Abstract
The outer capsid layer of Bluetongue virus, a member of the nonenveloped Reoviridae family, is composed of two proteins, a receptor-binding protein, VP2, and a second protein, VP5, which shares structural features with class I fusion proteins of enveloped viruses. In the replication cycle of Bluetongue virus VP5 acts as a membrane permeabilization protein that mediates release of viral particles from endosomal compartments into the cytoplasm. Here, we show that VP5 can also act as a fusion protein and induce syncytium formation when it is fused to a transmembrane anchor and expressed on the cell surface. Fusion activity is strictly pH-dependent and is triggered by short exposure to low pH. No cell-cell fusion is observed at neutral pH. Deletion of the first 40 amino acids, which can fold into two amphipathic helices, abolishes fusion activity. Syncytium formation by VP5 is inhibited in the presence of VP2 when it is expressed in a membrane-anchored form. The data indicate an interaction between the outer capsid protein VP2 and VP5 and show that VP5 undergoes pH-dependent conformational changes that render it capable of interacting with cellular membranes. More importantly, our data show that a membrane permeabilization protein of a nonenveloped virus can evolve into a fusion protein by the addition of an appropriate transmembrane anchor. The results strongly suggest that the mechanism of membrane permeabilization by VP5 and membrane fusion by viral fusion proteins require similar structural features and conformational changes.
Virus entry into the host cells involves a number of discrete steps, which result, ultimately, in the release of the viral genome into the cytosol. For the majority of viruses, the initial stage of the entry process is the binding of a viral attachment protein to a generalized receptor such as heparin sulfate, followed by interaction with a particular host cell receptor. For enveloped viruses, membrane fusion between the viral and cellular membranes occurs after receptor docking and before the virus core formally penetrates the cell. Envelope glycoproteins are typically synthesized as “inactive” precursors that undergo proteolytic cleavage to become fully active. After receptor interaction, a conformational change, sometimes pH-triggered, is necessary to expose a hydrophobic “fusion peptide,” which is able to interact with the cell membrane and mediate membrane fusion (1-4). Fusion appears to be driven, in many cases, by a coiled-coil structure intimately involved with the conformational changes that accompany the process (5-7). Whereas a detailed understanding of the entry process and fusion mechanisms are available for many enveloped viruses, much less is known on the entry processes of viruses lacking an envelope. Because these viruses have no virion membrane, the entry mechanism cannot involve membrane-membrane fusion. The penetration proteins of these viruses either possess a myristoyl group at their N terminus and/or undergo autolytic cleavage, as well as structural and conformational changes to trigger cell permeabilization (8-15). However, neither the nature of the conformational change nor the exact process of membrane interaction for these viruses is clear. We have initiated a study of the entry mechanism of the nonenveloped Bluetongue virus (BTV), an Orbivirus within the family Reoviridae. Like other members of the family, BTV consists of two concentric capsids enclosing the viral double-stranded RNA genome. The outer capsid is made up of two major proteins of virions, VP2 and VP5, which are responsible for virus entry process. The outer capsid in turn encapsidates the internal capsid, or core, which is composed of two major proteins (VP3 and VP7) and three minor proteins (VP1, VP4, and VP6), in addition to the genome of 10 segments of double-stranded RNA (16-18). The 110-kDa VP2 is the outermost viral protein and has a sail-shaped protruding triskelion spike-like structure (19, 20). It is the cellular receptor binding protein, elicits virus-neutralizing antibody, and is responsible for hemagglutination activity and serotype specificity (21-26). In contrast to VP2, much less is known of the role of the closely associated 60-kDa, VP5 protein, the inner adjacent layer to VP2, in virus infection. VP5 has an overall globular configuration and is less exposed than VP2 on the surface of the particles (19, 20). From our previous studies it appears that VP5 is likely to be involved in cell permeabilization and translocation of the internal core into the cytosol of the infected cells (27). However, VP5 does not have an N-terminal myristoyl group nor is it cleaved autocatalytically like the equivalent proteins of reoviruses and rotaviruses. BTV is internalized by receptor-mediated endocytosis forming clathrin-coated pits that contain virus particles, although subsequent events that allow the release of the transcriptionally active core into the host cell cytoplasm are currently obscure (28). Plausibly, the formation of intracellular endosome with low pH may trigger VP2 degradation and exposure of a functional form of VP5. VP5 possesses an N-terminal amphipathic helix, which is highly cytotoxic to the cells. Immediately adjacent to this peptide is a coiled-coil domain, which is connected to a more globular domain by means of a flexible short peptide (27). Thus, VP5 has certain features similar to the fusion proteins of many enveloped viruses. Because study on VP5 action within cells is difficult, we took advantage of the similarities between VP5 and fusion proteins of enveloped viruses and redesigned VP5 for expression on the surface of cells. We report here that expression of VP5 on the plasma membrane leads to syncytium formation in expressing cells resembling that shown by several fusion proteins of enveloped viruses. Physiologically, therefore, when translocated to the plasma membrane VP5 from a nonenveloped virus appears to behave as the equivalent of the fusion proteins of enveloped viruses.
