in contradistinction to other hypertrophic stimuli such as aortic banding (19).
Fig. 2.
Quantitative RT-PCR analysis of ET-1 mRNA in cardiac myocytes. (A) Autoradiogram of a Southern blot of ET-1 exon 2 amplicons from cardiomyocyte preparations of ET-1flox/flox;Cre+ and ET-1flox/flox;Cre- mice with and without 24 h of treatment with T3 in vivo. (B) Quantitation of radioactive signal from the gel in A (by PhosphorImager). (C) Analysis of the same RNA preparations assessed by real-time RT-PCR. Cardiac myocytes from two animals were independently assessed, and the mean value is provided.
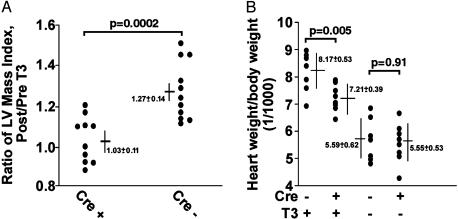
Resistance to Cardiac Hypertrophy Induced by Thyroid Hormone. We treated 13 ET-1flox/flox;Cre+ mice and 11 ET-1flox/flox;Cre- age- matched mice with T3 for 3 weeks to induce cardiac hypertrophy. Using MRI we determined left ventricular mass and ejection fraction before and after treatment with T3 (Fig. 3A). In the course of treatment with T3 we observed no obvious difference in the appearance of mice with either genotype. The body weight of these mice increased during treatment (3.63 ± 0.63 g increase in Cre+ and 3.12 ± 0.71 g in Cre- mice, P = 0.60), suggesting no serious morbidity from hyperthyroidism in either genotype. However, after treatment with T3, three Cre+ mice died, two during induction of anesthesia for MRI. We could not determine the cause of death in these mice despite necropsies. Statistical analysis was performed only on those mice that survived both MRI scans. In Cre- mice the left ventricular mass indexed for body weight (LVMI) increased by 27 ± 0.14% after T3 treatment as compared with pretreatment values. By contrast, average increase in LVMI in Cre+ mice was only 2.8 ± 0.11% (P = 0.0002). To exclude the possible confounding effect of the genotype on body weight we also calculated the absolute (unindexed) hypertrophy, which was 18 ± 17% for Cre+ animals and 42 ± 19% for Cre- mice (P = 0.005). Ejection fraction was 0.69 ± 0.015 (before T3) and 0.71 ± 0.13 (after) in Cre- mice and 0.65 ± 0.016 (before) and 0.68 ± 0.016 (after) in Cre+ mice (P > 0.3 for all comparisons). We also examined a subset of animals in both genotypes after 5 weeks of treatment with T3. We saw no further increase in hypertrophy compared to 3 weeks of treatment, and the difference in hypertrophy between the genotypes persisted (data not shown).
Fig. 3.
Analysis of T3-induced hypertrophy. (A) A scattergram of the data from ET-1flox/flox;Cre+ mice and age-matched ET-1flox/flox;Cre- mice that were tested for their capacity to develop cardiac hypertrophy in response to exogenous T3. Left ventricular (LV) mass was determined by cardiac MRI before and after 3 weeks of daily injection of T3. (B) An analysis of total cardiac mass measured at necropsy after 16 days of treatment with T3. Each filled circle represents a separate animal.
MRI allows us to measure ventricular function and mass sequentially in the living animal, but it is not appropriate for determination of total cardiac mass, because the atrial and right ventricular walls are too thin for accurate quantitation by this technique (16). We therefore repeated the study of thyroid-induced hypertrophy in a separate group of animals (Fig. 3B) by using necropsy determination of total cardiac mass. In this case, we found a greater degree of cardiac hypertrophy in the ET-1flox/flox;Cre+ animals (30%) but still significantly less than in the ET-1flox/flox;Cre- mice (46%, P = 0.002). Baseline studies, without thyroid treatment, showed no significant difference in cardiac mass index between the two genotypes.
