SUMMARY
Polycomb repressive complex 2 (PRC2) regulates pluripotency, differentiation and tumorigenesis through catalysis of histone H3 lysine 27 trimethylation (H3K27me3) on chromatin. However, the mechanisms that underlie PRC2 recruitment and spreading on chromatin remain unclear. Here we report that histone H3 lysine 36 trimethylation (H3K36me3)-binding activity is harbored in the Tudor motifs of PRC2-associated polycomblike (PCL) proteins PHF1/PCL1 and PHF19/PCL3. Ectopically expressed PHF1 induced Tudor-dependent stabilization of PRC2 complexes on bulk chromatin and mediated spreading of PRC2 and H3K27me3 into H3K36me3-containing chromatin regions. In murine pluripotent stem cells, we identified coexistence of H3K36me3, H3K27me3, and PHF19/PCL3 at a subset of ‘poised’ developmental genes, and demonstrated that PHF19/PCL3 Tudor function is required for optimal H3K27me3 and repression of these loci. Collectively, our data suggest that PCL recognition of H3K36me3 promotes intrusion of PRC2 complexes into active chromatin regions to promote gene silencing and modulate the chromatin landscape during development.
INTRODUCTION
Modulation of the chromatin landscape by covalent histone post-translational modifications (PTMs) represents a fundamental way of regulating DNA-templated processes such as gene transcription (Chi et al., 2010; Kouzarides, 2007). Histone H3 lysine 27 trimethylation (H3K27me3) promotes gene silencing, whereas promoter-associated histone H3 lysine 4 trimethylation (H3K4me3), together with gene body-associated H3 lysine 36 trimethylation (H3K36me3), demarcates active genes (Mikkelsen et al., 2007). PRC2 complexes are the major enzymatic machineries responsible for ‘writing’ H3K27me3, and PRC2-mediated gene silencing is involved in various biological processes, including stem cell pluripotency, differentiation, and cancer progression (Bracken and Helin, 2009; Margueron and Reinberg, 2011). While a reconstituted tetrameric PRC2 core complex (comprising EZH2, EED, SUZ12 and NURF55/RbAp48) is sufficient to induce H3K27me3 (Cao and Zhang, 2004; Margueron and Reinberg, 2011), a number of PRC2-associated factors, including EED (Margueron et al., 2009), JARID2 (Li et al., 2010; Pasini et al., 2010; Peng et al., 2009; Shen et al., 2009) and non-coding RNAs (Bracken and Helin, 2009; Margueron and Reinberg, 2011), have recently been identified that either modulate H3K27me3 catalysis or help to stabilize and tether PRC2 to appropriate genomic regions. Despite these recent advances, mechanisms responsible for PRC2 targeting and spreading on active chromatin to promote de novo repressed states remain poorly defined.
In this study, we show that two PCL family proteins (PHF1/PCL1 and PHF19/PCL3), accessory components of the PRC2 core complex (Cao et al., 2008; Hunkapiller et al., 2012; Sarma et al., 2008), harbor H3K36me3-‘reading’ activity within their N-terminal Tudor motifs. Biochemical, biophysical and structural analyses reveal tight binding to H3K36me3 through a conserved aromatic ‘cage’ formed by PCL Tudor domains. Our gene regulation and genomics analysis, using both overexpression and knockdown systems, define the PCL Tudor-H3K36me3 interaction as critical for both the targeting and spreading of PRC2 into active chromatin regions, and for the maintenance of optimal repression of ‘poised’ developmental genes where PCL, H3K36me3 and H3K27me3 co-exist. Our studies shed important light on the regulation of PRC2 by PCL proteins, implicating H3K36me3 ‘reading’ in this process, and contribute to our understanding of the dynamics of gene expression and chromatin remodeling associated with development and disease (Chi et al., 2010).
RESULTS
PHF1 directly binds to H3K36me3 through a conserved N-terminal Tudor motif
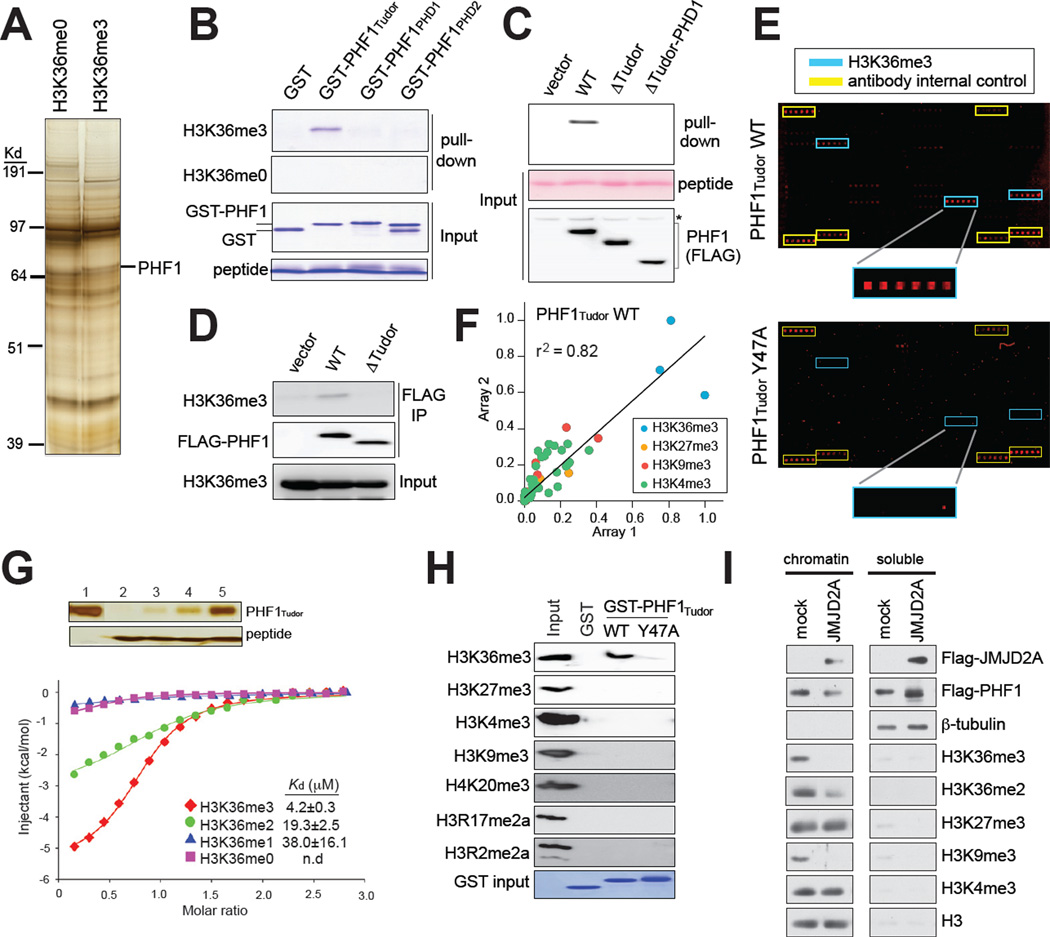
We began this study by attempting to identify by mass spectrometry H3K36me3-‘reader’ proteins using synthetic histone tail peptides as baits for nuclear extracts. In addition to the previously identified putative H3K36me3-‘reading’ effectors NSD2 and MSH6 (Vermeulen et al., 2010), we identified PHF1 (Figure 1A and S1A), a known accessory component of H3K27me3-promoting PRC2 complexes (Cao et al., 2008; Sarma et al., 2008). PHF1, and its homologues MTF2/PCL2 and PHF19/PCL3, have several putative chromatin-interacting domains (Cao et al., 2008), including a highly conserved N-terminal Tudor domain and two plant homeodomain (PHD) fingers (Figure S1B). In-solution peptide pull-down assays with recombinant GST-fusion proteins of each of these individual domains of PHF1 indicated that the Tudor domain facilitated H3K36me3 recognition (Figure 1B and S1C). To confirm this interaction, we established stable cell lines expressing wildtype or N-terminal truncated forms of PHF1 lacking either the Tudor domain or PHD fingers. Indeed, wildtype PHF1, but not that lacking the Tudor domain, pulled down with H3K36me3 peptides (Figure 1C and S1D). Furthermore, PHF1 coimmunoprecipitated (Co-IP) with endogenous histones that contained H3K36me3 in a Tudor-dependent manner (Figure 1D). Collectively, our data implicated the PHF1 Tudor domain (PHF1Tudor) as an H3K36me3-‘reading’ effector module.