Materials and Methods
Cells and Viruses. Spodoptera frugiperda (Sf9) cells were grown in Sf900 II medium (Invitrogen), as suspension cultures at 28°C in conical flasks. Recombinant baculoviruses were obtained as described by King and Possee (29, 30).
Generation of Constructs to Express BTV Proteins on the Cell Surface. Standard procedures were used for PCR and plasmid DNA manipulation (31). For construction of recombinant baculovirus transfer vectors, the coding regions of the VP2 and VP5 genes of BTV10 were amplified by PCR using pAcYM10.2 and pAcYM10.5 as templates (32). The primers were designed to introduce BamHI sites at both ends of the amplicons. After restriction digestion with BamHI, the amplicons were ligated into the BamHI site of the transfer vector pAcTM1 (33). The same procedure was used to generate a construct in which the VP5 amphipathic helices were deleted. Recombinant plasmids were identified by restriction digestion and verified by sequence analysis. The transfer plasmids were then used to generate recombinant baculoviruses as described.
SDS/PAGE and Western Blotting. Sf9 cells were infected with recombinant baculoviruses at a multiplicity of infection (moi) of 5. Cells were harvested 48-60 h after infection, and were lysed as described (27). Proteins were resolved by SDS/10% PAGE (34). The gels were either stained with Coomassie brilliant blue or were subjected to Western blotting as described by using either monoclonal mouse antibody against VP2 or polyclonal guinea pig anti-VP5 antiserum (27).
Surface Expression Density by Flow Cytometry. Sf9 cells were infected with recombinant baculoviruses at an moi of 3. After 16-60 h, the cells were harvested by centrifugation at 500 × g for 2 min, were washed twice in PBS containing 5% FCS and 0.1% sodium azide, and were incubated with polyclonal antisera specific for VP2 or VP5 for 4-5 h at room temperature. FITC rabbit anti-guinea pig Ig was used as a secondary antibody. Fluorescence intensity of 30,000 cells was measured by using a FACScan (Becton Dickinson) and analyzed by using cellquest software. As a control, uninfected Sf9 cells were incubated with the same antibodies and the fluorescence of these cells was compared with the infected sample.
Cell Viability Assay. Monolayers of Sf9 cells were infected with recombinant baculovirus at an moi of 5 and were incubated for 48 h at 28°C. The cells were washed with PBS, were stained for 2 min with 0.2% Trypan blue, and were examined by light microscopy.
Cell Fusion Assay. Monolayer cultures of Sf9 cells were infected at an moi of 0.1-2.5. At 24, 36, or 60 h after infection, the cells were washed in Sf900II medium and were incubated for 1 h with a monoclonal antibody against gp64 at 1:1,000 dilution (gift from I. Jones, Reading University, Reading, U.K.). To remove unbound antibody, the cells were washed with medium. The cells were then washed with low-pH buffer (PBS, pH 5.0) and were treated with the same buffer for a further 2 min. To restore the normal pH, the cells were washed two times with medium and then incubated in Sf900II medium at 28°C. Syncytium formation was observed by light microscopy at different time points after the pH shift.
Immunofluorescence. The recombinant baculoviruses expressing the chimeric proteins were used to infect a monolayer of Sf9 cells at an moi of 2.5 on 22- × 22-mm glass coverslips. After 42 h, the infected cells were fixed in 4% paraformaldehyde and processed for immunofluorescence assay by using a monoclonal VP2 and a polyclonal VP5 as primary antiserum and a FITC-conjugated secondary antibody (Sigma-Aldrich) as described (27). Samples were analyzed by using a Nikon TS 100 inverted microscope. Images were acquired by using a Nikon digital camera.
Confocal Microscopy. Insect cells were infected and prepared by using the same procedure as for immunolabeling. For VP5, the secondary antibody was the same as above, VP2 was labeled with an anti-mouse IgG tetramethylrhodamine isothiocyanate (TRITC)-conjugated antibody at 1:64 dilution (Sigma). Samples were analyzed on a Zeiss LSM 510 confocal microscope and images were obtained by using LSM 510 image browser software.
