Abstract
Human papillomavirus type 16 (HPV16) infection is a major risk factor for the development of squamous cell cancers of the cervix and of the head and neck. A major barrier to understanding the progression from initial infection to cancer has been the lack of in vitro models that allow infection, replication, and persistence of the viral genome as an episome in differentiated epithelial cells. To overcome this barrier, we designed an adenoviral delivery vector that contained a full HPV16 genome flanked by LoxP homologous recombination sites and a fluorescent reporter that was expressed only after the HPV genome was excised by Cre recombinase. This system delivered circular HPV16 genomes to cervical epithelial cells and well differentiated human airway epithelia. After delivery, the HPV16 genome replicated and persisted as an episome in cervical keratinocytes. These cells developed an immortalized phenotype and a dysplastic epithelial appearance. Moreover, induction of differentiation led to the expression of late genes and production of infectious HPV16 virions. This work provides a means of introducing biologically active HPV genomes into epithelial cells, which are normally difficult to transfect. These methods allow the study of HPV genome replication and gene expression in the earliest stages of HPV genome establishment, and they may provide a means to study nononcogenic HPV viral types.
Human papillomavirus type 16 (HPV16) is a small double-stranded DNA virus that infects mucosal epithelia (1). It is associated with cervical cancer (2, 3) and respiratory tract papillomatosis, and it is thought to contribute to the oncogenesis of head and neck squamous cell cancers. Of such cancers arising from the tonsil, 60% contain HPV16 (4, 5).
Current understanding of HPV pathogenesis related to cellular and viral gene interactions has come from studies using HPV infected keratinocytes obtained by one of two means. One method uses immortalized cell lines derived from in vivo lesions infected with HPV types 11, 16, 31, or 59 (6-11). A second method uses the transfection of full or partial HPV genomes, including 11, 16, 18, 31, and 45, as well as a chimeric HPV18/16, into low-passage human keratinocytes (12-17). After prolonged passaging of cells transfected with complete oncogenic HPV genomes, isolated immortalized cell lines containing episomal HPV DNA could be obtained. When these HPV-containing immortalized cells were grown under differentiating conditions, they produced infectious particles (6, 8, 12-14).
However, genome transfection methods used to produce infectious HPV particles have limitations. Even under optimal conditions, very few immortalized clones emerge, typically after weeks in culture. In addition, because of the requirement of selection and the relative paucity of HPV-containing cells in these models, it has not been possible to study early events in the viral lifecycle, such as HPV genome establishment and virus-cell interactions before cellular immortalization. Moreover, because cellular immortalization is required and induced by oncogenic HPV types, these methods have not been valuable for nononcogenic types. Finally, current transfection techniques have not been shown to deliver genomes to a differentiated epithelium.
Thus, the understanding of HPV lifecycle, the pathogenesis of HPV-related diseases, and the development of therapies could be enhanced by using an in vitro infection model that would allow for the exploration of events that occur early in the HPV lifecycle and for the study of other oncogenic and nononcogenic HPV types. Therefore, the goal of this study was to develop an in vitro model that would deliver a functional, circular HPV genome to low-passage keratinocytes or a differentiated epithelium.
Materials and Methods
Construction of Ad/HPV16/eGFP. Construction of pCMV-LoxP-HPV16-LoxP-eGFP required multiple cloning steps. A plasmid containing two LoxP (ATAACTTCGTATA ATGTATGC TATACGAAGTTAT) sites separated by an SphI (pLSL) site was constructed. To obtain a full-length HPV16 genome, pHPV16 (HPV16 from W12-E cells; inserted in pUC19 at the BamHI site) (ref. 18; a gift from Margaret Stanley, University of Cambridge, Cambridge) was digested with BamHI. The released linear HPV16 genome was recircularized by ligation; linearized at the SphI site at nucleotide 7,465; and ligated into the SphI site of pLSL such that LoxP sites flanked the HPV16 genome. This plasmid was cut with SacII and HindIII to release the HPV16 DNA flanked by LoxP sites on either side (LoxP-revHPV16-LoxP). This insert was ligated into the NheI and HindIII sites of peGFP-N1, containing the eukaryotic enhanced GFP (eGFP) (CLONTECH). This ligation produced pCMV-LoxP-revHPV16-LoxP-eGFP, which had a cytomegalovirus (CMV) promoter at the 5′ end and eGFP at the 3′ end of an HPV16 genome inserted in reverse orientation to the CMV promoter. pCMV-LoxP-revHPV16-LoxP-eGFP was cut with XhoI and NotI to release the HPV16 flanked insert, and this HPV16 genome was ligated into an adenovirus (Ad) shuttle plasmid to produce pAd/HPV16/eGFP. Direct sequencing was used to ensure fidelity for each step of the cloning process. pAd/HPV16/eGFP was used to produce high titer (1 × 1010 plaque-forming units per ml), replication-defective E1/E3-deleted Ad (19). Because of potential risks of increased infection capability, all experiments that used the Ad/HPV16/eGFP virus were performed in a biosafety level 3 facility. Previously characterized replication-defective Ad stocks expressing Cre recombinase (Ad/Cre) were supplied by the Gene Transfer Vector Core facility (University of Iowa, Iowa City) (20).
Isolation and Growth of Primary Cervical Epithelial Cells. Primary, HPV-negative cervical keratinocytes were collected from a noncancerous routine hysterectomy and cultured on irradiated murine J2 3T3 fibroblast feeder cells in E media containing FBS, as described (21). Keratinocytes were serially passed at a 1:4 split and maintained on fresh irradiated fibroblast feeders in 100-mm dishes containing E medium.