Figure 1. The N-terminal Tudor motif of PHF1 specifically recognizes H3K36me3.
(A) Enrichment of PHF1 in pull-downs using biotinylated histone tail peptides that contain H3K36me3, as compared to that using unmodified H3K36. See also Figure S1.
(B) Pull-down of GST recombinant proteins fused to different PHF1 domains using H3K36me3 or non-modified H3K36 peptides (top two panels). Input was shown in bottom panels.
(C) Immunoblot following H3K36me3 peptide pull-down (top panel) from extracts of cells transduced with vector or that encoding the full-length PHF1 or deletion forms. Input of peptides and PHF1 was shown in middle and bottom panels.
(D) Co-immunoprecipitation (CoIP) using FLAG antibodies detecting a Tudor-dependent association of PHF1 (middle) to H3K36me3-containing histones (top) in 293 cells.
(E) Representative scan images of histone peptide microarrays probed with either wild-type or mutant (Y47A) GST-PHF1 Tudor proteins. Highlighted are positions of H3K36me3- containing peptides (blue) and IgG control (yellow). See also Table S1.
(F) Scatter plot of two peptide arrays probed with the PHF1 Tudor motif. Peptides are colored according to the legend. All other peptides are shown in black. Correlation coefficient was derived from linear regression analysis using GraphPad Prism.
(G) ITC measurements of binding affinities of PHF1 Tudor to H3 peptides containing different H3K36 methylation states. Insert represents silver staining of input (lane 1) and pull-down samples of PHF1 Tudor (top panel) using peptides harboring un-, mono-, di-, or trimethylated H3K36 (lane 2–5, bottom panel).
(H) Immunoblot using various histone methylation antibodies on mononucleosomes pulled down by either wild-type (WT) or mutant (Y47A) GST-PHF1 Tudor proteins.
(I) Immunoblot of chromatin-bound and soluble cell fractions following 48-hour transient expression of JMJD2A/KDM4A. Mock indicates vector control.
PHF1Tudor binds specifically and with high affinity to H3K36me3
We next sought to determine the histone-binding specificity of PHF1Tudor in an unbiased manner by using a recently developed high-density histone peptide microarray platform (Fuchs et al., 2011; Rothbart et al., 2012b). Arrays were printed with a library of unmodified and modified peptides (Table S1) containing known single and combinatorial histone PTMs (methylation, acetylation, and phosphorylation) on all of four core histones (H2A, H2B, H3, and H4). Interrogation of the GST-tagged PHF1Tudor by peptide arrays (Figure 1E, top panel) revealed a strong preference for various H3K36me3-containing peptides (Figure 1E and S1E). In addition, PHF1Tudor displayed weak interactions to other methylated histone peptides such as H3 lysine 9 trimethylation (H3K9me3) and H3K27me3 (Figure 1F and S1F–G). Similar results were obtained using another peptide array platform (Figure S1H–I). Using isothermal titration calorimetry (ITC), we next quantified these observed interactions. Consistent with array results and peptide pull-downs, ITC measurements of the PHF1Tudor demonstrated high affinity and specificity for H3K36me3, with a dissociation constant (Kd) of ~4.2 µM, 19 µM and 38 µM for tri-, di- and mono-methylated H3K36 states, respectively (Figure 1G, Table 1, and Figure S1L–O). Compared to H3K36me3, interactions to H3K9me3 and to H3K27me3 were significantly weaker, with a Kd of ~38 µM and ~168 µM, respectively (Figure S1P–Q and Table 1). We further determined the histone-binding specificity towards nucleosomal substrates in a GST pull-down assay using GST-PHF1Tudor and mononucleosomes purified from 293 cells. Again, PHF1Tudor preferentially bound to nucleosomes containing H3K36me3, but not other examined methylations, including those marked with H3K27me3, H3K9me3, H3K4me3, trimethylation of H4 lysine 20, or asymmetric dimethylation of H3 arginine 2 and arginine 17 (Figure 1H, lane of WT). Collectively, this data showed that PHF1Tudor motif demonstrates tight affinity and high selectivity towards H3K36me3.
Table 1.
Summary of thermodynamic and curve fitting parameters for ITC assays using recombinant PHF1 or PHF19 Tudor domain proteins and various histone peptides. See also Figures S1L-S and S2G.
| Protein | Peptide | ΔH(cal/mol) | ΔS (cal/mol/deg) |
Ka(M-1) | N | Chi2/ DoF |
Kd (µM) |
|---|---|---|---|---|---|---|---|
| PHF1Tudor, wt | H3(28–43)K36me3 | −5626 ± 108 | 4.45 | 2.38E5 ± 1.91E4 | 0.79 ± 0.01 | 5415 | 4.2 ± 0.3 |
| PHF1Tudor, wt | H3(28–43)K36me2 | −3994 ± 261 | 7.26 | 5.18E4 ± 6.79E3 | 0.94 ± 0.04 | 4338 | 19.3 ± 2.5 |
| PHF1Tudor, wt | H3(28–43)K36me1 | −817 ± 342 | 17.3 | 2.63E4 ± 1.12E4 | 0.70 ± 0.23 | 658.3 | 38.0 ± 16.1 |
| PHF1Tudor, wt | H3(28–43)K36me0 | -- | -- | -- | -- | -- | N.D |
| PHF1Tudor, wt | H3(19–35)K27me3 | −5141 ± 469 | −1.21 | 5.93E3 ± 486 | 1.03 ± 0.07 | 347.7 | 168 ± 13.8 |
| PHF1Tudor, wt | H3(1–15)K9me3 | −817 ± 343 | 17.3 | 2.63E4 ± 1.12E4 | 0.70 ± 0.23 | 658.3 | 38.0 ± 16.2 |
| PHF1Tudor, W41A | H3(28–43)K36me3 | −1355 ± 457 | 15.8 | 3.32E4 ± 1.20E4 | 0.68 ± 0.18 | 1456 | 30.1 ± 10.9 |
| PHF1Tudor, Y47A | H3(28–43)K36me3 | -- | -- | -- | -- | -- | N.D. |
| PHF1Tudor, F65A | H3(28–43)K36me3 | -- | -- | -- | -- | -- | N.D. |
| PHF1Tudor, F71A | H3(28–43)K36me3 | -- | -- | -- | -- | -- | N.D. |
| PHF1Tudor, E66K | H3(28–43)K36me3 | −6471 ± 229 | −1.94 | 4.39E4 ± 3.76E3 | 0.60 ± 0.02 | 5055 | 22.8 ± 2.0 |
| PHF19Tudor, wt | H3(27–46)K36me3 | −6610 ± 267 | 0.466 | 1.60E5 ± 1.65E4 | 0.58 ± 0.02 | 7436 | 6.2 ± 0.6 |
| PHF19Tudor, wt | H3(27–46)K36me1 | -- | -- | -- | -- | -- | N.D. |
| PHF19Tudor, Y56A | H3(27–46)K36me3 | -- | -- | -- | -- | -- | N.D. |
wt, wildtype; N.D, not determined.