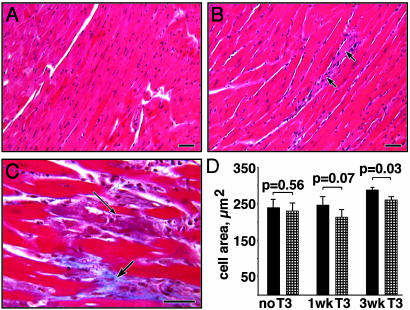
We examined the histology of the hearts of treated and untreated animals of each genotype. Data were combined for all animals treated similarly (n = 3-5) to obtain an average cardiomyocyte area. Comparison of grouped data showed no significant difference without T3 treatment (240 μ2 for Cre-, 230 μ2 for Cre+, P = 0.56) and confirmed hypertrophy after 3 weeks of treatment for the Cre- mice (284 μ2, P = 0.01) but not for the Cre+ (253 μ2, P = 0.13). Less hypertrophy was seen in the Cre+ compared to the Cre- animals at 3 weeks (P = 0.03, Fig. 4D). Thyroid treatment produced a mild degree of myocardiocyte loss and early fibrosis after 3 weeks (illustrated in Fig. 4 B and C) but with no difference between strains as assessed by two independent observers, blinded to genotype.
Fig. 4.
Histology of ET-1flox/flox;Cre- mouse hearts. (A) The typical morphology of myocardium from an untreated animal (hematoxylin and eosin). (B) Myocardiocyte hypertrophy seen after 3 weeks of treatment with T3. The arrows identify a degenerating myofiber with an associated inflammatory infiltrate. (C) A higher magnification of a trichrome stain of a treated heart showing an apoptotic figure within a degenerating myofiber (thin arrow) and evidence of early collagen deposition (thick arrow). (D) A histogram of average cell area is shown with comparison of ET-1flox/flox;Cre- (filled bars) and ET-1flox/flox;Cre+ (hatched bars) mice. (Scale bars = 40 μm.)
Discussion
Genetic Analysis of ET-1 Function in Adult Mice. Pharmacological inhibition of ETA, the principal endothelin receptor in the cardiac myocytes, has been shown to reduce the hypertrophic response to various stimuli in vivo. However, this approach is limited by both pharmacological considerations (tissue availability, half-life of the drug, incomplete blockade, and possibly questionable specificity of drug action), and more importantly, by the systemic effects of ET-1 blockade on blood pressure, plasma volume, and coronary perfusion. Also, pharmacological manipulation at the receptor level does not provide information regarding the locations of ET-1 production relevant for cardiac hypertrophy. Some of these issues can be addressed through genetically altered animal models. Because the neonatal lethality of conventional ET-1 knockout mice, caused by embryonic malformations, prevents analysis in adult mice, we generated a mouse strain in which ET-1 can be disrupted in a tissue-specific manner. ET-1flox/flox mice were born with no abnormalities and appeared healthy into adulthood. The level of mature ET-1 peptide from several different tissues of ET-1flox/flox mice was not significantly different from the tissues from WT mice (data not shown). This finding indicates that the floxed ET-1 allele functions normally. Furthermore, ET-1dlox/dlox mice showed the identical phenotype as conventional ET-1-deficient mice (20), indicating that Cre-mediated deletion produces a null ET-1 allele as predicted.
Generation of the Cardiac-Specific ET-1 Knockout Mice. Previous studies indicate that expression of the α-MHC-Cre transgene is restricted to postmitotic cardiac myocytes (11). ET-1flox/flox;Cre+ mice develop normally and are normally active into adulthood. MRI analysis demonstrated that neither cardiac morphology nor cardiac function was affected by the presence of the Cre transgene under baseline conditions. Genomic Southern blot analysis agreed with histological analysis, showing that expression of the α-MHC-Cre transgene leads to tissue-specific, Cre-mediated recombination in a majority of postmitotic cardiac myocytes. As expected from previous studies (6, 8), the cardiac ET-1 level was the same for ET-1flox/flox;Cre+ mice and ET-1flox/flox;Cre- mice under normal physiological conditions.