Ad Infections. Cervical keratinocytes at passage 8 were coinfected with Ad/Cre and Ad/HPV16/GFPeGFP at a multiplicity of infection of 25 and 250 plaque-forming units per cell, respectively. Control infections included either Ad/HPV16/GFPeGFP alone or, in certain cases, a coinfection with Ad/LacZ and Ad/HPV16/GFPeGFP. As an additional uninfected control, identical passage 8 keratinocytes were serially passed 1:4 to determine normal life span in culture before senescence. Cultures were serially passed 1:4 when they became ≈80% confluent.
Differentiated airway epithelia were infected as described (22). Recombinant adenoviral stocks Ad/Cre and Ad/HPV16/eGFP were added to basolateral surfaces by inverting the epithelia and applying 50 and 100 plaque-forming units per cell, respectively, in a volume of 25 μl to the bottom of the filter for 1 h. Images were obtained 16 h later on an inverted fluorescent microscope (Nikon).
Quantitative PCR. Total HPV16 genomes, whether linear or circular, were quantified with the primers and probes described in ref. 23. The primers used to quantify recombined genomes were 5′-CAGTGCAGGTCAGGAAAACA-3′ and 5′-AGCGGCCATTTTGTAGCTT-3′, and the probe was 5′-TTCCTGCTTGCCATGCGTGC-3′. Ad genomes were quantified by using primers 5′-GCCGCTGCCCTGATAACA-3′ and 5′-ACTCGCAGAACGAATGTGTAGG-3′ and the probe 5′-CGCAGAATAAGCCACACCCAGCCA-3′. For quantification of the L1 transcript, we used the primers and probes described in ref. 24.
To create a standard curve of recombined HPV16 genomes, pAd/HPV16/GFPeGFP was exposed to Cre recombinase overnight, quantified by using spectrophotometry, and serially diluted. A standard curve was created for Ad type 5 (Ad5) by serially diluting a known quantity of isolated Ad5 genomes. Total DNA from cell samples was prepared by using a PureGene DNA isolation kit (Gentra Systems). Amplification reactions (25 μl total volume) contained TaqMan Universal PCR master mix or TaqMan One-Step RT-PCR master mix (Applied Biosystems), 900 nM primers, 200 nM probe, and 200 ng of template nucleic acid. Preincubation PCR conditions were 50°C for 2 min and 95°C for 10 min. Cycling conditions were 95°C for 15 sec and 60°C for 1 min for a total of 40 cycles. Reverse transcription of RNA included an additional initial incubation at 48°C for 30 min with Moloney murine leukemia virus reverse transcriptase. Internal controls for sample variation used for RNA-expression analysis included amplification of and standardization to GAPDH. Samples for each experiment were tested in triplicate, and results shown are averages obtained from all points in repeated experiments.
Total and Hirt Supernatant DNA Extraction and Analyses. Cellular DNA (3 μg) was digested with BamHI, separated by electrophoresis on a 1% agarose gel, transfered onto a Zeta Probe membrane (Bio-Rad), and hybridized with a radioactively labeled, equimolar mixture of four PCR fragments spanning the HPV16 genome. Extraction of low-molecular-weight DNA was performed on 107 cells by following a modified Hirt protocol (25). Radiographic images were obtained by using a Storm phosphorimager (Molecular Dynamics).
Organotypic (Raft) Epithelial Tissue Culture for Virion Production, Virion Quantification, and Experimental Infections. Epithelial organotypic (raft) tissue cultures for in vitro differentiation were maintained as described (26, 27). Virus particles were extracted from 14-day epithelial raft tissues by using a modified protocol of Favre et al. (28, 29). To quantify the number of viral DNA-containing particles in the extracts, 20 and 40 μl of each extract or purified, cloned vDNA copy number controls were subjected to dot blot hybridization as described (28). HaCaT cells (a generous gift from N. Fusenig, Deutsche Krebsforschungszentrum, Heidelberg, Germany) are a spontaneously immortalized epithelial cell line established from normal adult skin (30). HaCaT cells were seeded at 60-80% confluence in 4 cm2. Virion stocks were thawed at room temperature and sonicated for 20-30 sec at 0°C. Virus dilutions were added to each well in 0.25 ml of HaCaT medium, and plates were rocked 1 h at 4°C. Unbound viral inocula were removed from the cells by washing an excess of normal media, refed, and moved to 37°C.
RNA Extraction and RT-PCR Analyses of HaCaT HPV16 Infections. Total RNAs were extracted by using TRIzol reagent (Invitrogen) and reverse-transcribed by using random hexamer primers, and PCR was performed by using a GeneAmp RNA PCR kit and an AmpliTaq Gold DNA polymerase with 2.5 mM MgCl2 (Applied Biosystems).
For the first round of amplification of E1^E4 cDNAs, the primers were 5′-ACA AGCAGA ACCGGAC-3′ and 5′-CTCTGATCTTGGTCGCTG-3′. For nested PCR, the primers were 5′-CGCTTCGGTTGTGCGTAC-3′ and 5′-TTTGGTATGGGTCGCGGC-5′. The annealing temperatures were, e.g., 57°C and 62°C, respectively. Primers for cellular β-actin were described (6, 31). The thermocycling profile was 10 min at 95°C; 50 cycles of 95°C for 45 sec, 57°C or 62°C for 30 sec, and 72°C for 30 sec; and 7-min extension at 72°C.