Full-length PHF1 localizes exclusively in the nucleus (Figure 3D). We therefore asked whether association of full-length PHF1 with chromatin is dependent on H3K36me3. To do this, we diminished global levels of H3K36me3/2 on chromatin by overexpressing an H3K36me3/2 demethylase JMJD2A/KDM4A in 293 cells stably expressing a FLAG-tagged form of PHF1. Consistent with previous finding (Klose and Zhang, 2007), we observed a dramatic reduction in H3K36me3/2 and H3K9me3 after biochemical separation of JMJD2A/KDM4A-overexpressing cells into chromatin-bound and soluble cytoplasmic/ nucleoplasm fractions, with no change detected in H3K4me3 or H3K27me3 (Figure 1I). Importantly, compared to control, a reduction was observed in the chromatin-bound fraction of PHF1, concurrent with an increase in its soluble pool (Figure 1I), thus demonstrating that PHF1 is less stable on chromatin in the absence of global H3K36me3/2. Collectively, our in vitro and cellular biochemical data comprehensively showed that PHF1Tudor specifically and strongly binds to H3K36me3, and this interaction is necessary to target PHF1 to chromatin. In addition, the PHF1Tudor-H3K36me3 interaction represents the first high-affinity effector interaction to H3K36me3, which is several orders of magnitude tighter than all of the previously reported H3K36me3-reading motifs (Sun et al., 2008; Vezzoli et al., 2010; Xu et al., 2008).
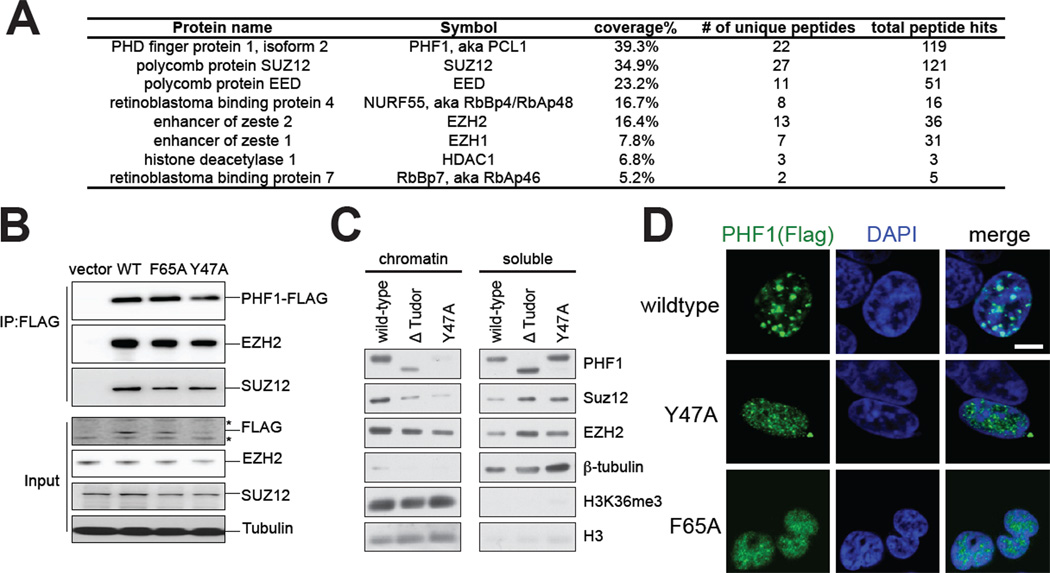
Figure 3. PHF1Tudor alters sub-nuclear localization of PHF1-PRC2 complexes, but not PRC2 core structure formation.
(A) Summary of protein peptides identified by mass spectrometry that specifically associates with Flag-tagged PHF1 complexes purified from 293 stable cell lines. Numbers shown in the table are those of coverage percentage of amino acid sequences, unique peptides, and total peptide hits. See also Table S2.
(B) Co-IP of FLAG-tagged wild-type (WT) or Tudor-mutated forms of PHF1 with PRC2 core complex components EZH2 and SUZ12. Bottom panels are immunoblots of input. Asterisk represents nonspecific bands. See also Figure S3.
(C) Immunoblot of the indicated FLAG-PHF1 stable expression cell lines separated into chromatin and soluble fractions.
(D) Representative confocal immunofluorescence images revealing localization of wild-type versus Tudor mutant form of PHF1 proteins (Flag tagged, green) and chromatin (stained by DAPI, blue) in transiently transfected 293 cells. Scale bar, 5 micrometer.
Structural analysis of PHF1Tudor revealed a unique H3K36me3-‘reading’ pocket
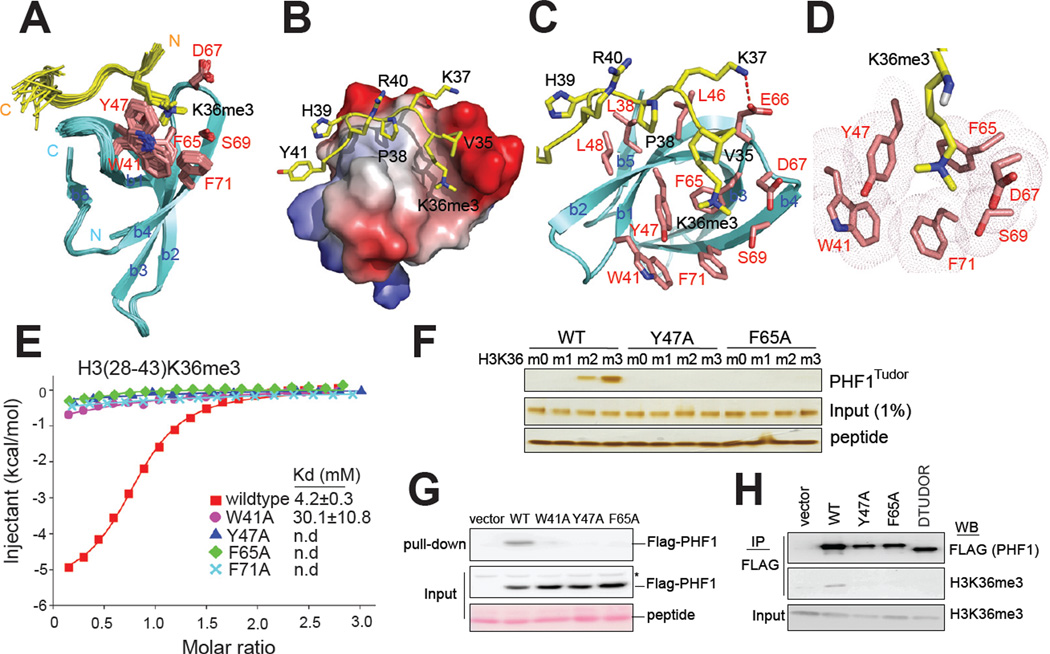
To gain insight into the molecular basis for recognition of H3K36me3 by PHF1Tudor, we performed NMR spectroscopy and obtained the solution structure of PHF1Tudor (residues 7–83) in complex with an H331–41K36me3 peptide. Initial comparison of 1H,15N-HSQC spectra of the unbound PHF1Tudor with those in the presence of H3K36me3 indicated that, while the majority of PHF1Tudor residues exhibited no detectable change in chemical shifts, several residues showed a large perturbation, indicating a role for these residues in histone tail engagement (Figure S2A). The NMR structures of the PHF1Tudor-H3K36me3 complex revealed residues 31–81 of PHF1Tudor adopted a classic Tudor domain fold (Figure 2A and Table 2), as initially observed in the Survival of Neuron Tudor domain (Selenko et al., 2001), whereas flanking residues were disordered, evidenced by poor chemical shift dispersion and lack of NOE connectivity. We also found that the structure of PHF1Tudor is dominated by a β- barrel formed by five antiparallel β-strands, and that association of PHF1Tudor with H3K36me3 is mediated by direct contacts to both H3K36me3 and several surrounding histone tail residues (Figure 2B–C). The trimethylammonium side chain of H3K36me3 fits into an aromatic ‘cage’ at one end of the β-barrel formed by residues Trp41, Tyr47, Phe65, Ser69, Asp67 and Phe71 (Figure 2D). Notably, Trp41 and Phe71 are not conserved in the Tudor domain of drosophila polycomb-like (Pcl) protein (Figure S2B), which was reported to lack ability to bind to histone methylation (Friberg et al., 2010), suggesting the role of these aromatic residues in formation of a closed cage composition. The overall structure of H3K36me3-binding ‘cage’ found in PHF1Tudor is reminiscent of those previously reported for the BRPF1 PWWP domain (Vezzoli et al., 2010) and the Eaf3 chromodomain (for comparison, see Figure S2C–E) (Sun et al., 2008; Xu et al., 2008). Several distinct features of PHF1Tudor were also revealed by our analysis, mainly occurring at contacts to histone residues surrounding H3K36me3. First, a salt bridge formed between the H3K37 side chain and the carboxylate of Glu66 of PHF1Tudor (Figure 2C, a dashed line). Second, the H3P38 side chain interacted with a hydrophobic patch generated by Leu38, Leu46 and Leu48 of PHF1Tudor (Figure 2B–C). Third, hydrophobic contacts were observed between the H3H39 side chain and Leu38 and Leu48 of PHF1Tudor (Figure 2B–C).