Induction of Cardiac Hypertrophy by Thyroid Hormone. There are many experimental models of exogenous hypertrophic stimuli in rodents. We selected hyperthyroidism for several reasons: (i) it rapidly produces a substantial degree of hypertrophy (21), (ii) it is simple to apply with low morbidity and, (iii) it closely recapitulates a form of cardiac hypertrophy found in humans. Each experimental model is likely to have its own idiosyncratic characteristics. A possible limitation of hyperthyroidism for these studies is that it is a multifactorial stimulus that affects afterload (pressure overload), preload (volume overload), contractility, heart rate, and metabolic rate in addition to the direct actions of T3 on cardiomyocytes (22). This pleiotropic quality of the hypertrophic stimulus from thyroid hormone complicates the identification of the specific signaling pathways that ET-1 may impinge on. It is possible that other hypertrophic stimuli, for example, pure pressure overload provided by aortic banding, or adrenergic stimulation with infusion of an agonist, might be influenced by the cardiac myocyte ET-1 axis in a different manner or to a different degree.
Cardiomyocyte-Specific ET-1 Knockout Mice Are Resistant to Hypertrophy Induced by Thyroid Hormone. Because disruption of ET-1 is tightly restricted to cardiac myocytes in these mice, our result indicates that ET-1 mediates cardiac hypertrophy in a paracrine/autocrine manner within the heart and is a crucial downstream effector for the hypertrophic response to thyroid stimulation. Neither Cre+ mice nor Cre- mice showed obvious signs of cardiac failure during treatment with T3. However, three ET-1flox/flox;Cre+ mice died unexpectedly during this study; it is possible that inadequate cardiac hypertrophy or the loss of the inotropic stimulation (23) offered by ET-1 may have produced intolerance of the peripheral vasodilation and inotropic depression caused by anesthesia. Alternatively, Cre recombinase, which is thought to be a nontoxic enzyme in mammalian cells, may have unexpected effects on cardiac myocytes. Transcription of transgenes under the control of the α-MHC promoter may be up-regulated in response to thyroid hormone (24). However, we detected no differences in ventricular function in otherwise normal mice expressing Cre compared to WT animals (data not shown).
Mechanistic Implications. The role of ET-1 as a potential central actor in cardiac hypertrophy is supported by the finding that inactivation of the genes for Gα11 and Gαq, which transduce the signal from the endothelin receptor (among others) produces mice that are resistant to the hypertrophic stress of aortic banding (25). One likely node of interaction of ET-1 and T3 signaling is intracellular calcium. ET-1 receptors, coupled to Gq, are thought to generate their signal in part through an increase in intracellular Ca2+ concentrations. For example, ET-1 produces a >50% increase in the amplitude of calcium transients, perhaps via an effect on the Na/Ca exchanger (26), the Na/H exchanger (27), or the L-type calcium channel (28). T3 regulates several proteins involved in intracellular calcium metabolism, including the sarcoplasmic reticulum Ca2+-ATPase (29) and its regulator phospholamban (30, 31), Ca2+ channels (32), the ryanodine receptor (33), and protein kinase C (34). If the principal hypertrophic stimulus of T3 is mediated by changes in cytosolic calcium concentration or flux, then it seems likely that the effect of ET-1 on systolic calcium transients could be the mechanism of its action in facilitating T3-induced hypertrophy.