Results
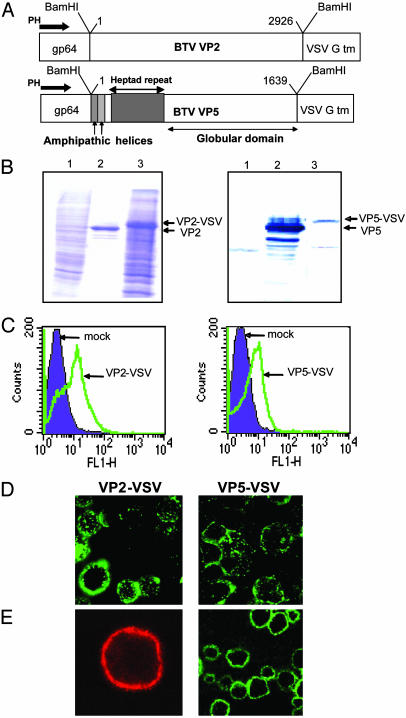
Expression of VP2 and VP5 on the Surface of Cells. Several studies have confirmed that the fusogenic activity of enveloped viruses lies in particular features of their envelope proteins. Two particular characteristic features of these proteins are coiled-coil domains (responsible for protein oligomerization and conformational changes) and amphipathic helices (responsible for cell membrane destabilization). VP5, an outer capsid protein of BTV, possesses both of these structural features, suggesting that it may have similar membrane fusion activity. In a previous report (27), using VP5 expressed by a recombinant baculovirus in the cytoplasm of insect cells, we showed that VP5 was cytotoxic and that cytotoxicity resided within the amphipathic helices, which is consistent with a role in membrane insertion. However, this source of VP5 was not suitable for further studies of activity. To provide a more suitable source of VP5 to investigate possible VP5-membrane interaction, we sought to express VP5 on the cell surface in a form analogous to the membrane fusion proteins of enveloped viruses. To this end, a recombinant baculovirus was constructed by using pAcVSVGTM (33) that expressed VP5 after a signal peptide derived from the baculovirus gp64 signal peptide and fused at the C terminus to the transmembrane domain of the vesicular stomatitis virus (VSV) G protein (Fig. 1A). In addition, and because the second outer capsid VP2 is juxtaposed to VP5 on the virion particle and is the cellular receptor binding protein of BTV, a second construct was similarly prepared that expressed VP2 on the cell surface (Fig. 1 A). Both chimeric proteins were synthesized in infected cells 24 h after infection under polyhedrin promoter control. Recombinant baculoviruses were prepared as described in Materials and Methods and the expression of chimeric proteins in infected cells were examined by SDS/10% PAGE. The Coomassie blue-stained gel showed proteins generated by each recombinant virus that were slightly larger in apparent molecular mass than the native BTV counterpart, confirming the presence of the VSV G protein TM domain on each BTV protein (Fig. 1B Left). The authenticity of BTV proteins was confirmed by Western blot analysis using an anti-BTV serum (Fig. 1B Right). To verify that the recombinant proteins were displayed on the cell surface we used fluorescence-activated cell sorter analysis of infected cells after staining with a BTV serum and FITC conjugate. The infected cells were treated with nonpermeabilizing buffers (see Materials and Methods) so that the fluorescence-activated cell sorter profile represented the proteins expressed on the cell surface. Both chimeric proteins were expressed on the cell surface, demonstrating that the gp64 signal peptide and the VSV G transmembrane domain retained their biological function, and that both BTV proteins were displayed on the cell surface (Fig. 1C). To monitor further the sites of protein localization in infected cells, we performed both immunofluorescence and confocal microscopy analysis of nonpermeabilized infected cells. Whereas immunofluorescence data of VP2-VSV and VP5-VSV (Fig. 1D) showed the localization of both proteins all around the cells, the confocal studies exhibited distinct fluorescence rings around the cell membranes confirming the cell surface display (Fig. 1E).
Fig. 1.
Construction and expression of membrane-anchored VP2 and VP5. (A) Baculovirus transfer vectors were constructed in which the coding sequences of VP2 and VP5 of BTV10 were fused in-frame to the signal peptide of the baculovirus gp64 and the C-terminal part of VSV G. (B) SDS/10% PAGE of insect cells infected with AcVP2-VSV (Left) and AcVP5-VSV (Right). Lanes: 1, cell lysate of uninfected Sf9 cells; 2, purified VP2 or VP5; 3, lysate of insect cells infected for 42 h with AcVP2-VSV or AcVP5-VSV. (C) Flow cytometric analysis. Sf9 cells were infected with AcVP2-VSV and AcVP5-VSV, were stained with polyclonal antisera and FITC-conjugated secondary antibodies, and were analyzed on a FACScan flow cytometer. Mock-infected cells were stained with the same antibodies as a control for nonspecific binding. (D) Immunofluorescence assay. Sf9 cells were coinfected at an moi of 2.5 for 42 h. The cells were then labeled under nonpermeabilizing conditions with polyclonal antisera against VP2 and VP5, followed by FITC-conjugated secondary antibodies, and were visualized on a Nikon fluorescence microscope. (E) Confocal microscopy. Sf9 cells were infected and stained as described above, except that tetramethylrhodamine isothiocyanate-conjugated secondary antibodies were used to label VP2. Pictures were taken on a Zeiss LSM510 microscope.