Figure 2. Structural and biophysical analysis revealed intermolecular interaction between PHF1TUDOR and H3K36me3-containing histones.
(A) Backbone superposition of 20 energy-minimized conformers representing the NMR structure of PHF1 Tudor in complex with an H3K36me3 peptide. For clarity, residues 1–30 and 82–83 of PHF1 Tudor and 31–34 of H3K36me3 peptide are not shown due to disorder. The PHF1 Tudor (cyan) and bound H3K36me3 peptide are shown in ribbon representation, with side chains of H3K36me3-binding cage (red) and H3K36me3 (yellow) in stick representation. See also Figure S2.
(B) Surface electrostatic representation of PHF1 Tudor bound to H3K36me3 peptide.
(C) Representative structure of PHF1 Tudor-H3K36me3 complex, with side chains involved in intermolecular contacts. The salt bridge formed between PHF1 Tudor Glu66 and H3K37 is depicted as a red dashed line.
(D) Positioning of the H3K36me3 side chain within an aromatic cage of the indicated residues (pink) on the surface of PHF1 Tudor.
(E) ITC measurements of binding affinities of wild-type or mutant PHF1 Tudor to H3K36me3 peptides.
(F) Silver staining of pull-down samples (top panel) of wild-type (WT) or mutant (Y47A and F65A) forms of PHF1 Tudor using histone tail peptides that harbor either un-, mono-, di-, or tri-methylated H3K36. Inputs of protein and peptide used were shown in middle and bottom panels.
(G) Anti-FLAG immunoblot following peptide pull-down (top panel) from extracts of cells transduced with either empty vector or that encoding a wild-type (WT) or mutant form of FLAG-tagged PHF1.
(H) Western blots (WB) examining association of PHF1 (top) to endogenous H3K36me3- containing histones (middle) in HeLa cells that expressed a wild-type (WT) or mutant form of FLAG-tagged PHF1.
Table 2.
Statistics for the 20 energy-minimized conformers of PHF1Tudorin complex with H331–41K36Me3
| Distance constraints | |
|---|---|
| Long [(i–j)>5] (intramolecular) | 250 |
| Medium [1<(i–j)≤5] | 106 |
| Sequential [(i–j)=1] | 234 |
| Intraresidue [i=j] | 462 |
| Intermolecular | 38 |
| Dihedral angle constraints (φ and ψ) | 96 |
| Hydrogen bond constraints | 16 |
| Average pairwise R.M.S.D. to the mean structure (Å)a | |
| Backbone (Cα, C′, N, O) | 0.26 ± 0.05 |
| Heavy atoms | 0.72 ± 0.10 |
| Deviations from idealized covalent geometry | |
| Bond (Å) | 0.008 ± 0.002 |
| Angles (°) | 0.744 ± 0.012 |
| Impropers (°) | 0.479 ± 0.010 |
| R.M.S.D. from experimental distance restraints (Å) | 0.032 ± 0.001 |
| R.M.S.D. from experimental dihedral restraints (°) | 0.578 ± 0.027 |
| Ramachandran statistics (% of all residues)a | |
| Most favored | 91.3 |
| Additionally allowed | 8.7 |
| Generously allowed | 0.0 |
| Disallowed | 0.0 |
Statistics are given for residues 31–81 of PHF1Tudorand residues 35–40 of the H331–41K36Me3 peptide.
To examine the functional relevance of observed contacts to H3K36me3 recognition, we introduced individual alanine substitutions to each of the H3K36me3-‘caging’ residues. We found that these single point mutations abrogated the binding of PHF1Tudor to H3K36me3 peptides on arrays (Figure 1E, Y47A, bottom panel), by ITC (Figure 2E; Table 1; Figure S1R), and by peptide pull-downs (Figure 2F, and Figure S2F, left panel). Point mutations also abrogated binding of full-length PHF1 to H3K36me3 peptides (Figure 2G) and to H3K36me3- containing histones as assayed by GST-pull downs (Figure 1H, Y47A) and Co-IPs (Figure 2H). Furthermore, disruption of the salt bridge to H3K37 by mutating Glu66 of PHF1Tudor to Lys, also caused a significant reduction in H3K36me3 binding (Kd= ~22.8 µM, Figure S1S and Table 1). Collectively, our structural and mutational analysis demonstrated that, unlike other known H3K36me3 effector proteins, numerous Van de Waals contacts and electrostatic interactions contribute to a high-affinity association of PHF1Tudor with H3K36me3.
PHF1Tudor alters localization of PHF1-PRC2 complexes, but not PRC2 core structure
PHF1 was reported to interact with PRC2 core complexes where PHF1 modulates the enzymatic activity of associated PRC2 core complexes in vitro (Cao et al., 2008; Sarma et al., 2008). To determine whether PHF1 primarily associates with PRC2, we used mass spectrometry analysis to identify PHF1-associated factors from 293 stable cell lines. The top unique hits of identified proteins were the four subunits of tetrameric PRC2 core complexes (SUZ12, EED, NURF55/RbAp48, and EZH2 or EZH1) (Figure 3A and Table S2), consistent with co-migration of PHF1 and PRC2 core components observed in gel filtration chromatography analyses (Cao et al., 2008; Sarma et al., 2008). We next sought to determine whether the PHF1Tudor motif was needed for PRC2 complex formation. Co-IPs with EZH2 and SUZ12 in cells stably expressing wildtype or Tudor-mutated forms of PHF1 showed that membership in the PRC2-PHF1 complex was not dependent on PHF1Tudor (Figure 3B and S3A–B). Notably, while JARID2 was previously identified to associate with the PRC2 core complex (Li et al., 2010; Pasini et al., 2010; Peng et al., 2009; Shen et al., 2009), we did not detect JARID2 in either mass spectrometry analysis (Table S2) or in CoIP experiments with PHF1 (Figure S3C). These results are consistent with previous proteomics analysis of JARID2-associated complexes that failed to identify PCL proteins such as PHF1 or MTF2 (Shen et al., 2009). These studies collectively indicate that PHF1 primarily assembles in protein complexes with PRC2, potentially to modulate PRC2 targeting via its histone-‘reading’ ability, and that PCL proteins and JARID2 reside in distinct PRC2 subcomplexes.
We next sought to determine whether the PHF1Tudor domain was required for PRC2 recruitment to chromatin. To this end, we fractionated PHF1 stable expression cells into chromatin and soluble fractions, and found that, consistent with our observation following JMJD2A/KDM4A overexpression (Figure 1I), chromatin association of PHF1 was perturbed in the absence of Tudor recognition of H3K36 methylation (Figure 3C, lane 1 versus lane 2–3). Notably, PRC2 association with bulk chromatin was also hindered in the absence of a functional PHF1Tudor in our PHF1 overexpression system, evidenced by a loss of SUZ12 and EZH2 from the chromatin fraction and a concurrent increased soluble pool of these PRC2- associated factors (Figure 3C). Consistent with these results, confocal immunofluorescence microscopy of wild-type PHF1 showed punctate nuclear foci overlapping with DAPI-dense regions of chromatin, while Tudor mutant forms of PHF1 showed diffuse distribution throughout the nucleus with accumulations in the chromatin-free nucleoplasm (Figure 3D and Figure S3D). Collectively, our data suggests that PHF1 alters chromatin localization or stabilization of PHF1 and associated PRC2 proteins via an H3K36me3/2-recognizing PHF1Tudor, a function separated from assembly of a higher-order PHF1-PRC2 complex.