Pharmacological Implications. Sustained cardiac hypertrophy may lead to ventricular dilation and eventually cardiac failure. One could extrapolate from our study that the previously reported beneficial effects of ETA blockade may not be limited to a lowering of systemic vascular resistance, but also, and perhaps predominately, be caused by blockade of the local action of myocyte-derived ET-1. Thus, ETA blockade could interfere with the hypertrophic response of the cardiac myocyte, and pharmacological strategies should incorporate consideration of the direct effects of ETA blockade on the cardiac myocyte.
Acknowledgments
We thank John Shelton and James Richardson for expert advice and assistance with histology. This work was supported by National Institutes of Health Grant RO-1 HL64041 and a Texas Higher Education Coordinating Board grant (to R.V.S.), and by grants from the Perot Family Foundation, the W. M. Keck Foundation, and the Exploratory Research for Advanced Technology (Japan)/Japan Science and Technology Agency (to M.Y.). Y.Y.K. was an Associate and M.Y. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: ET-1, endothelin 1; T3, tri-iodothyronine; α-MHC, α-myosin heavy chain.
References
- 1.Frey, N. & Olson, E. N. (2003) Annu. Rev. Physiol. 65, 45-79. [DOI] [PubMed] [Google Scholar]
- 2.Hunter, J. J. & Chien, K. R. (1999) N. Engl. J. Med. 341, 1276-1283. [DOI] [PubMed] [Google Scholar]
- 3.Shubeita, H. E., McDonough, P. M., Harris, A. N., Knowlton, K. U., Glembotski, C. C., Brown, J. H. & Chien, K. R. (1990) J. Biol. Chem. 265, 20555-20562. [PubMed] [Google Scholar]
- 4.Ito, H., Hirata, Y., Hiroe, M., Tsujino, M., Adachi, S., Takamoto, T., Nitta, M., Taniguchi, K. & Marumo, F. (1991) Circ. Res. 69, 209-215. [DOI] [PubMed] [Google Scholar]
- 5.Yorikane, R., Sakai, S., Miyauchi, T., Sakurai, T. & Goto, K. (1994) Arzneimittelforschung 44, 412-415. [PubMed] [Google Scholar]
- 6.Arai, M., Yoguchi, A., Iso, T., Takahashi, T., Imai, S., Murata, K. & Suzuki, T. (1995) Am. J. Physiol. 268, H2084-H2091. [DOI] [PubMed] [Google Scholar]
- 7.Ito, H., Hirata, Y., Adachi, S., Tanaka, M., Tsujino, M., Koike, A., Nogami, A., Murumo, F. & Hiroe, M. (1993) J. Clin. Invest. 92, 398-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito, H., Hiroe, M., Hirata, Y., Fujisaki, H., Adachi, S., Akimoto, H., Ohta, Y. & Marumo, F. (1994) Circulation 89, 2198-2203. [DOI] [PubMed] [Google Scholar]
- 9.Sakai, S., Miyauchi, T., Kobayashi, M., Yamaguchi, I., Goto, K. & Sugishita, Y. (1996) Nature 384, 353-355. [DOI] [PubMed] [Google Scholar]
- 10.Hosoda, K., Hammer, R. E., Richardson, J. A., Baynash, A. G., Cheung, J. C., Giaid, A. & Yanagisawa, M. (1994) Cell 79, 1267-1276. [DOI] [PubMed] [Google Scholar]
- 11.Agah, R., Frenkel, P. A., French, B. A., Michael, L. H., Overbeek, P. A. & Schneider, M. D. (1997) J. Clin. Invest. 100, 169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Araki, K., Araki, M., Miyazaki, J. & Vassalli, P. (1995) Proc. Natl. Acad. Sci. USA 92, 160-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolska, B. M. & Solaro, R. J. (1996) Am. J. Physiol. 271, H1250-H1255. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto, H., Suzuki, N., Onda, H. & Fujino, M. (1989) Biochem. Biophys. Res. Commun. 164, 74-80. [DOI] [PubMed] [Google Scholar]