Results
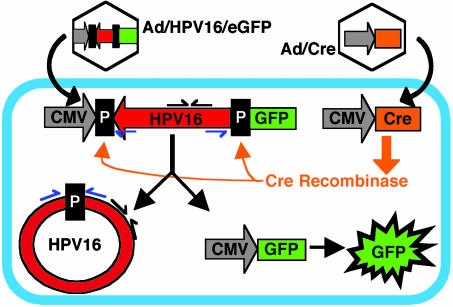
Delivery, Replication, and Persistence of Episomal HPV16 Genomes in Cervical Keratinocytes. We designed a strategy to deliver episomal HPV16 genomes to cells. Fig. 1 illustrates this strategy schematically. For a vector, we used E1/E3 replication-defective Ad5. The vector contained the full-length HPV16 genome flanked by LoxP recombination sites inserted between a CMV promoter and the cDNA for eGFP (Ad/HPV16/eGFP). The HPV16 genome was inserted in reverse orientation to the CMV promoter to avoid CMV-driven expression of HPV16 genes. Also, we used a replication-defective Ad expressing Cre recombinase under control of the CMV promoter (Ad/Cre). Cre recombinase excises and induces homologous recombination of the intervening DNA sequence between two LoxP recognition sites (32, 33). The Cre/LoxP system has been used extensively to produce controlled and reliable recombination events (34).
Fig. 1.
Strategy to deliver the HPV16 genome to cells by using an Ad and Cre/LoxP system. Homologous recombination catalyzed by Cre recombinase should result in cells that contain a circular HPV16 genome and express eGFP. LoxP sites (P) flank the SphI site at nucleotide 7,465 in the HPV16 LCR. PCR primers designed to detect recombined HPV16 are indicated by blue arrows, and nonrecombination sensitive primers are shown as black arrows.
To ascertain whether our strategy would allow the generation, replication, and persistence of episomal HPV16 genomes, we infected HPV-negative, low-passage cervical epithelial cells with both Ad/HPV16/eGFP and Ad/Cre. We found, 3 days later, that ≈5-10% of cells were eGFP-positive in cultures infected simultaneously with Ad/HPV16/eGFP and Ad/Cre but not when Ad/LacZ substituted for Ad/Cre (Fig. 2). These data indicated that recombination of the Ad/HPV16/eGFP yielded eGFP expression. The presence of the viral genome was assayed by using quantitative PCR with primer sets that could detect the HPV16 genome in any form (non-recombination-sensitive primers) or the homologously recombined circular HPV16 genome (recombination-sensitive primers) (Fig. 1). We detected, 2 days after infection, HPV16 DNA in cells infected with Ad/HPV16/eGFP, but only cells also coinfected with Ad/Cre contained recombined HPV16 genomes (Table 1).
Fig. 2.
Intracellular recombination of Ad/HPV16/eGFP in human cervical cells. Cells were mock infected with media alone (A), coinfected with Ad/LacZ and Ad/HPV16/eGFP (B), or coinfected with Ad/Cre and Ad/HPV16/eGFP (C). Images were obtained by fluorescence microscopy (×40 magnification) when the cells reached confluence at 3 days later. Images are representative of an experiment performed in triplicate and repeated multiple times.
Table 1. Relative copy number per cell of HPV and adenoviral genomes in cervical keratinocytes quantified by PCR.
| Day
|
|||||
|---|---|---|---|---|---|
| 2 | 15 | 30 | 40 | 60 | |
| Ad/HPV16EGFP alone* | 0.7 | 0 | 0 | ND | ND |
| Ad/HPV16EGFP + Ad/Cre* | 3.6 | 11 | 55 | 88 | 84 |
| Ad/HPV16EGFP alone† | 0 | 0 | 0 | ND | ND |
| Ad/HPV16EGFP + Ad/Cre† | 0.12 | 1.8 | 52 | 58 | 55 |
| Ad/HPV16EGFP alone‡ | 6.5 | 0.1 | 0 | ND | ND |
| Ad/HPV16EGFP + Ad/Cre‡ | 17.1 | 0.88 | 0 | ND | ND |
Passage 8 keratinocytes were coinfected with Ad/HPV16/eGFP and Ad/Cre or Ad/HPV/eGFP alone. On the indicated days, total cellular DNA was isolated. The number of viral genomes per cell was calculated by completing quantitative PCR on identical quantities of cellular DNA. The values indicate the average of samples run in triplicate. Similar results were obtained on samples from a repeated experiment. ND, not done because the cells infected only with Ad/HPV16/eGFP, senesced, and could not be passed.
Non-recombination-sensitive primers.
Recombination-sensitive primers.
Ad5 genome primers.
Episomal HPV16 genomes in cervical cells are expected to persist if the HPV genome replicates. To test for persistence, we passaged the cells and performed quantitative PCR by using the two different HPV16 primer sets. Table 1 shows that in cells infected with Ad/HPV16/eGFP alone, HPV16 DNA was no longer detected by day 15. Consistent with the disappearance of the nonrecombined HPV16 genome, quantitative PCR showed that adenoviral DNA also was lost from the cells. In contrast, HPV genomes persisted when Cre recombinase was delivered. By 30-60 days after infection (after five to nine passages at a 1:4 split), we detected 50-80 copies of the HPV16 genomes per cell. Even at passage 44, the copies per cell remained in the same range.