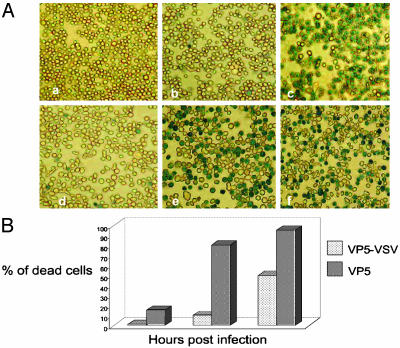
Cytotoxic Effects of VP5 Expressed Intracellularly Versus on the Cell Surface. VP5 has been shown to be highly toxic for Sf9 cells when expressed in the absence of other BTV proteins (32). Direct evidence of membrane leakage caused by purified VP5 was obtained from an in vitro assay system (27). To verify whether this feature of VP5 is affected when the protein is displayed on the cell surface, we performed a cell viability assay by using Trypan blue staining of cells infected with VP5-VSV recombinant virus. Whereas neither the recombinant VP2 nor the VP2-VSV had any apparent cytotoxic activity on the infected cells (Fig. 2 Ab and Ad), at 48 h after infection not only did the unmodified VP5 (Fig. 2 Ac) cause cell death as seen previously (27, 32) but some cell death was also caused by VP5-VSV (Fig. 2 Ae). However, the numbers of dead cells by VP5-VSV were much less than that seen by normal VP5 protein and scored only 55% in comparison to 95% caused by the normal VP5 (Fig. 2 Ac with Fig. 2 Ae). The data were further confirmed by a viability assay performed at different time points after infection (Fig. 2B). In this assay, the infected cells were stained 24, 48, and 60 h after infection. A fixed number of cells were counted and the percentage of dead cells was calculated for each sample. The data show that only 10% of the cells infected with AcVP5-VSV were dead 48 h after infection, as compared with cells infected with AcVP5, which caused 80% cell death after the same time, and 100% after 60 h. These data suggest that although the overall function of the chimeric VP5 was not altered, the altered localization of the recombinant protein reduced the toxic effect of VP5.
Fig. 2.
Cell viability assay. (A) Sf9 cells were infected at an moi of 5 for 48 h and were then incubated in medium containing 0.2% Trypan blue. Pictures were taken on a Nikon light microscope to visualize dead cells. (Aa) Uninfected cells. (Ab) Cells expressing cytoplasmic VP2. (Ac) Cells expressing cytoplasmic VP5. (Ad) Cells expressing membrane-anchored VP2. (Ae) Cells expressing membrane-anchored VP5. (Af) Cells expressing both VP2-VSV and VP5-VSV. (B) The experiment described in A was repeated and the number of dead cells was counted 24, 48, and 60 h after infection. The percentage of dead cells is shown in the graph.
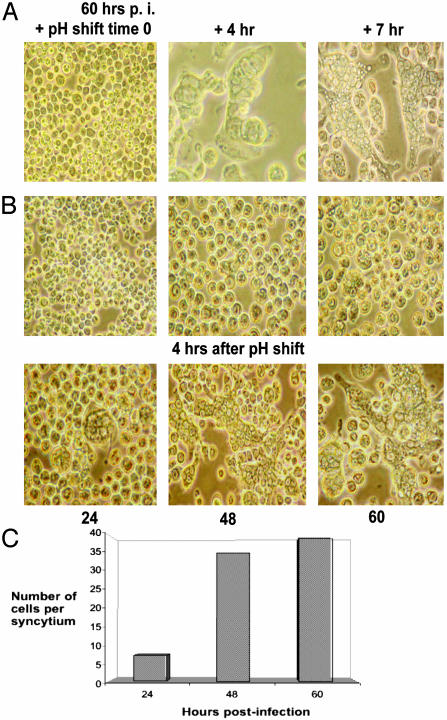
VP5 Is Responsible for Membrane Fusion and Induces Syncytium Formation in Cells. For some enveloped viruses (e.g., influenza), viral fusion proteins undergo conformational changes to become functional for fusion only in a low-pH environment. Because BTV enters the cell by means of endocytosis, it is presumed that the interaction of VP5 with the endosomal membrane and membrane destabilization may be a low-pH-dependent process. To examine this possibility, the activity of VP5 was assessed after incubation at pH 5.0. After a pH shift, cells infected with VP5-VSV virus exhibited a high number of syncytia within 4 h (Fig. 3A Center) and both the numbers and sizes of the syncytia increased over a period of 7-8 h post-pH shift (Right). However, no cell-cell fusion was observed when the cells were incubated over a long period (up to 60 h after infection) in the presence of normal growth medium at pH 6.5 (Fig. 3B Upper). In contrast, high numbers of syncytia were detected when these infected cells were shifted to low pH for a short period (Fig. 3B Lower). Time course studies showed that the number of cells per syncytium increased gradually up to 5-fold, when they were exposed to acidic pH at later times after infection (24-60 h after infection; Fig. 3C). These results demonstrate that exposure to low pH is essential for the formation of syncytia by VP5-VSV. No syncytia were induced in cells infected with recombinant viruses expressing native VP2 or VP5 protein with or without the low pH exposure (data not shown). To confirm that syncytium formation was due to the activity of VP5 but not due to the baculovirus expressed gp64, as controls, infected cells were incubated for 1 h with a gp64 monoclonal antibody before each experiment. No reduction in syncytia formation was observed in the presence of anti-gp64 antibody (data not shown), confirming that the baculovirus protein was not the primary cause for the observed fusion activity.