PHF1 induces spreading of PRC2 and H3K27me3 into H3K36me3-marked loci
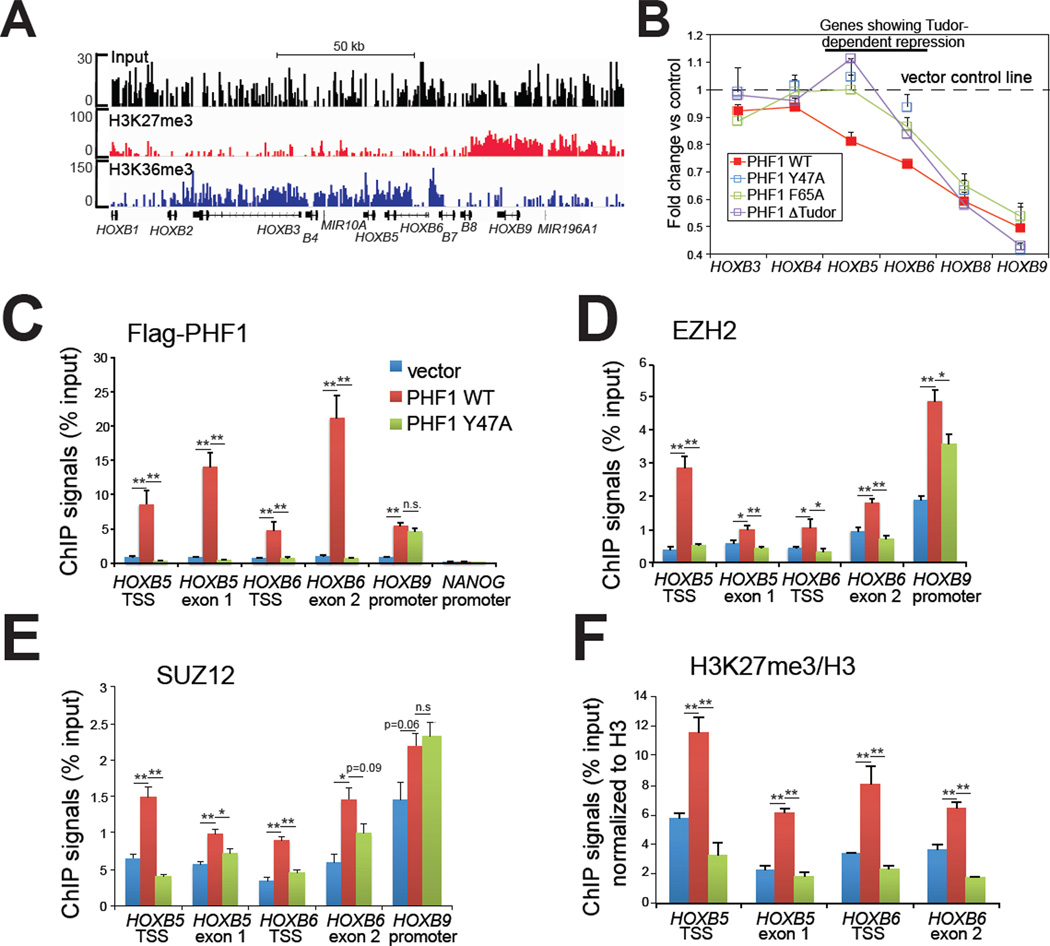
Our data presented thus far suggest that PCL family proteins may link PRC2 core complexes to active chromatin via H3K36me3 recognition, and potentially modulate PRC2 stabilization and spreading during conversion of transcriptionally active/permissive states to repressed ones – a process often observed in cell differentiation and lineage specification (Mikkelsen et al., 2007). We next tested this hypothesis in HeLa cells, whose endogenous PHF1 expression is low (Boulay et al., 2011), and asked whether ectopic expression of PHF1 could shift chromatin boundaries that separate H3K27me3 from established H3K36me3 domains. Profiling of HeLa cells by chromatin immunoprecipitation coupled with deep sequencing (ChIP-Seq) revealed that the HOX-B gene clusters, known to be regulated by PRC2 (Cao et al., 2008), exhibited a bimodal distribution of H3K27me3 and H3K36me3 (Figure 4A). H3K27me3 covered the distal HOX-B genes (from HOXB13 to HOXB7) and H3K36me3 decorated the proximal ones (from HOXB6 to HOXB1). We first confirmed this bimodal pattern using directed ChIPs (Figure S4A–B). In examination of changes in HOX-B gene expression before and after ectopic PHF1 expression, we found that wildtype and Tudor-mutant forms of PHF1 demonstrated a chromatin environment-dependent effect on HOX repression. First, both wildtype and Tudor-mutant PHF1 repressed HOXB9, HOXB8 and HOXB7, three distal HOXs marked by high H3K27me3 and low H3K36me3 (Figure 4A–B and Figure S4C). Using ChIP assays, we detected exogenous FLAG-tagged PHF1, as well as an increase in PRC2 residence and H3K27me3 at HOXB9 and HOXB8 (Figure 4C–F and Figure S4D), suggesting that, independent of the Tudor motif, pre-existing PRC2 may recruit PHF1 to H3K27me3-positive regions where PHF1 either enhances PRC2 enzymatic activities as previously reported (Cao et al., 2008; Sarma et al., 2008), or further stabilizes the chromatin binding of PRC2. While we detected no change in expression of proximal HOXs positioned deep within the H3K36me3 domain, including HOXB4, HOXB3 and HOXB1 (Figure 4A–B), wild-type, but not Tudor-mutant PHF1, was able to repress HOXB6 and HOXB5 (Figure 4B, red line versus green, blue or purple), two HOX genes situated exactly on the junction of H3K36me3 and H3K27me3 domains (Figure 4A). This data suggested a Tudor-dependent mechanism for PRC2 targeting to regions of HOXB6 and HOXB5. Indeed, PHF1 residence at HOXB6 and HOXB5 was completely dependent on Tudor function (Figure 4C), as was the residence of the PRC2 core complex components EZH2 and SUZ12 and the spread of H3K27me3 into these regions (Figure 4D–F). No dramatic change in H3K36me3 was detected at examined HOX-B loci (Figure S4E). A similar Tudor-dependent H3K27me3 spreading was also observed at PBX1 and TADA1/ADA1, two other developmentally critical genes that also exhibit bimodal distribution of H3K36me3 (at gene bodies) and H3K27me3 (at promoters) (Figure S4F–G). Taken together, our data demonstrates that, through Tudormediated recognition of H3K36me3, ectopically expressed PHF1 is able to mediate targeting and spreading of associated PRC2 core complexes to H3K36me3-demarcated active chromatin regions to modulate gene expression.
Figure 4. Ectopic expression of PHF1 induced a Tudor-dependent spreading of PRC2 complexes and H3K27me3 into an adjacent H3K36me3 region within HOX-B gene clusters.
(A) ChIP-Seq revealing a bimodal distribution of H3K27me3 and H3K36me3 at HOX-B gene clusters. Position of each HOX is indicated at the bottom. See also Figure S4.
(B) Change in expression of HOX-B genes in HeLa stable cells with ectopic expression of either wild-type or Tudor-mutant form of PHF1. Overexpression of the wild-type, but not Tudor-mutant form of PHF1 led to repression of HOXB5, and HOXB6 (highlighted by a line on the top), two genes situated at the H3K36me3-H3K27me3 conjunction (panel A). Data of relative mRNA levels (y axis) from three independent experiments were normalized against vector control (shown as a dash line), and then presented as mean ± SD.
(C–F) ChIP for FLAG-tagged PHF1 (C), EZH2 (D), SUZ12 (E) and H3K27me3 (F) across different HOX-B genes in HeLa cells overexpressing either wild-type PHF1 or its Tudormutant form. Data of ChIP signals (y axis) from three independent experiments were normalized to 1% of input used, data of histone modification (F) further normalized to those of total histone H3, and then presented as mean ± SD. Statistics shown are t-test comparisons of ChIP in cells transduced with wild-type (WT) PHF1 to that with control or the mutant (Y47A) form. *, p<0.05; **, p<0.005; n.s., not significant.