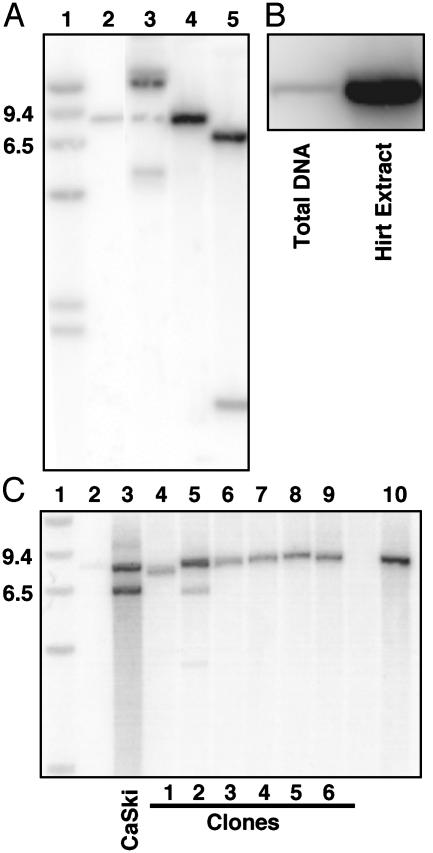
Southern blot analysis of restriction digests of DNA extracted from cells 44 days after infection suggested that the HPV16 genomes were maintained episomally (Fig. 3A). The uncut HPV16 DNA showed a pattern of open circle, nicked circular, and supercoiled DNA forms expected for episomal genomes (Fig. 3A, lane 3) (35). Digestion with BamHI (lane 4) or SphI plus BamHI (Fig. 3A, lane 5), each of which cut the HPV16 genome once, produced the results predicted for an episomal genome. To assess the physical state of the HPV16 DNA further, we compared total and Hirt supernatant DNA; the Hirt procedure preferentially isolates small extra-chromosomal DNA (25). Equal amounts of total and Hirt supernatant DNA were analyzed. The HPV16 genomes were highly enriched in the Hirt preparation, indicating that HPV16 DNA persisted predominantly in an episomal form (Fig. 3B).
Fig. 3.
Southern blot hybridization of total and Hirt supernatant DNA extracted from cervical cells containing HPV16. (A) Establishment of episomal HPV16 genomes in cervical cells. Southern blot of restriction enzyme digests of total cellular DNA harvested 44 days after infection with Ad/HPV16/eGFP and Ad/Cre. Lane 1, 9.4- and 6.5-kb molecular weight markers. Lane 2, 3 pg of BamHI linearized HPV16 DNA. Lanes 3-5, total DNA from HPV16-containing cervical cells digested with PacI (no restriction sites in HPV16), BamHI (one restriction site in HPV16), and SphI plus BamHI (one site each at nucleotides 7,465 and 6,152), respectively. The majority of the uncut DNA form in lane 3 is present as an open circular DNA form, which is attributed to the DNA isolation technique because the amount of each DNA form varied among isolations (data not shown). (B) DNA from cervical cells isolated as total DNA or as a Hirt supernatant, which enriches episomal DNA. Equal quantities of DNA were digested with BamHI and loaded in each lane. (C) Assay for integration of HPV16 in isolated cell clones. Lanes 2 and 10 show 3 and 30 pg of linearized HPV16 DNA, respectively, as described in A. Lane 3-9, BamHI restriction enzyme digests of total cellular DNA isolated from CaSki cells (a positive control for integrated HPV16 DNA) and compared with DNA from single-cell clones 1-6.
As an additional assessment of episomal HPV16 persistence vs. integration, we expanded six single-cell clones from the original coinfected population after eight cell passages. To make certain we could detect a single integration event, we loaded 3 pg of linearized HPV16 genomes (half of the expected copy number for a single integration event) as a control. Total DNA was isolated from the clones and subjected to BamHI digestion. Southern blot hybridization revealed a single band for five of the six clones (Fig. 3C). A multiple-band pattern was detected only for clone 2, consistent with the pattern of integrated viral genomes observed in DNA from the CaSki cervical carcinoma cell line (Fig. 3C). Thus, these data suggest that although the HPV16 genome persists episomally in clonal cell lines, integration can occur.
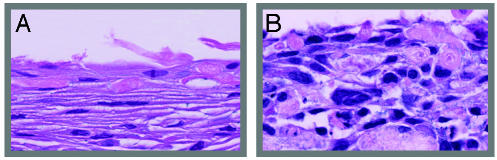
HPV16-Dependent Immortalization and Dysregulated Epithelial Differentiation. Studies have shown that after transfection of genomes from oncogenic HPV types 18, 31, and 45, cells developed an immortal phenotype with continuous cell passage (12, 13, 15, 16). Similarly, the cervical epithelial cells infected simultaneously with Ad/HPV16/eGFP and Ad/Cre could be passaged for multiple generations (Table 1). To date, the lines have been in culture for >100 cell doublings without senescence. In contrast, uninfected cells and cells infected with Ad/HPV16/eGFP alone underwent senescence 10 cell doublings after infection. In vivo HPV16 infection is associated with altered epithelial morphology (36), and in vitro transfection of other oncogenic HPV types to anogenital keratinocytes causes dysplastic cell growth (12, 13, 15, 16). Compared with uninfected low-passage cells, we found that cervical keratinocytes containing episomally replicating HPV16 genomes developed a strikingly dysplastic morphology compared with HVP16-negative cells when grown as organotypic tissues (Fig. 4). Thus, delivery of the episomal HPV16 genome induced both immortal and dysplastic phenotypes.
Fig. 4.