- 15.Xu, D., Emoto, N., Giaid, A., Slaughter, C., Kaw, S., deWit, D. & Yanagisawa, M. (1994) Cell 78, 473-485. [DOI] [PubMed] [Google Scholar]
- 16.Franco, F., Dubois, S. K., Peshock, R. M. & Shohet, R. V. (1998) Am. J. Physiol. 274, H679-H683. [DOI] [PubMed] [Google Scholar]
- 17.Adler, C. P., Friedburg, H., Herget, G. W., Neuburger, M. & Schwalb, H. (1996) Virchows Arch. 429, 159-164. [DOI] [PubMed] [Google Scholar]
- 18.Kedzierski, R. M., Grayburn, P. A., Kisanuki, Y. Y., Williams, C. S., Hammer, R. E., Richardson, J. A., Schneider, M. D. & Yanagisawa, M. (2003) Mol. Cell. Biol. 23, 8226-8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izumo, S., Nadal-Ginard, B. & Mahdavi, V. (1988) Proc. Natl. Acad. Sci. USA 85, 339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurihara, Y., Kurihara, H., Suzuki, H., Kodama, T., Maemura, K., Nagai, R., Oda, H., Kuwaki, T., Cao, W. H., Kamada, N., et al. (1994) Nature 368, 703-710. [DOI] [PubMed] [Google Scholar]
- 21.Lortet, S., Zimmer, H. G. & Rossi, A. (1989) J. Cardiovasc. Pharmacol. 14, 707-712. [DOI] [PubMed] [Google Scholar]
- 22.Klein, I. & Ojamaa, K. (2001) N. Engl. J. Med. 344, 501-509. [DOI] [PubMed] [Google Scholar]
- 23.Rossmanith, G. H., Hoh, J. F., Turnbull, L. & Ludowyke, R. I. (1997) J. Physiol. (London) 505, 217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rindt, H., Subramaniam, A. & Robbins, J. (1995) Transgenic Res. 4, 397-405. [DOI] [PubMed] [Google Scholar]
- 25.Wettschureck, N., Rutten, H., Zywietz, A., Gehring, D., Wilkie, T. M., Chen, J., Chien, K. R. & Offermanns, S. (2001) Nat. Med. 7, 1236-1240. [DOI] [PubMed] [Google Scholar]
- 26.Yang, H. T., Sakurai, K., Sugawara, H., Watanabe, T., Norota, I. & Endoh, M. (1999) Br. J. Pharmacol. 126, 1785-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo, S. H. & Lee, C. O. (1999) J. Mol. Cell Cardiol. 31, 631-643. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe, T. & Endoh, M. (1999) Naunyn-Schmiedeberg's Arch. Pharmacol. 360, 654-664. [DOI] [PubMed] [Google Scholar]
- 29.Muller, A., Zuidwijk, M. J., Simonides, W. S. & van Hardeveld, C. (1997) Am. J. Physiol. 272, H1876-H1885. [DOI] [PubMed] [Google Scholar]
- 30.Kiss, E., Jakab, G., Kranias, E. G. & Edes, I. (1994) Circ. Res. 75, 245-251. [DOI] [PubMed] [Google Scholar]
- 31.Kiss, E., Brittsan, A. G., Edes, I., Grupp, I. L., Grupp, G. & Kranias, E. G. (1998) Circ. Res. 83, 608-613. [DOI] [PubMed] [Google Scholar]
- 32.Kim, D., Smith, T. W. & Marsh, J. D. (1987) J. Clin. Invest. 80, 88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, M., Xu, A., Tokmakejian, S. & Narayanan, N. (2000) Am. J. Physiol. 278, H1429-H1438. [DOI] [PubMed] [Google Scholar]
- 34.Hamasaki, Y., Shinohara, O., Ishida, H., Hayashi, Y. & Nakazawa, H. (2000) Life Sci. 67, 1859-1868. [DOI] [PubMed] [Google Scholar]