PHF19/PCL3 genetic complementation in murine pluripotent stem cells demonstrates a Tudor-dependent repression of differentiation-associated genes
We next sought to examine the role of the PCL Tudor motif in PRC2-mediated gene silencing under physiological conditions. Endogenous expression of PHF1 in HeLa and HEK293 cells is low (Boulay et al., 2011), and its knockdown only induced subtle changes in global H3K27me3 levels and expression of several tested PRC2-regulated genes (data not shown). Since endogenous PHF1 expression was only found highly expressed in germ cells (Kawakami et al., 1998), we focused on a murine PHF1 homologue, PHF19 (also known as PCL3) (Figure S1B), which is highly expressed in murine ES cells and an ES cell-like embryonic carcinoma cell line, F9 (Figure S5A). Importantly, the Tudor motif of PHF19 showed similar preference and affinity towards H3K36me3 and demonstrated a similar requirement of aromatic ‘caging’ residues for binding to H3K36me3 peptides (Table 1, Figure S1J-K, and Figure S2F–G), indicating a functional redundancy among these proteins in recognition of H3K36 methylation. We also found that, unlike a recent study showing that Phf19 knockdown in ES cells also led to down-regulation of Suz12 (Hunkapiller et al., 2012), Phf19 knockdown in F9 cells did not cause any detectable change in expression of all examined PRC2 complex genes including Ezh2, Suz12 and Mtf2/Pcl2 as examined by RTPCR (Figure S5A) and by immunoblot (Figure S5B). Knocking down Phf19 in F9 embryonic carcinoma cells led to derepression of numerous PRC2 direct target genes as revealed by gene array analysis (Table S3) and RT-qPCR (Figure S5A). Consistent with a Phf19 knockdown experiment performed in E14 murine ES cells (Hunkapiller et al., 2012), the upregulated, but not down-regulated genes found in Phf19-knockdown cells were enriched with pathways associated with organismal development and cell differentiation (Table S3). Among the top 100 up-regulated genes, greater than 40% are H3K27me3 positive at their promoters, demonstrating a significant overlap between H3K27me3 and Phf19-repressed genes (with pvalue of <2.2e–16, data not shown). Such observed premature derepression of development programs in pluripotent cells is similar to phenotypes observed in Jarid2-null ES cells (Li et al., 2010; Pasini et al., 2010; Peng et al., 2009; Shen et al., 2009). Thus, F9 embryonic carcinoma cells serve as a suitable model to study functions of the Phf19 Tudor motif.
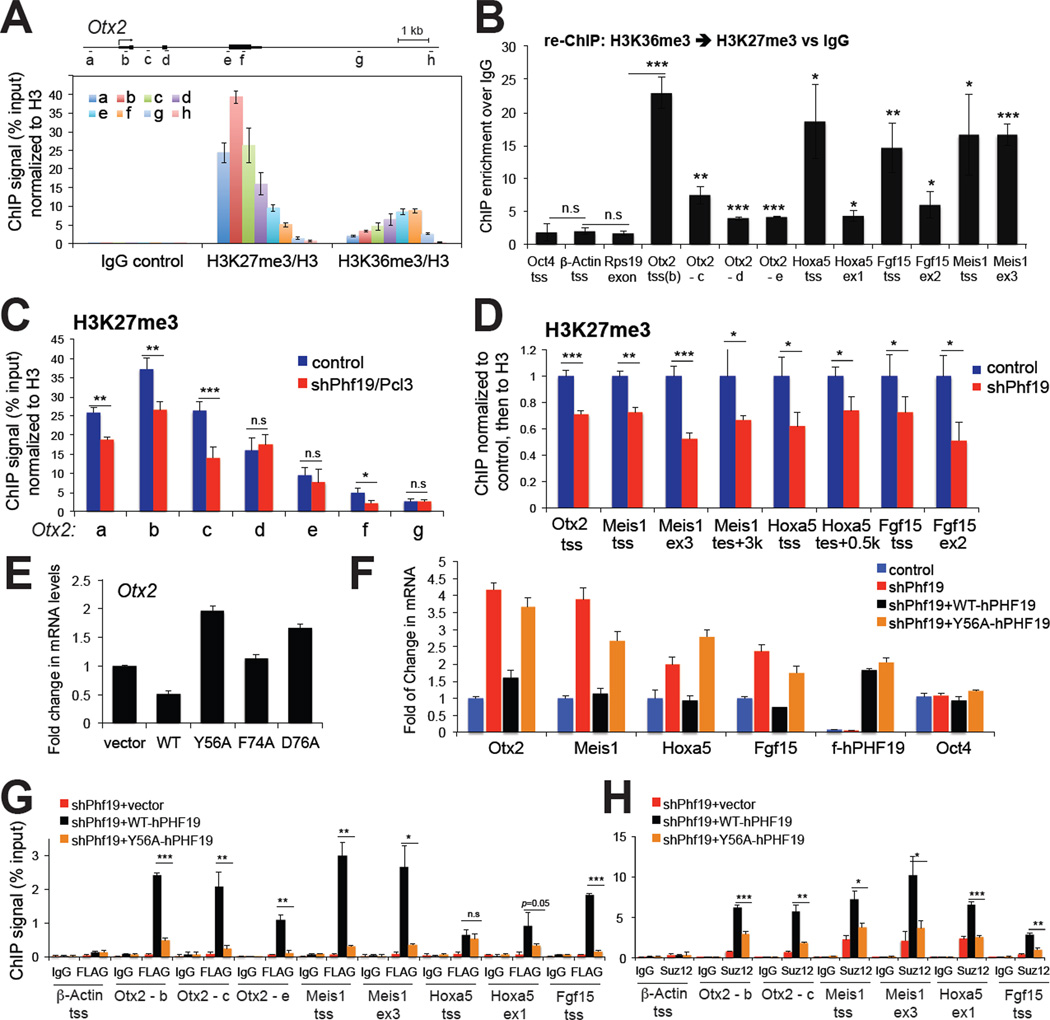
Previously, it has been shown that Phf19 binding sites strongly correlated to those of PRC2 complexes and H3K27me3 peaks (Hunkapiller et al., 2012). Our analysis of ChIP-Seq profiling data obtained from murine E14 ES cells (Hunkapiller et al., 2012; Xiao et al., 2012) further revealed a moderate genome-wide association between Phf19 and H3K36me3, where higher levels of Phf19 binding were found at promoters of genes with higher levels of H3K36me3 among genes (n=19,387) containing intermediate to low levels of H3K27me3 at promoters (Figure S5C). Analyses of these ES cell ChIP-Seq data also revealed a subset of PRC2-targeted genes marked by H3K36me3 which coexisted with H3K27me3 and Phf19, as exemplified by Otx2 (see an boxed region, Figure S5D). Using directed ChIP (Figure 5A and S5E-H) and sequential re-ChIP analyses (Figure 5B), we confirmed such co-existence of H3K36me3 and H3K27me3 at PRC2 target genes including Otx2, Meis1, Hoxa5 and Fgf15, in both ES and F9 cells. At these ‘bivalent’ PRC-targeted genes, H3K36me3 tends to increase towards gene bodies at the 3’-end, and H3K27me3 is enriched at promoter regions in the 5’-end. In addition, H3K36me3 was typically detected to a lesser extent when compared to that found at a locus of the housekeeping gene Rps19 (Figure S5E). Detection of such H3K27me3-H3K36me3 ‘bivalent’ genes is consistent with recent genomic studies showing co-existence of H3K36me3 and H3K27me3 at ‘multivalent’ development genes in zebrafish sperm cells (Wu et al., 2011), as well as a proteomic study detecting a small percentage of H3 containing both H3K27 and H3K36 methylation in mammalian cells (Voigt et al., 2012).
Figure 5. Phf19/Pcl3 demonstrates a Tudor-dependent repression of differentiation-associated genes.
(A) ChIP for H3K36me3 and H3K27me3 across the Otx2 gene in F9 embryonic carcinoma cells. The diagram on top depicts the genomic organization of Otx2, with positions of each ChIP PCR amplicon marked as a to h (not drawn to scale). Data of ChIP signals (y axis) from three independent experiments were normalized to 1% of input and to histone H3, and presented as mean ± SD. See also Figure S5.
