Abstract
Sulfide:quinone oxidoreductases (SQRs) are ubiquitous enzymes which have multiple roles: sulfide detoxification, energy generation by providing electrons to respiratory or photosynthetic electron transfer chains, and sulfide homeostasis. A recent structure-based classification defines 6 groups of putative SQRs (I – VI), and representatives of all but group III have been confirmed to have sulfide oxidase activity. In the current work, we report the first characterization of a predicted group III SQR from Caldivirga maquilingensis, and confirm that this protein is a sulfide oxidase. The gene encoding the enzyme was cloned, and the protein was expressed in E. coli and purified. The enzyme oxidizes sulfide using decylubiquinone as an electron acceptor, and is inhibited by aurachin C and iodoacetamide. Analysis of the amino acid sequence indicates that the C. maquilingensis SQR has two amphiphilic helices at the C-terminus but lacks any transmembrane helices. This suggests that C. maquilingensis SQR interacts with the membrane surface and that the interactions are mediated by the C-terminal amphiphilic helices. Mutations within the last C-terminal amphiphilic helix resulted in a water-soluble form of the enzyme which, remarkably, retains full SQR activity using decylubiquinone as the electron acceptor. Mutations at one position, L379, also located in the C-terminal amphiphilic helix, inactivated the enzyme by preventing the interaction with decylubiquinone. It is concluded that the C-terminal amphiphilic helix is important for membrane binding and for forming part of the pathway providing access of the quinone substrate to the protein-bound flavin at the enzyme active site.
Keywords: sulfide:quinone oxidoreductase, Caldivirga maquilingensis, monotopic membrane protein, amphiphilic helix
1. Introduction
Sulfide (S2-, HS− and H2S) is found in marine and soil environments and is endogenously produced by eukaryotic and prokaryotic cells as a product of cysteine catabolism. Although highly toxic, particularly as an inhibitor of aerobic respiration (via the terminal oxidases), sulfide is an important source of electrons in prokaryotes that can grow under phototrophic or chemotrophic conditions [1–5], as well as in mitochondria [6, 7]. In addition, sulfide (H2S) has been shown to be important in some bacteria as a universal defense against antibiotics [8] and as an endogenous gaseous signaling molecule [9, 10], and as a neuromodulator [11] in mammals.
A key enzyme in the maintenance of sulfide homeostasis and bioenergetics is sulfide:quinone oxidoreductase (SQR), which is present in many eubacteria, archaea and in the mitochondria of eukaryotic cells. SQR catalyzes the two-electron oxidation of sulfide to elemental sulfur and reduces quinone in the membrane. The sulfur is released either as a highly insoluble octameric ring, S8, or as short chains of polysulfide (HS-(Sn)-SH), which results from the reaction of elemental sulfur with H2S. The resulting sulfur is stored in cytoplasmic or periplasmic globules [2, 12]. It is not unusual for archaea or bacteria to encode more than one SQR in its genome, exemplified by Chlorobaculum tepidum, a green sulfur bacterium, which encodes at least three SQRs or SQR-like proteins in its genome [13–15].
SQR proteins are single-subunit flavoproteins with a molecular mass of about 50 kDa, associated with the prokaryotic cytoplasmic membrane or the mitochondrial inner membrane. It has been suggested that there is a single phylogenetic origin of all SQRs [16]. X-ray structures are available for SQRs from Aquifex aeolicus (a hyperthermophilic bacterium) [13, 17, 18], Acidithiobacillus ferrooxidans (an acidophilic and chemolithotrophic bacterium) [19, 20] and Acidianus ambivalens (a thermoacidophilic archaeon of the order Sulfolobales) [21]. None of these enzymes contains a transmembrane helical anchor and they are designated as monotopic membrane proteins, associated with one leaflet of the membrane bilayer [17]. Monotopic membrane proteins are generally attached to the surface of the membrane by interactions via basic residues with the phospholipid headgroups along with hydrophobic interactions with the bilayer core [22]. The extent of penetration into the membrane bilayer differs for different monotopic proteins though the amount of experimental data is quite limited [22]. Generally, SQRs have two short amphiphilic helices at their C-terminus and it is proposed that the membrane attachment is via one or both of these helices [18].