Dysregulated epithelial organotypic tissue growth after HPV16 genome delivery to anogenital keratinocytes. (A) Uninfected low-passage anogenital keratinocytes. (B) Cervical keratinocytes (C-100) infected with Ad/HPV16/eGFP and Ad/Cre and grown as an epithelium at passage 20 (×40 magnification).
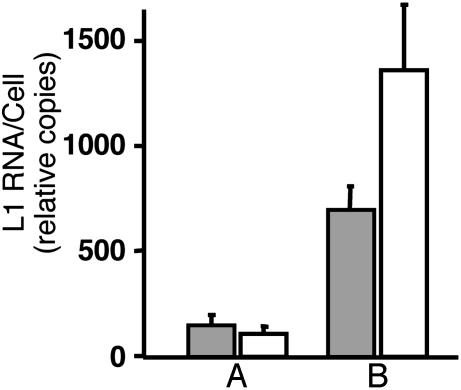
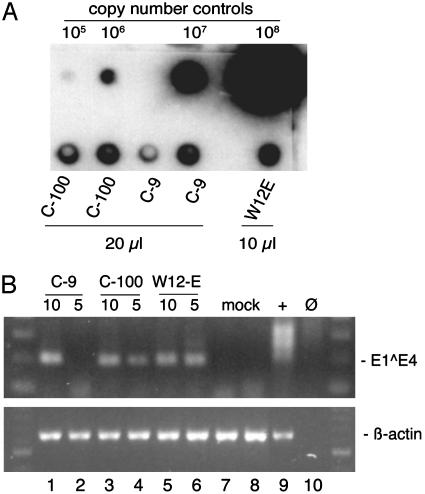
HPV16 Late Gene Expression and Virion Production in Cells Grown as Organotypic Tissues. In differentiated epithelia, HPV16 expresses late viral genes and produces virions (36). To test whether differentiation would up-regulate late gene expression and result in viral production, we grew the HPV16-infected cervical epithelial cells (C-100) and one of the isolated cell clones (C-9) for 2 weeks as organotypic epithelial raft tissues. Quantitative RT-PCR of the viral L1 gene showed a 5- to 10-fold increase in L1 transcripts compared with undifferentiated monolayer cells (Fig. 5). To test for virion production, the HPV16-containing cervical epithelial cells and one of the clones were cultured for 14 days as raft tissues. Virions were extracted from organotypic tissues and quantified by dot blot hybridization to determine the number of viral genome equivalents (vge) in each preparation (6, 28). The cervical epithelial cell population C-100 produced a mean of 4.97 ± 0.70 × 106 vge per raft, and clone C-9 produced 3.58 ± 1.40 × 106 vge per raft (n = 2 for each cell line; Fig. 6A). The number of HPV16 vge per raft was similar to that obtained from W12-E rafts grown from a cell line derived from a cervical biopsy (ref. 18; M.A.O., unpublished data).
Fig. 5.
Differential expression of L1 transcripts in HPV16-containing cell lines grown as undifferentiated monolayers or as differentiated organotypic epithelial tissues. Total RNAs were isolated and HPV16 L1 RNAs were quantified by using RT-PCR and real-time quantitative PCR. HPV16 L1 transcripts were normalized to those of GAPDH. L1 RNA quantities are shown for HPV16 infected cervical keratinocytes at similar passage. Shaded bars represent copies of L1 in a clonal population (C-9), and unshaded bars represent L1 in total population (C-100). (A) RNA isolated from undifferentiated cells grown as a monolayer. (B) RNA isolated from cells grown as differentiated epithelial raft tissues.
Fig. 6.
Quantification of virion production and analysis of reinfection in HaCaT cells. (A) Virions were extracted from organotypic tissues and analyzed by dot blot hybridization. Copy number control HPV16 DNA from cloned genomes ranging from 105 to 108 vge and 10 or 20 μl of virion preparations were denatured with sodium hydroxide, transferred to a nylon membrane, and then hybridized with a radioactively labeled HPV16 genome probe and exposed to a phosphorimage screen for quantification. (B) HPV16 virions isolated from a cloned line (C-9), a total cell population (C-100), and W12-E cells grown as raft tissues were used to infect HaCaT cells. The HaCaT cells were harvested 2 days after infection. Total RNAs (3 μg) were reverse-transcribed. (Upper) RNAs were analyzed from W12-E cells known to contain E1^E4 message (+, lane 9); mock-infected HaCaT cells (mock, lanes 7-8); HaCaT cells infected with a multiplicity of infection corresponding to ≈10 and 5 vge per cell, respectively, from C-9 (lanes 1-2); C-100 (lanes 3-4); and W12-E (lanes 5-6). No RNA input (Ø, lane 10) served as a negative amplification control. (Lower) β-actin primers were used to detect a 641-bp amplimer derived from spliced β-actin RNA in one round of PCR. The input RNA corresponded to 2.7 μg for HPV16 E1^E4 and 0.3 μg for β-actin. Molecular size standards (100-bp ladder) are shown in the lateral lanes.
We tested whether these isolated HPV16 virions were infectious by incubating the preparations with a subconfluent monolayer of HaCaT cells. Previous studies used RT-PCR detection of newly synthesized spliced viral RNAs to demonstrate that HaCaT cells are susceptible to infection by HPV types 11, 16, and 31 (6, 31, 37). Infectious virions extracted from W12-E raft tissue were included as a positive control. At 2 days after infection, we detected spliced HPV16 E1^E4 transcripts in the cells infected with each of the HPV16 virion preparations (Fig. 6B). Sequencing verified that the amplimers were derived from the spliced E1/E4 message of HPV16. These results confirm that epithelial differentiation induced transcription of late genes and these differentiated epithelia produced infectious viral particles.