(B) Sequential re-ChIP showing fold of enrichment by comparing ChIP signals of H3K27me3 over IgG control in a sequential IP following the first IP using H3K36me3 antibodies. Signal of ChIP (y axis) from three independent experiments were normalized to those of 1% of input and then to nonspecific IgG control, and presented as mean ± SD. Statistics shown are t-test comparisons of each tested loci to three negative control loci, the transcriptional starting site (tss) of Oct4 and β-Actin, and gene body of Rps19. *, p<0.05; **, p<0.01, ***, p<0.005; n.s., not significant.
(C–D) ChIP examining H3K27me3 levels associated with Otx2 (panel C) and other examined development genes (panel D) in F9 cells transduced with control or Phf19-specific hairpins. Data of ChIP signals (y axis) from three independent experiments were normalized to 1% of input and to histone H3, and presented as mean ± SD. ex, exon; tes, transcriptional ending site.
(E) Real-time PCR detecting Otx2 expression levels in F9 cells following overexpression of wild-type (WT) and Tudor mutant forms of PHF19/PCL3. Data of relative mRNA levels (y axis) from three independent experiments were normalized to vector transduced cells and presented as mean ± SD.
(F) Real-time PCR of Otx2, Meis1, Hoxa5, Fgf15 and Oct4 expression in F9 cells following Phf19 knockdown (sh_Phf19) and re-introduction of hairpin-resistant wild-type (WT) and Tudor mutant forms of FLAG-tagged human PHF19/PCL3 (F-PHF19). Y-axis represents fold change after normalization of data from three independent experiments to control cells (presented as mean ± SD). See also Table S3.
(G–H) ChIP examining the residence of FLAG-tagged PHF19/PCL3 (panel G) and SUZ12 (panel H) at Otx2, Meis1, Hoxa5 and Fgf15 genes after transduction of hairpin-resistant wildtype or Tudor mutant PHF19/PCL3 into knockdown cells. Signal of ChIP (y axis) from three independent experiments were normalized to those of 1% of input and presented as mean ± SD. IgG and tss of β-Actin were used as antibody and locus control, respectively. Statistics shown are t-test comparisons of ChIP in cells transduced with wild-type PHF19 to that with Tudor mutant forms. *, p<0.01; **, p<0.001, ***, p<0.0001.
Next, we used Phf19-expressing F9 cells to examine the function of the H3K36me3- ‘recognizing’ Tudor motif in regulating these bivalent genes described above. Concurrent with derepression of these development genes following Phf19 knockdown (Figure S5A), we detected a significant reduction in levels of H3K27me3 at all examined target genes (Figure 5C–D). No measurable change in H3K36me3 was found at tested loci (Supplementary Fig. S5I). Similar to PHF1 (Figure 3B) and findings of a previous report (Hunkapiller et al., 2012), both wildtype and Tudor-mutant forms of PHF19 efficiently associated with PRC2 complexes (Figure S5J). Introduction of a wild-type hairpin-resistant PHF19 into F9 cells repressed Otx2 and other tested target genes (Meis1, Hoxa5, Fgf15), either in parental F9 cells (Otx2 as tested in Figure 5E) or cells with endogenous Phf19 knocked down (Figure 5F), while the mutants that failed to engage H3K36me3 binding either led to Otx2 derepression in a dominant negative fashion in parental F9 cells (Figure 5E) or failed to restored gene repression (Figure 5F). The effect of PHF19 on gene expression was found correlated to the binding of wildtype but not Tudor-mutant PHF19 (Figure 5G) to all tested genes, the recruitment of PRC2 core components (Figure 5H), and the level of H3K27me3 (Figure S5K) as examined by ChIP assays. Collectively, our data suggests that PHF19, another member of PCL family proteins, also regulates repression of PRC2 targets via a functional Tudor motif that recognizes H3K36me3.
DISCUSSION
PCL family identified as high-affinity H3K36me3 effector proteins
In this study, we have identified the PCL family of Tudor motifs as H3K36me3 effector modules. Our structural, biochemical and biophysical studies of these Tudor motifs revealed an aromatic H3K36me3-‘caging’ pocket that forms direct contacts to H3K36me3, as well as to the surrounding H3 residues, which explained our observed high affinity and selectivity. In comparison to the previously identified H3K36me3-‘reading’ effectors BRPF1 and EAF3, whose affinities to H3K36me3 were measured in the high µM to low mM range (Kd = ~0.3–3 mM), both the PHF1 and PHF19 Tudor motifs bind to H3K36me3 with affinities in the low µM range (Kd = ~5–7 µM). These Kd measurements are similar to affinities measured for effectors of other common histone methylation marks, including H3K4me3-recognizing PHD fingers (Kd=~1–10 µM) (Li et al., 2006; Wang et al., 2009), H3K27me3- and H3K9me3- recognizing chromo domains (Kd=~5–50 µM) (Bernstein et al., 2006; Kaustov et al., 2011), and an H3K9me3-recognizing Tudor domain (Rothbart et al., 2012a). Notably, PCL family proteins also contain two PHD fingers, one of which is in close proximity to the Tudor motif (Figure S1B), and the histone-binding activities of these domains, as well as their influence of PCL functions, remains to be determined.
PCL proteins serve as a bridging and/or targeting mechanism for PRC2 to intrude into active chromatin region
Our collective biochemical and genetic studies demonstrate a PRC2-targeting mechanism wherein the cofactor PCL proteins either stabilize, tether, or establish intrusion of PRC2 core components into the H3K36me3-containing chromatin via Tudor-mediated recognition of H3K36me3 and histone H3. Co-IP studies of PHF1 showed this protein associates with PRC2 core components and assembles a higher-order complex efficiently, a process independent of the Tudor motif. We further show that PCL proteins promote PRC2 chromatin association and catalysis of H3K27me3, consistent with previous observations (Cao et al., 2008; Hunkapiller et al., 2012; Sarma et al., 2008). Our studies of ectopic PHF1 expression, modulation of global H3K36me3, as well as knockdown of endogenous Phf19 (without changing PRC2 core complexes per se), demonstrated a role of PCL proteins in the bridging of PRC2 to H3K36me3-containing nucleosomes, a mechanism that may act together with those provided by other PRC2-targeting mechanisms such as EED (Margueron et al., 2009), JARID2 (Li et al., 2010; Pasini et al., 2010; Peng et al., 2009; Shen et al., 2009) and noncoding RNAs (Bracken and Helin, 2009; Margueron and Reinberg, 2011). As we did not detect association of PHF1 or PHF19 to non-PRC2 target genes that are highly H3K36me3- positive (exemplified by CD44 in Figure S4H and β-Actin in Figure 5G), an intriguing model would be that, non-coding RNA, DNA-binding, and chromatin-binding activities associated with PRC2 complexes and cofactors (for example, EED and JARID2) collectively mediate initial PRC2 recruitment to H3K27me3-positive promoters/nucleosomes, while PCL proteins further modulate PRC2 stabilization and tether PRC2 to adjacent H3K36me3/2-positive nucleosomes towards gene bodies. Although it has been previously shown that active histone methylation marks, including H3K36me3, inhibit PRC2-mediated H3K27me3 (Schmitges et al., 2011; Yuan et al., 2011), our studies demonstrate that PRC2 spreads and intrudes into active chromatin during gene silencing and chromatin remodeling, a critical process concurrent with development and differentiation (Mikkelsen et al., 2007).
Coexisting together: how do seemingly biologically opposing chromatin marks work together in the chromatin landscape?
Our genomics and ChIP analysis revealed that relatively lower levels of H3K36me3 exist in previously described ‘poised’ bi- or multivalent chromatin state genes found in ES cells (Mikkelsen et al., 2007; Xiao et al., 2012). Such detected H3K36me3 may represent residual species of H3K36me3 docked by Phf19 in processes of PRC2-mediated chromatin remodeling associated with lineage differentiation and development. Indeed, a recent mass spectrometry based analysis identified asymmetrically methylated, ‘bivalent’ mononucleosomes that carry H3K36me3/2 along with H3K27me3 (Voigt et al., 2012). We suspect that these asymmetrically methylated mononucleosomes are more likely to be found at conjunction regions of H3K27me3 and H3K36me3 in those ‘bivalent’ or ’bimodal’ genes or loci as examined in our study (exemplified by HOX-B gene cluster in Figure 4A, PBX1 in Figure S4F, and Otx2 in Figure 5A), where PCL proteins serve a ‘bridging’ and stabilizing mechanism for PRC2 complex encroachment and spreading.