Fig. 3.
Fusogenic activity of VP5-VSV. (A) Sf9 cells were infected for 48 h at an moi of 2.5 and were then exposed to pH 5.0 for 2 min, after which the low-pH buffer was replaced by normal growth medium. Pictures were taken on an inverted light microscope at different time points after the pH shift. Uninfected cells (Left) and infected cells after 4 h (Center) and 7 h (Right) pH shift are shown. (B) Insect cells were infected for 24, 48, and 60 h, after which they were exposed to pH 5.0 as described above. Pictures were taken before and 4 h after the pH shift, and the number of cells per syncytium were counted at each time point (C).
Is the Fusion Activity of VP5 Influenced by the BTV Receptor-Binding Protein VP2? Viral entry mechanisms are commonly multistep processes, and many viruses use two or more proteins in concert to achieve cell entry (35, 36). In BTV, VP2 protein has been shown to be responsible for receptor binding and has the viral hemagglutination activity (26). A 3D structural study of BTV particles by cryoelectron microscopy revealed that the receptor-binding VP2 protein is organized in trimers and forms triskelion-like structures that are closely associated with the globular VP5 oligomers on the virion surface (19). Hence, VP2 may have direct influence on the fusogenic activity of VP5. To investigate this possibility, we performed the following experiments using VP2-VSV recombinant virus. Insect cells were infected with a virus expressing either membrane-anchored chimeric VP2 alone or coinfected with viruses expressing both chimeric VP2 and VP5 on the cell surface. At 48 h after infection, the infected cells were exposed at pH 5.0 for 2 min and were then shifted back to a normal pH growth medium. Formation of syncytia was monitored by light microscopy after 4-6 h. Unlike the results obtained with the VSV-VP5, no syncytia could be detected with or without a pH shift in the cells infected withVP2-VSV, demonstrating that VP2 is not able to induce cell-cell fusion activity (Fig. 4A). In addition, most strikingly, when insect cells were coinfected with both VP5-VSV and VP2-VSV, the fusogenic activity of VP5-VSV was drastically suppressed with a resulting reduction both in sizes and numbers of the syncytia formed (Fig. 4B; compare Center with Right). The data indicate that VP2 clearly influences fusion activity of VP5 in this context.
Fig. 4.
Membrane-anchored VP2 lacks fusogenic activity and inhibits VP5 activity. (A) Sf9 cells were infected with AcVP2-VSV at an moi of 2.5. The infected cells were exposed to pH 5.0 at 24, 48, and 60 h after infection, and pictures were taken before and 4 h after the pH shift as described above. (B) Insect cells were coinfected with AcVP2-VSV and AcVP5-VSV each at an moi of 2.5. The fusogenic activity was checked at 48 h after infection as described above, and pictures were taken before and 4 h after the pH shift.
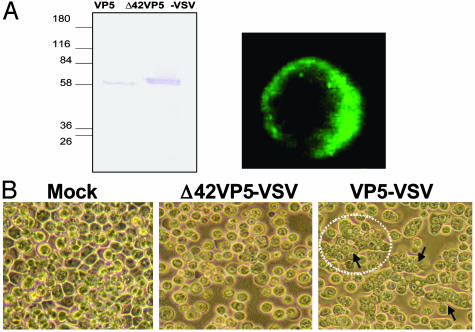
Deletion of N-Terminal Putative Amphipathic Helical Peptide Abrogates the VP5 Fusion Activity. Fusion peptides generally can fold into amphipathic structures, which can bind to and alter the structure of lipid bilayers and are a characteristic feature of membrane permeabilizing peptides. The N-terminal 42 residues of VP5 could fold into two amphipathic helices and have been shown both by deletion mutant analysis and by using synthetic peptides encompassing each helix to be responsible for the cytotoxic activity of VP5 (27). In terms of position and biological activity, this region of VP5 resembles class I fusion proteins, in which the fusion peptide is localized at the N terminus of the fusion-promoting subunit (37). It is therefore likely that the N-terminal sequences play an essential role for the VP5 fusogenic activity. To obtain direct evidence, we constructed a recombinant DNA in which the sequences encoding the first 42 residues of VP5 were deleted. The truncated DNA was placed between the gp64 signal peptide and the C-terminal region of VSV G protein as described above for the full-length VP5 (Fig. 5A). A recombinant baculovirus was subsequently generated and shown to express a truncated form of VP5. The truncated VP5 showed a protein of the expected size that was produced at a very high level at 24 h after infection (Fig. 5A Left Upper). The surface display of the truncated VP5 was also confirmed by confocal microscopy (Fig. 5A Left Lower) and fluorescence-activated cell sorter analyses (data not shown). The fusion activity of the Δ42VP5-VSV protein was examined in Sf9 cells infected with the recombinant virus as described above. As shown in Fig. 5B (compare Center with Lower), the inhibition of cell-cell fusion was observed after exposure to pH 5, indicating that the N-terminal amphipathic helices participate in the fusogenic activity of VP5.