A structure-based classification system of SQRs based on sequence fingerprints has been proposed [13], which results in the definition of six groups within the SQR family (I, II, III, IV, V and VI). These groups largely correspond to a phylogenetically defined nomenclature that was recently suggested [5] : Type I = SqrA; Type II = SqrB; Type III = SqrC; Type IV = SqrD plus SqrX; Type V = SqrE and Type VI = SqrF. The greatest amount of information is available for the Type I SQRs (SqrA), which includes those from A. aeolicus and A. ferrooxidans, for which structures are known. It also includes the SQR from R. capsulatus, which has been characterized using site-directed mutagenesis [23]. The structure of a truncated version of a Type V/SqrE SQR, from A. ambivalens, has also been determined [21]. The least amount of information is available for the putative SQRs of Type III/SqrC. Whereas representatives of each of the other five SQRs have been demonstrated to have sulfide:quinone oxidoreductase activity, no Type III SQR has been shown to have this enzymatic activity. Genes encoding Type III SQR genes are found in both archaea (e.g., Vulcanisaeta distribute and Pyrobaculum aerophilum) and in bacteria (e.g., Magnetospirillum magnetotaticum and Chlorobaculum tepidum).
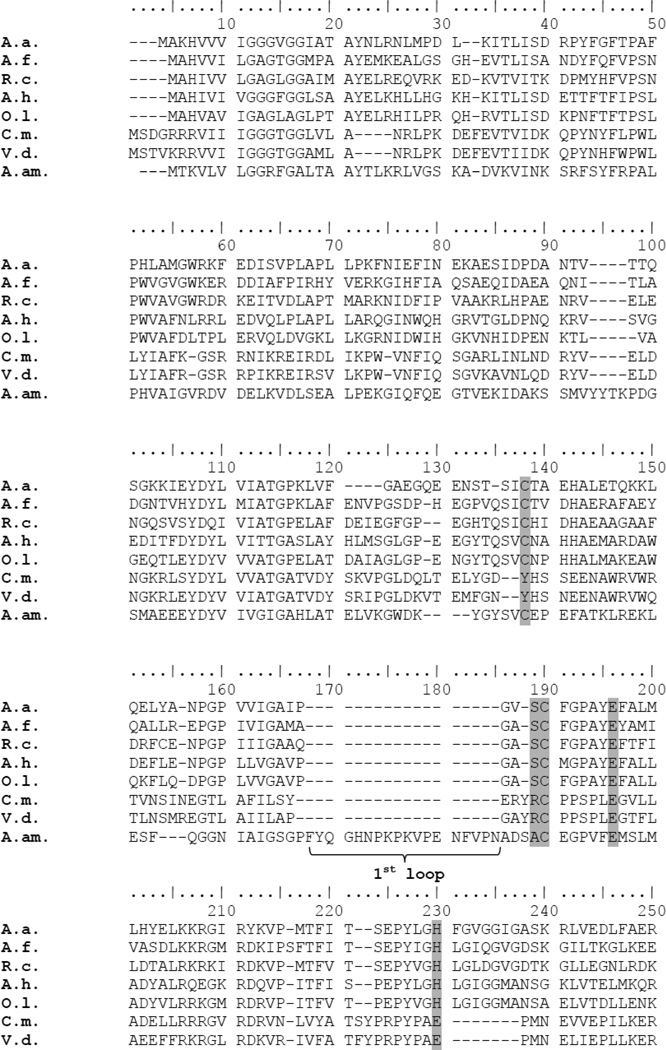
Figure 1 compares the sequences of several Type I and Type III SQRs, along with the sequence of the Type V SQR from A. ambivalens. Among the features [13] that distinguish the Type III SQRs are 1) the absence of a cysteine residue which, in Type I SQRs is implicated in flavin binding (C124 in the A. aeolicus SQR); 2) the absence of two “capping loops” (designated loop 1 and loop 2), one of which (loop 2) is present in the Type I SQRs and the other in Type V SQRs; 3) absence of an elongated C-terminus, which characterizes the Type I SQRs.
Figure 1. Amino acid sequence alignment of several SQRs, including that from C. maquilingensis.