We also observed overall separation of high peaks/domains of H3K27me3 and H3K36me3 in ChIP-Seq as previously reported (Mikkelsen et al., 2007; Xiao et al., 2012) (data not shown), and we suspect that PCL proteins may be involved in a dynamic process aimed at conversion of active chromatin to the poised/repressed one by recruiting additional factors such as H3K36 demethylases to achieve completion of remodeling. We also hypothesize that residual H3K36me3 marks retained at these ‘poised’ development genes might serve as a ‘memory’ of cellular and epigenetic states, a mechanism that separates them from those silenced permanently. In accordance, expression of PCL family genes is dynamically regulated during organismal development, with PHF1 specifically expressed in murine germ cells (Kawakami et al., 1998), and Mtf2/Pcl2 preferentially expressed in undifferentiated ES cells (Walker et al., 2010). Both PHF1 and PHF19 were reported to be translocated or overexpressed in various human tumor types (Micci et al., 2006; Wang et al., 2004). Collectively, our studies strongly indicate that the chromatin- and H3K36me3- recognizing activities harbored in mammalian PCL proteins are critical for PRC2 targeting and regulation of developmentally critical genes. They further shed light on the modulation of the chromatin landscape and gene expression dynamics during processes of development and oncogenesis.
EXPERIMENTAL PROCEDURES
Plasmid construction
Plasmids containing cDNA of PHF1 (also known as polycomb-like 1 or PCL1, NCBI accession NM_024165) and PHF19 (also known as polycomb-like 3 or PCL3, NCBI accession BC125076) were purchased from Open Biosystems. cDNAs were fused to either HA-FLAG or 2xFLAG tag by PCR and cloned into MSCV retroviral expression vectors (Clontech) and pGEX GST-fusion plasmids (GE Life Science). Deletion and mutation were introduced by PCR and site-directed mutagenesis. All plasmids used were confirmed by sequencing.
Recombinant glutathione S-transferase (GST) protein production
GST-fusion proteins were generated and purified as previously described (Ruthenburg et al., 2011; Wang et al., 2009).
Histone peptide microarrays
Methods for fabrication of the histone peptide array platform (with peptide identities listed in Table S1) and analysis of effector protein binding were previously described (Fuchs et al., 2011; Rothbart et al., 2012b). A second histone peptide array platform was also obtained and used according to the manufacture’s specifications (Active Motif).
Chromatin fractionation
Chromatin and associated proteins were isolated from asynchronously growing 293 cells as described (Rothbart et al., 2012a).
ChIP and sequential re-ChIP assays
ChIP was performed using a previously described protocol (Wang et al., 2009). ChIP signals were represented as a percentage of input chromatin, and fold of enrichment was calculated by normalizing against signals of nonspecific IgG. Sequential re-ChIP was performed using a previously described protocol (Voigt et al., 2012). Information of primers used is described in Table S4.
NMR spectroscopy
Collection of NMR spectra and structural calculations are detailed in Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Tudor motifs of PHF1/PCL1 and PHF19/PCL3 bind histone H3 lysine 36 methylation
The PHF1 Tudor domain forms a unique H3K36me3-recognizing “aromatic” cage
PHF1 induces a Tudor-dependent spread of H3K27me3 into H3K36me3-marked genomic loci
Phf19 promotes Tudor-dependent H3K27me3/repression at ‘poised’ genes in stem cells
ACKNOWLEDGEMENTS
We thank K. Krajewski for synthesizing peptides, M. Vernon for gene microarray analysis, Y. Zhang for providing JMJD2 plasmids, and T. Magnuson for anti-EED antibodies used in this study. L.C. and S.B.R. are supported by postdoctoral fellowships awarded by the US Army- DOD Prostate Cancer Research Program and the UNC Lineberger Comprehensive Cancer Center Basic Sciences Training Program (T32CA09156), respectively. This research was also supported by grants from the Leukemia & Lymphoma Society and STARR Foundation to D.J.P. and C.D.A., and National Institutes of Health (NIH) grants to D.Z. and B.D.S. (GM085394 and GM068088). J.S. is supported by funds of UC-Riverside, and G.G.W. supported by NIH/NCI ‘Pathway to Independence’ Award in Cancer Research (CA151683) and University Cancer Research Fund (UCRF) of the NC State. G.G.W. is a Martin D. Abeloff, M.D. V Scholar of the V Foundation for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The structural coordinates of PHF1 Tudor domain protein in the complex with H331–41K36me3 peptide have been deposited in the Protein Data Base under accession number 2M0O.
REFERENCES
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Molecular and cellular biology. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay G, Rosnoblet C, Guerardel C, Angrand PO, Leprince D. Functional characterization of human Polycomb-like 3 isoforms identifies them as components of distinct EZH2 protein complexes. Biochem J. 2011;434:333–342. doi: 10.1042/BJ20100944. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Molecular and cellular biology. 2008;28:1862–1872. doi: 10.1128/MCB.01589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Molecular cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friberg A, Oddone A, Klymenko T, Muller J, Sattler M. Structure of an atypical Tudor domain in the Drosophila Polycomblike protein. Protein Sci. 2010;19:1906–1916. doi: 10.1002/pro.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Current biology : CB. 2011;21:53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller J, Shen Y, Diaz A, Cagney G, McCleary D, Ramalho-Santos M, Krogan N, Ren B, Song JS, Reiter JF. Polycomb-like 3 promotes polycomb repressive complex 2 binding to CpG islands and embryonic stem cell self-renewal. PLoS Genet. 2012;8:e1002576. doi: 10.1371/journal.pgen.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaustov L, Ouyang H, Amaya M, Lemak A, Nady N, Duan S, Wasney GA, Li Z, Vedadi M, Schapira M, et al. Recognition and specificity determinants of the human cbx chromodomains. The Journal of biological chemistry. 2011;286:521–529. doi: 10.1074/jbc.M110.191411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami S, Mitsunaga K, Kikuti YY, Ando A, Inoko H, Yamamura K, Abe K. Tctex3, related to Drosophila polycomblike, is expressed in male germ cells and mapped to the mouse t-complex. Mammalian genome : official journal of the International Mammalian Genome Society. 1998;9:874–880. doi: 10.1007/s003359900886. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes & development. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer research. 2006;66:107–112. doi: 10.1158/0008-5472.CAN-05-2485. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nature structural & molecular biology. 2012a;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Strahl BD, Fuchs SM. Peptide microarrays to interrogate the "histone code". Methods Enzymol. 2012b;512:107–135. doi: 10.1016/B978-0-12-391940-3.00006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, Allis CD. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Molecular and cellular biology. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Molecular cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Hong J, Zhang P, Dong X, Shen X, Lin D, Ding J. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. The Journal of biological chemistry. 2008;283:36504–36512. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nature structural & molecular biology. 2010;17:617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- Voigt P, Leroy G, Drury WJ, 3rd, Zee BM, Son J, Beck DB, Young NL, Garcia BA, Reinberg D. Asymmetrically modified nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell stem cell. 2010;6:153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Robertson GP, Zhu J. A novel human homologue of Drosophila polycomblike gene is up-regulated in multiple cancers. Gene. 2004;343:69–78. doi: 10.1016/j.gene.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Wu SF, Zhang H, Cairns BR. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011;21:578–589. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Xie D, Cao X, Yu P, Xing X, Chen CC, Musselman M, Xie M, West FD, Lewin HA, et al. Comparative epigenomic annotation of regulatory DNA. Cell. 2012;149:1381–1392. doi: 10.1016/j.cell.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Cui G, Botuyan MV, Mer G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure. 2008;16:1740–1750. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. The Journal of biological chemistry. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.