Fig. 5.
Expression and fusion activity of membrane-anchored VP5 lacking the amphipathic helices. A recombinant baculovirus AcΔ42VP5-VSV was generated expressing a membrane-anchored mutant of VP5 in which the first 42 amino acids were deleted. (A) Sf9 cells were infected with the recombinant virus and protein expression was monitored by SDS/PAGE 24 h after infection (Left). Confocal microscopy confirmed membrane display on infected Sf9 cells (Right). (B) Insect cells were infected with AcΔ42VP5-VSV for 48 h, and syncytium formation was monitored 4 h after exposure to pH 5. Note that AcΔ42VP5-VSV exhibited no detectable fusion, in comparison with full-length AcVP5-VSV.
Discussion
To infect host cells, both enveloped and nonenveloped viruses need to transport their genomic material across a lipid bilayer. Animal viruses do so by means of special structural proteins that act either at the plasma membrane or the membrane of intracellular vesicular compartments (15). The currently accepted model on how the core particles of BTV gain access to the cytoplasm, where they become transcriptionally active, involves receptor mediated endocytosis and pH-triggered conformational change inside endosomal vesicles, which activates a membrane-permeabilizing protein. Previous studies (26, 28) have shown that the outer capsid protein VP2 is the BTV receptor-binding protein that mediates attachment to the cell surface and receptor-mediated endocytosis and appears to be proteolytically degraded once the virions reach endosomal compartments. The second outer capsid protein, VP5, is believed to act as the membrane-permeabilizing protein, based on its structural features and cytotoxic activity (27). By using an experimental system for cell-surface expression of the outer capsid proteins of BTV, we provide further evidence to support this model. First, we show that VP5 but not VP2 has the ability to interact with host cell membranes, as demonstrated by its ability to induce cell-cell fusion when expressed on the cell surface. Second, we show that VP5 only exhibits its membrane-interacting properties after it has undergone a low-pH-triggered activation step, which presumably alters the conformation of VP5, rendering it fusion-competent.
Our data provide indirect evidence that VP2 and VP5 can interact with each other in the absence of other viral structural proteins, as demonstrated by the ability of VP2 to prevent VP5 from acting as a fusion protein. In the normal replication cycle, VP2 appears to be degraded once the virions reach endosomal compartments. The specific proteases involved in this degradation are presumably not present when the protein is expressed on the cell surface, which could explain why VP5 becomes active inside endosomes, but not on the cell surface when VP2 is present. Fusion proteins of enveloped viruses can be grouped into several classes based on their 3D structure. Despite having a different overall structure, all fusion proteins have certain features in common, most notably the presence of heptad repeat regions that mediate oligomerization of the proteins and the presence of a hydrophobic region termed “fusion peptide” that can insert into lipid bilayers (38). VP5 of BTV shares both of these features, having an N-terminal membrane-inserting region followed by a long heptad repeat region. It is unclear at present whether VP5 contains additional membrane-interacting regions similar to the pretransmembrane regions identified in most fusion proteins. These additional regions, which play an essential role in the fusion reaction, might be dispensable for the normal function of VP5. The fact that VP5 is still capable of inducing fusion in our experimental setting can then be explained by the fact that the VSV G fragment provides the missing element; i.e., the association with the donor membrane (39). Additional experiments will have to be carried out to address this question. Notwithstanding the precise mechanism, it is clear from our data that VP5 can functionally substitute for a typical viral fusion protein.
VP5 shows the same arrangement of membrane-inserting peptide and heptad repeat region as class I fusion proteins (37). Unlike class I fusion proteins, however, VP5 does not require a proteolytic activation step to render it fully functional, because the fusion-peptide analogue is already located at the N terminus of the native protein. The maturation of the functional homologue of VP5 in the related reovirus, however, includes a proteolytic activation step of a precursor protein, which results in exposure of an internal fusion peptide at the N terminus of the C-terminal cleavage product very similar to the situation observed in class I fusion proteins.