The residues and regions of the sequences that are used to distinguish the 6 classes of SQRs are highlighted in gray. The “1st and 2nd capping loops” and the extended C-terminal are also indicated. Asterisks mark the mutations made in the C. maquilingensis SQR. A.a., Aquifex aeolicus Type I SQR; A.f., Acidithiobacillus ferrooxidans Type I SQR; R.c., Rhodobacter capsulatus Type I SQR; A.h. Anphotece halophytica Type I SQR; O.l., Oscillatoria limnetica Type I SQR; C.m., Caldivirga maquilingensis Type III SQR; V.d., Vulcanisaeta distributa Type III SQR; A.am., Acidianus ambivalens Type V SQR.
The primary goal of the current work was to obtain a protein designated as a type III SQR and verify that the protein, in fact, has the predicted enzymatic sulfide:quinone oxidoreductase activity. In order to address it, the Type III/SqrC SQR from Caldivirga maquilingensis [24], a hyperthermophilic crenarchaeon, was characterized. This organism grows either anaerobically or microaerophilically optimally at 85° C under acidic conditions (pH 4). The chromosomal gene encoding this enzyme was cloned and expressed in E. coli. The enzyme is associated with the bacterial membrane and purified following detergent solubilization. The C. maquilingensis SQR oxidizes sulfide, using decylubiquinone as the electron acceptor. Since relatively little is known about the details of how monotopic membrane proteins bind to the membrane [22], a secondary goal was to define which portion(s) of the polypeptide are required for membrane binding. For that, site-directed mutagenesis was used to demonstrate that the C-terminal amphiphilic helix is both necessary and sufficient for membrane binding.
2. Materials and Methods
2.1. Sequence analysis
Genes encoding SQRs were retrieved from the National Center for Biotechnology Information (NCBI) and Department of Energy Joint Genome Initiative (JGI) databases. The sequences were aligned using MUSCLE [25]. Conserved residues were identified with BIOEDIT (Hall TA, 1999). Secondary structure was predicted by using PSIPRED v3.0 software [26].
2.2. Construction of expression plasmids and site-directed mutagenesis
The SQR from Caldivirga maquilingensis IC-167 [24] strain was cloned into pET22b (Apr, Novagen). A Quick Change site-directed mutagenesis kit (Stratagene) was used to construct the different point mutations. The His-tag (8 histidines) was introduced in the N-terminal and the final plasmids were transformed into E. coli C43 (DE3) strain (Avidis, France) containing pRARE (Cmr, Novagen).
2.3. Cell growth, enzyme expression and purification
E. coli C43 cells were grown in LB medium with 100 µg/ml ampicillin and 20 µg/ml chloramphenicol at 37 °C. Gene expression was induced by addition of 1 mM IPTG (isopropyl-D-thiogalactoside) when cells reached an OD600 ~ 0.7. All purification procedures were carried out at 0–4° C. Cells were harvested and resuspended in buffer A (50 mM sodium phosphate, pH 7.5, 300 mM NaCl) with 5 mM MgSO4, DNase I and a protease inhibitor cocktail (Sigma). The cells were then disrupted by passing twice through a microfluidizer at a pressure of 80,000 psi. The cell extract was centrifuged at 14,000×g for 10 min to remove the unbroken cells. Membranes were obtained after centrifugation at 230,000×g for 4 h. Pellets were resuspended in buffer A plus the protease inhibitor cocktail, and then solubilized by the addition of a stock solution of 20% DDM (dodecyl-β-D-maltoside) dropwise to a final concentration of 1%. The suspension was incubated at 4° C for 2 h with mild agitation and then cleared by centrifugation at 230,000×g for 1 h. The supernatant was added to 5 ml Ni-NTA resin (Qiagen) pre-equilibrated with buffer A plus 0.05% DDM and 10 mM imidazole. The protein bound to the resin was washed with buffer A plus 0.05% DDM and 50 mM imidazole and then eluted with buffer A with 0.05% DDM and 200 mM imidazole. Fractions were concentrated by filtration, and the imidazole was removed by dialysis against buffer A plus 0.05% DDM. The purified protein could be stored frozen at −80°C after the addition of glycerol to a final concentration of 10%.