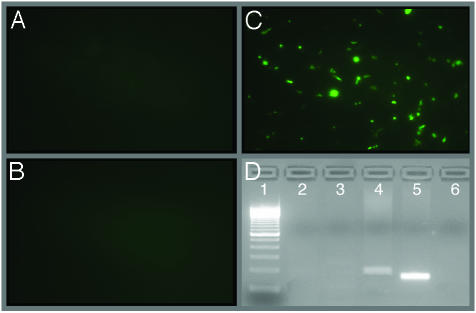
Delivery of Circular HPV16 Genomes to Well Differentiated Human Airway Epithelia. HPV16 causes disease in airway epithelia also (38-40). Although variation in expression patterns and latent infections have been reported (41), relatively little is known regarding HPV pathogenesis in this pseudostratified ciliated epithelia. An in vitro method to deliver HPV to differentiated airway epithelia could facilitate investigation of the pathogenesis. We asked whether our method would deliver circular HPV to airway epithelia. We studied primary cultures of human airway epithelial cells grown at the air-liquid interface. After 2 weeks, the epithelia develop a ciliated morphology, differentiation-dependent markers, a transepithelial resistance, and polarized distribution of receptors that is similar to in vivo airway epithelia (22). These differentiated epithelia were coinfected with Ad/HPV16/eGFP and Ad/Cre. As controls, the epithelia were either infected with Ad/HPV16/eGFP alone or coinfected with Ad/HPV16/eGFP and Ad/LacZ. At 16 h after infection, ≈8% of the coinfected cells were positive for eGFP fluorescence (Fig. 7C); however, no green cells were observed in the control groups (Fig. 7 A and B). These data suggest that there was no expression of eGFP from the CMV promoter without recombination and that recombination did not occur in the absence of Cre recombinase. Recombination sensitive PCR and sequence verification of the PCR amplicons confirmed that accurate recombination occurred over the LoxP sites only in the cells infected with Ad/Cre and Ad/HPV16/eGFP simultaneously (Fig. 7D). Thus, the Ad system successfully delivered circular HPV16 to differentiated airway epithelia. Long-term studies examining viral message expression and changes in cellular morphology in these cells remain to be studied.
Fig. 7.
Intracellular recombination of Ad/HPV16/eGFP in differentiated human airway epithelia. Epithelia were infected with Ad/HPV16/eGFP (A), Ad/HPV16/eGFP plus Ad/LacZ (B), or Ad/HPV16/eGFP plus Ad/Cre (C). Images (×10 magnification) are representative of triplicate infections, with the experiment repeated multiple times. (D) PCR results demonstrating intracellular recombination of Ad/HPV16/eGFP. PCR amplification was performed over the recombination site for HPV. The 100-bp ladder (lane 1), uninfected HAE control (lane 2), HAE infected with Ad/HPV16/eGFP alone (lane 3), HAE infected with Ad/HPV16/eGFP and Ad/Cre (lane 4; amplimer is 35-bp larger because it contains the LoxP sequence as compared with the cloned HPV16 positive control), cloned HPV16 DNA as a positive control (lane 5), and no template control (lane 6).
Discussion
The Ad Cre/LoxP system for HPV genome delivery described here provides a method for initiating HPV viral synthesis, especially for HPV16. Studies using transfection techniques have been invaluable for our understanding of HPV pathogenesis. Our method of adenoviral delivery has the potential to augment this knowledge in several ways. First, recombinant, replication defective Ads are easy to construct, grow, and purify to high titers, and we anticipate that this system will be useful for other HPV types for which cloned viral genomes are available. Second, in many cases, recombinant Ads can be used to deliver genes to cells that are difficult to transfect. For example, we were able to detect eGFP expression indicating HPV16 delivery and homologous recombination in 5-10% of the cervical epithelial cells (Fig. 2C), whereas typical stable transfections yielded <0.01% of cells containing episomal HPV genomes (M.A.O. and J.H.L., unpublished observations). Also, we have found that airway epithelia are very resistant to transfection. For example, our initial attempts using multiple transfection techniques to differentiated epithelia yielded no evidence of HPV delivery, but our adenoviral-shuttling mechanism delivered a recombined genome to ≈8% of cells. Third, because expression of eGFP coincides with HPV16 recombination, this model will allow separation of HPV16-infected cells from noninfected cells by fluorescence-activated cell sorting. Such a model will be valuable for obtaining a large population of cells immediately after the delivery of an HPV16 episomal genome. These cells could possibly be used to study the events that are important for the initial establishment of HPV episomal genomes. Fourth, current DNA transfection methods cannot analyze the establishment of HPV genomes because they rely on selection with an antibiotic resistance gene and outgrowth of very few immortalized clones. Expansion of sufficient transfected cells for passing and analysis after DNA transfection and selection takes weeks. Our method could possibly circumvent this limitation. Finally, because we can isolate a large population of cells immediately after the viral gene is recombined, we expect this Ad-mediated method will be also particularly useful for studying the life cycles of HPV types that are not able to immortalize or provide significant growth advantage to cells.