In light of the data described here and the striking similarities between class I viral fusion proteins and membrane permeabilizing proteins of members of the Reoviridae family, it is tempting to speculate that these two classes of proteins have evolved from a common ancestor. However, it is also possible that the proteins have evolved into similar structures independent of each other.
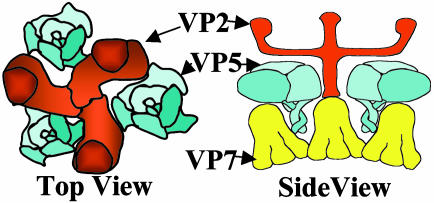
It is interesting to note in this context that there also appears to be a functional analogue in orbiviruses, protein NS3, for the matrix protein of negative-strand RNA viruses, which is involved in virus release (40). Intriguingly the matrix protein analogue carries two transmembrane domains, whereas proper matrix proteins lack transmembrane domains. It seems possible to us that exchange of transmembrane domains between the matrix-protein and fusion-protein analogues of BTV might be one step in an evolutionary process that could convert an enveloped virus into a nonenveloped virus or vice versa. Support for this hypothesis is provided by the fact that rotavirus maturation involves the transient formation of an enveloped particle (41). Although it is not yet clear whether fusion induced by VP5 follows the same mechanisms proposed for enveloped viruses (2), it seems probable that the amphipathic helices insert into the target membrane as they would do in the context of the BTV particle. This early interaction and the formation of the initial pore might be mechanistically different for VP5 and proper fusion proteins, considering the differences between fusion peptides and the amphipathic N-terminal part of VP5 that more closely resembles pore-forming antimicrobial peptides than a typical fusion peptide (42, 43). In conclusion, our data provide support for the protein-coat hypothesis (44), according to which the driving force for several consecutive stages of the fusion process is generated by a dense interconnected layer of proteins, including those outside the initial contact zone. The dense layer of fusion proteins has an intrinsic tendency to bend away from the viral particle and to adopt a curvature that effectively results in the formation of an outside-in proteinaceous vesicular structure. The protein-coat hypothesis is consistent with our study, which shows highly localized expression of VP5 on the cell surface as revealed by confocal microscopy. In the BTV particle, the VP5 protein layer makes contact with the underlying VP7 layer and also with the spike protein VP2, which protrudes above VP5 (see Fig. 6). Combining the available structural and biochemical data and applying the protein coat model to BTV, we propose that the low pH inside endosomes induces a rearrangement and conformational changes of VP5, thereby loosening the interactions of VP5 and VP7 and simultaneously allowing VP5 to form a protein layer with intrinsic outside-in curvature. Proteolytic removal, or pH-induced conformational change of VP2, allows the VP5 amphipathic helices to freely interact with membranes and initiate the permeabilization process.
Fig. 6.
Schematic diagrams of a section of a BTV particle. The schematics show the organizations of the three trimeric major proteins, VP2, VP5, and VP7, depicted from image reconstruction of cryo-electron microscopy analysis and the arrangement of VP5 in relation to VP2 (top view) or VP2 and VP7 (side view). Note that VP2 shrouds VP5 under normal physiological conditions.
Acknowledgments
We thank I. M. Jones (Reading University, Reading, U.K.) for providing us with reagents and fruitful discussions during this study. This work was partly supported by the National Institutes of Health (U.S.A.) and the Wellcome Trust Fund (U.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BTV, Bluetongue virus; Sf9, Spodoptera frugiperda; moi, multiplicity of infection; VSV, vesicular stomatitis virus.
References
- 1.Cohen, F. S. & Melikyan, G. B. (2001) Nat. Struct. Biol. 8, 653-655. [DOI] [PubMed] [Google Scholar]
- 2.Peisajovich, S. G. & Shai, Y. (2002) Trends Biochem. Sci. 27, 183-190. [DOI] [PubMed] [Google Scholar]
- 3.Tamm, L. K., Han, X., Li, Y. & Lai, A. L. (2002) Biopolymers 66, 249-260. [DOI] [PubMed] [Google Scholar]
- 4.Jahn, R., Lang, T. & Sudhof, T. C. (2003) Cell 112, 519-533. [DOI] [PubMed] [Google Scholar]
- 5.Weissenhorn, W., Dessen, A., Calder, L. J., Harrison, S. C., Skehel, J. J. & Wiley, D. C. (1999) Mol. Membr. Biol. 16, 3-9. [DOI] [PubMed] [Google Scholar]
- 6.Matthews, J. M., Young, T. F., Tucker, S. P. & Mackay, J. P. (2000) J. Virol. 74, 5911-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentz, J. (2000) Biophys. J. 78, 886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossman, M. G., Bella, J., Kolatkaer, P. R., He, Y., Wimmer, E., Kuhn, R. J. & Baker, T. S. (2000) Virology 269, 239-247. [DOI] [PubMed] [Google Scholar]
- 9.Hogle, J. M. (2002) Annu. Rev. Microbiol. 56, 677-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nemerow, G. R. & Stewart, P. L. (1999) Microbiol. Mol. Biol. Rev. 63, 725-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greber, U. F., Willetts, M., Webster, P. & Helenius, A. (1993) Cell 75, 477-486. [DOI] [PubMed] [Google Scholar]
- 12.Greber, U. F. (2002) Cell. Mol. Life Sci. 59, 608-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jane-Valbuena, J., Breun, L. A., Schiff, L. A. & Nibert, M. L. (2002) J. Virol. 76, 5184-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liemann, S., Chandran, K., Baker, T. S., Nibert, M. L. & Harrison, S. C. (2002) Cell 108, 283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandran, K. & Nibert, M. L. (2003) Trends Microbiol. 11, 374-382. [DOI] [PubMed] [Google Scholar]
- 16.Verwoerd, D. W., Els, H. J., De Villiers, E. M. & Huismans, H. (1972) J. Virol. 10, 783-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy, P. (1995) in Field's Virology, ed. Fields, B. N. (Lippincott-Raven, Philadelphia), Vol. 1, pp. 1709-1734. [Google Scholar]
- 18.Roy, P. (1996) Virology 216, 1-11. [DOI] [PubMed] [Google Scholar]
- 19.Hewat, E. A., Booth, T. F. & Roy, P. (1992) J. Struct. Biol. 109, 61-69. [DOI] [PubMed] [Google Scholar]
- 20.Nason, E., Rothnagel, R., Muknerge, S. K., Kar, A. K., Forzan, M., Prasad, B. V. & Roy, P. (2004) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 21.Huismans, H. & Erasmus, B. J. (1981) Onderstepoort J. Vet. Res. 48, 51-58. [PubMed] [Google Scholar]
- 22.Kahlon, J., Sugiyama, K. & Roy, P. (1983) J. Virol. 48, 627-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huismans, H., van der Walt, N. T., Cloete, M. & Erasmus, B. J. (1987) Virology 157, 172-179. [DOI] [PubMed] [Google Scholar]
- 24.Eaton, B. T. & Crameri, G. S. (1989) J. Gen. Virol. 70, 3347-3353. [DOI] [PubMed] [Google Scholar]
- 25.Roy, P., Urakawa, T., Van Dijk, A. A. & Erasmus, B. J. (1990) J. Virol. 64, 1998-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassan, S. H. & Roy, P. (1999) J. Virol. 73, 9832-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassan, S. H., Wirblich, C., Forzan, M. & Roy, P. (2001) J. Virol. 75, 8356-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eaton, B. T., Hyatt, A. D. & Brookes, S. M. (1990) Curr. Top. Microbiol. Immunol. 162, 89-118. [DOI] [PubMed] [Google Scholar]
- 29.King, L. A. & Possee, R. D., eds. (1992) The Baculovirus Expression System: A Laboratory Guide (Chapman and Hall, London).
- 30.Kitts, P. A. & Possee, R. D. (1993) BioTechniques 14, 810-817. [PubMed] [Google Scholar]
- 31.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 32.Marshall, J. J. & Roy, P. (1990) Virus Res. 15, 189-195. [DOI] [PubMed] [Google Scholar]
- 33.Chapple, S. D. & Jones, I. M. (2002) J. Biotechnol. 95, 269-275. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 35.Eckert, D. M. & Kim, P. S. (2001) Annu. Rev. Biochem. 70, 777-810. [DOI] [PubMed] [Google Scholar]
- 36.Klasse, P. J., Bron, R. & Marsh, M. (1998) Adv. Drug Delivery Rev. 34, 65-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colman, P. M. & Lawrence, M. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 309-319. [DOI] [PubMed] [Google Scholar]
- 38.Peisajovich, S. G. & Shai, Y. (2003) Biochim. Biophys. Acta 1614, 122-129. [DOI] [PubMed] [Google Scholar]
- 39.Jeetendra, E., Robison, C. S., Albritton, L. M. & Whitt, M. A. (2002) J. Virol. 76, 12300-12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaton, A. R., Rodriguez, J., Reddy, Y. K. & Roy, P. (2002) Proc. Natl. Acad. Sci. USA 99, 13154-13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrie, B. L., Graham, D. Y. & Estes, M. K. (1981) Intervirology 16, 20-28. [DOI] [PubMed] [Google Scholar]
- 42.Merrifield, R. B., Vizioli, L. D. & Boman, H. G. (1982) Biochemistry 21, 5020-5031. [DOI] [PubMed] [Google Scholar]
- 43.Putsep, R. A., Branden, C. I., Boman, H. G. & Normark, S. (1999) Nature 398, 671-672. [DOI] [PubMed] [Google Scholar]
- 44.Kozlov, M. M. & Chernomordik, L. V. (2002) Traffic 3, 256-267. [DOI] [PubMed] [Google Scholar]