A potential limitation of our strategy was the insertion of the 35-bp LoxP site into the HPV16 genome. We strove to use a region of the viral genome that would be least likely to alter the normal viral life cycle in vitro. The HPV long control region (LCR) is known to play an important role in the viral replicative cycle (42-49). However, we modified an area of the LCR that has not been implicated in the viral replicative cycle and frequently shows sequence variation in clinical isolates (50-52), suggesting that it can tolerate change without altering function. Our data indicate that the LoxP insertion likely did not alter the function of the virus in a detectable manner. The genital epithelial cells containing the HPV16 genome with the insertion of the LoxP site in the LCR behaved in a manner consistent with genital epithelial cells containing wild-type genomes of HPV16, 18, and 31: they maintained episomal genomes, showed dysplastic growth in organotypic culture, had differentiation-dependent expression of late genes, and produced infectious virions on epithelial differentiation.
In summary, the strategy we describe offers opportunities to better understand viral synthesis and episomal persistence of HPV16, which has been particularly difficult to cultivate and study in vitro. It may also provide a better understanding of the pathogenesis of HPV16 and other medically important HPV infections. Finally, by providing a model that may mimic in vivo infections, it may be of value in developing therapeutic options.
Acknowledgments
We thank Nicole A. Ruple and the Airway Cell Culture Core for excellent technical assistance; members of the Lee, Welsh, Klingelhutz, and Ozbun laboratories, as well as Drs. Cosette M. Wheeler, Lubomir Turek, and Tom Haugen for helpful discussions and critical comments on the manuscript; Norbert Fusenig for the HaCaT cells; and Margaret Stanley for the W12 cells. We also thank the In Vitro Models and Cell Culture Core, supported by National Institutes of Health, National Heart, Lung, and Blood Institute Grants HL51670, HL61234, and DK54759. This work was supported by National Institutes of Health Career Development Award K08 DC005627-01 and a Parker B. Francis Fellowship Award (to J.H.L.); National Institutes of Health Public Health Service Grants CA-85747 and AI-052049 (to M.A.O.); National Institutes of Health Grant R01AG18265 (to A.J.K.); National Institutes of Health Training Grant T32AI07533 (to K.L.B.); and National Institutes of Health Research Training Program for Otolaryngology Grant T32DC000040 (to S.M.P.Y.). M.J.W. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: Ad, adenovirus; CMV, cytomegalovirus; eGFP, enhanced GFP; HPV16, human papillomavirus type 16; LCR, long control region; vge, viral genome equivalents.
References
- 1.Laimins, L. (1993) Infect. Agents Dis. 2, 74-86. [PubMed] [Google Scholar]
- 2.Zur Hausen, H. (2000) J. Natl. Cancer Inst. 92, 690-698. [DOI] [PubMed] [Google Scholar]
- 3.Walboomers, J. M. M., Jacobs, M. V., Manos, M. M., Bosch, F. X., Kummer, J. A., Shah, K. V., Snijders, P. J. F., Peto, J., Meijer, C. J. L. M. & Muñoz, N. (1999) J. Pathol. 189, 12-19. [DOI] [PubMed] [Google Scholar]
- 4.Smith, E. M., Pignatari, S. S., Gray, S. D., Haugen, T. H. & Turek, L. P. (1993) Arch. Otolaryngol. Head Neck Surg. 119, 554-557. [DOI] [PubMed] [Google Scholar]
- 5.McKaig, R. G., Baric, R. S. & Olshan, A. F. (1998) Head Neck 20, 250-265. [DOI] [PubMed] [Google Scholar]
- 6.Ozbun, M. A. (2002) J. Virol. 76, 11291-11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyers, C., Frattini, M. G., Hudson, J. B. & Laimins, L. A. (1992) Science 257, 971-973. [DOI] [PubMed] [Google Scholar]
- 8.Lehr, E. E., Qadadri, B., Brown, C. R. & Brown, D. R. (2003) Virology 314, 562-571. [DOI] [PubMed] [Google Scholar]
- 9.Sterling, J., Stanley, M., Gatward, G. & Minson, T. (1990) J. Virol. 64, 6305-6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreider, J. W., Howett, M. K., Leure-Dupree, A. E., Zaino, R. J. & Weber, J. A. (1987) J. Virol. 61, 590-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnez, W., DaRin, C., Borkhuis, C., de Mesy Jensen, K., Reichman, R. C. & Rose, R. C. (1998) J. Virol. 72, 5256-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLaughlin-Drubin, M. E., Wilson, S., Mullikin, B., Suzich, J. & Meyers, C. (2003) Virology 312, 1-7. [DOI] [PubMed] [Google Scholar]
- 13.Meyers, C., Mayer, T. J. & Ozbun, M. A. (1997) J. Virol. 71, 7381-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyers, C., Bromberg-White, J. L., Zhang, J., Kaupas, M. E., Bryan, J. T., Lowe, R. S. & Jansen, K. U. (2002) J. Virol. 76, 4723-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frattini, M. G., Lim, H. B. & Laimins, L. A. (1996) Proc. Natl. Acad. Sci. USA 93, 3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frattini, M. G., Lim, H. B., Doorbar, J. & Laimins, L. A. (1997) J. Virol. 71, 7068-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mungal, S., Steinberg, B. M. & Taichman, L. B. (1992) J. Virol. 66, 3220-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanley, M. A., Browne, H. M., Appleby, M. & Minson, A. C. (1989) Int. J. Cancer 43, 672-676. [DOI] [PubMed] [Google Scholar]
- 19.Anderson, R. D., Haskell, R. E., Xia, H., Roessler, B. J. & Davidson, B. L. (2000) Gene Ther. 7, 1034-1038. [DOI] [PubMed] [Google Scholar]
- 20.Stec, D. E., Davisson, R. L., Haskell, R. E., Davidson, B. L. & Sigmund, C. D. (1999) J. Biol. Chem. 274, 21285-21290. [DOI] [PubMed] [Google Scholar]
- 21.Sprague, D. L., Phillips, S. L., Mitchell, C. J., Berger, K. L., Lace, M., Turek, L. P. & Klingelhutz, A. J. (2002) Virology 301, 247-254. [DOI] [PubMed] [Google Scholar]
- 22.Karp, P. H., Moninger, T. O., Weber, S. P., Nesselhauf, T. S., Launspach, J. L., Zabner, J. & Welsh, M. J. (2002) Methods Mol. Biol. 188, 115-137. [DOI] [PubMed] [Google Scholar]
- 23.Tucker, R. A., Unger, E. R., Holloway, B. & Swan, D. (2001) Mol. Diagn. 6, 39-47. [DOI] [PubMed] [Google Scholar]
- 24.Lanham, S., Herbert, A. & Watt, P. (2001) J. Clin. Pathol. 54, 304-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen, A. P., Reid, R., Campion, M. & Lorincz, A. T. (1991) J. Virol. 65, 606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gewin, L. & Galloway, D. A. (2001) J. Virol. 75, 7198-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCance, D. J., Kopan, R., Fuchs, E. & Laimins, L. A. (1988) Proc. Natl. Acad. Sci. USA 85, 7169-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozbun, M. A. (2002) J. Gen. Virol. 83, 2753-2763. [DOI] [PubMed] [Google Scholar]
- 29.Favre, M. (1975) J. Virol. 15, 1239-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boukamp, P., Petrussevska, R. T., Breitkreutz, D., Hornung, J., Markham, A. & Fusenig, N. E. (1988) J. Cell Biol. 106, 761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, L. H., Foster, C., Hitchcock, M. E., Leiserowitz, G. S., Hall, K., Isseroff, R., Christensen, N. D. & Kreider, J. W. (1995) J. Invest. Dermatol. 105, 438-444. [DOI] [PubMed] [Google Scholar]
- 32.Hasan, N., Koob, M. & Szybalski, W. (1994) Gene 150, 51-56. [DOI] [PubMed] [Google Scholar]
- 33.Abremski, K., Hoess, R. & Sternberg, N. (1983) Cell 32, 1301-1311. [DOI] [PubMed] [Google Scholar]
- 34.Sauer, B. (2002) Endocrine 19, 221-228. [DOI] [PubMed] [Google Scholar]
- 35.Wettstein, F. O. & Stevens, J. G. (1982) Proc. Natl. Acad. Sci. USA 79, 790-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howley, P. (1990) Fields Virology (Raven, New York), 2nd Ed., pp. 1625-1676.
- 37.White, W. I., Wilson, S. D., Bonnez, W., Rose, R. C., Koenig, S. & Suzich, J. A. (1998) J. Virol. 72, 959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore, C. E., Wiatrak, B. J., McClatchey, K. D., Koopmann, C. F., Thomas, G. R., Bradford, C. R. & Carey, T. E. (1999) Otolaryngol. Head Neck Surg. 120, 698-705. [DOI] [PubMed] [Google Scholar]
- 39.Mounts, P. & Kashima, H. (1984) Laryngoscope 94, 28-33. [DOI] [PubMed] [Google Scholar]
- 40.Mounts, P., Shah, K. V. & Kashima, H. (1982) Proc. Natl. Acad. Sci. USA 79, 5425-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maran, A., Amella, C. A., Di Lorenzo, T. P., Auborn, K. J., Taichman, L. B. & Steinberg, B. M. (1995) Virology 212, 285-294. [DOI] [PubMed] [Google Scholar]
- 42.Chan, W.-K., Klock, G. & Bernard, H.-U. (1989) J. Virol. 63, 3261-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gloss, B., Chong, T. & Bernard, H.-U. (1989) J. Virol. 63, 1142-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fontaine, V., Van der Meijden, E., De, G. J., Ter, S. J. & Struyk, L. (2000) Virology 272, 40-49. [DOI] [PubMed] [Google Scholar]
- 45.Smola-Hess, S., De Silva, U. S., Hadaschik, D. & Pfister, H. J. (2001) J. Gen. Virol. 82, 2335-2339. [DOI] [PubMed] [Google Scholar]
- 46.Sibbit, G. J., Cuthill, S. & Campo, M. S. (1995) J. Virol. 69, 4006-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Carranca, A., Thierry, F. & Yaniv, M. (1988) J. Virol. 62, 4321-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fergusson, D. & Campo, M. S. (1998) J. Gen. Virol. 79, 2753-2760. [DOI] [PubMed] [Google Scholar]
- 49.Chen, Y. H., Huang, L. H. & Chen, T. M. (1996) Biochem. Biophys. Res. Commun. 224, 651-659. [DOI] [PubMed] [Google Scholar]
- 50.Smits, H. L., Traanberg, K. F., Krul, M. R. & al., e. (1994) J. Gen. Virol. 75, 2457-2462. [DOI] [PubMed] [Google Scholar]
- 51.Chen, Z., Storthz, K. A. & Shillitoe, E. J. (1997) 57, 1614-1619. [PubMed]
- 52.Tornesello, M. L., Buonaguro, F. M., Meglio, A., Buonaguro, L., Beth-Giraldo, E. & Giraldo, G. (1997) J. Gen. Virol. 78, 2199-2208. [DOI] [PubMed] [Google Scholar]