2.4. Analytical methods
Protein concentration was determined using the BCA protein assay (Thermo Scientific, Pierce Protein Research Products). The protein purity was evaluated by SDS-PAGE using a 4–20 % gradient gel (NuSep). The FAD content of the pure protein was determined spectroscopically with an Agilent Technologies spectrophotometer (model 8453), using an extinction coefficient of 10.8 cm−1 mM−1 for the oxidized flavin [27] after extraction from the protein by treatment of the sample with 5 % trichloroacetic acid [28]. No flavin was present in the precipitated protein pellet. Additionally, fluorescence emission and excitation spectra of the supernatant and the resuspended pellet were recorded using a fluorescence spectrophotometer (Cary Eclipse, Agilent Technologies). The samples were excited at 365 nm and the emission spectra were recorded from 480 to 630 nm. The redox state of the flavin in the intact protein was monitored by the fluorescence excitation spectrum (emission wavelength of 520 nm), recorded at room temperature before and after the addition of different concentrations of sulfide (reductant) and quinone (oxidant).
2.5. Enzyme activity assays
SQR activity was measured at 60° C. The 200 µL reaction mixture contained 50 mM sodium phosphate pH 7.5, 300 mM NaCl, 0.05% DDM, 10 µM to 200 µM decylubiquinone (Sigma), and 5 µg (0.55 µM) of the purified enzyme. The reaction was started with the addition of 25 µM to 250 µM sodium sulfide, prepared fresh with N2-flushed 50 mM sodium phosphate, pH 7.5. The reaction progress was monitored for 3 min by the decrease in absorption of decylubiquinone at 275 nm using a Shimadzu spectrophotometer UV-2101PC, following a procedure modified from Shahak et al [29]. An extinction coefficient of 12.4 cm−1 mM−1 at 275 nm was used to determine the extent of reduction of decylubiquinone [30].
2.6. Determination of kinetic constants
Kinetic parameters were determined using nonlinear least square analysis (Origin8.0) of the data fitted to the Michaelis–Menten rate equation (v = Vmax (S) / (Km + S), where v is the velocity, Vmax is the maximum velocity, S is the substrate concentration and Km is the Michaelis-Menten. The enzyme rates are expressed as a turnover number (kcat) based on the concentration of flavin, nmol quinone reduced s−1 nmol FAD−1.
3. Results
3.1. Protein expression and characterization
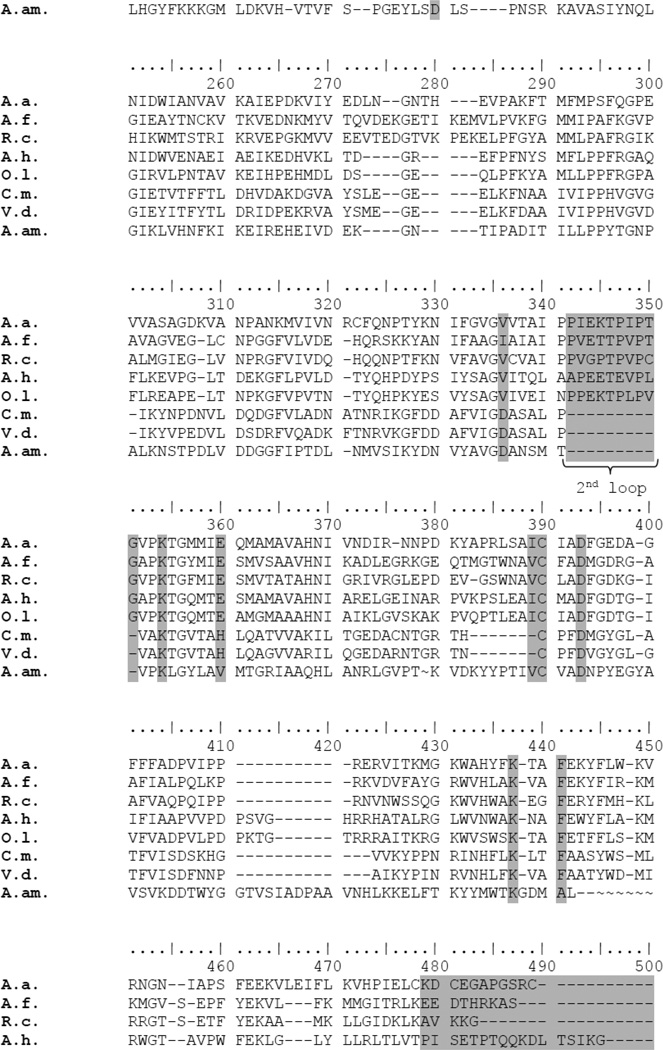
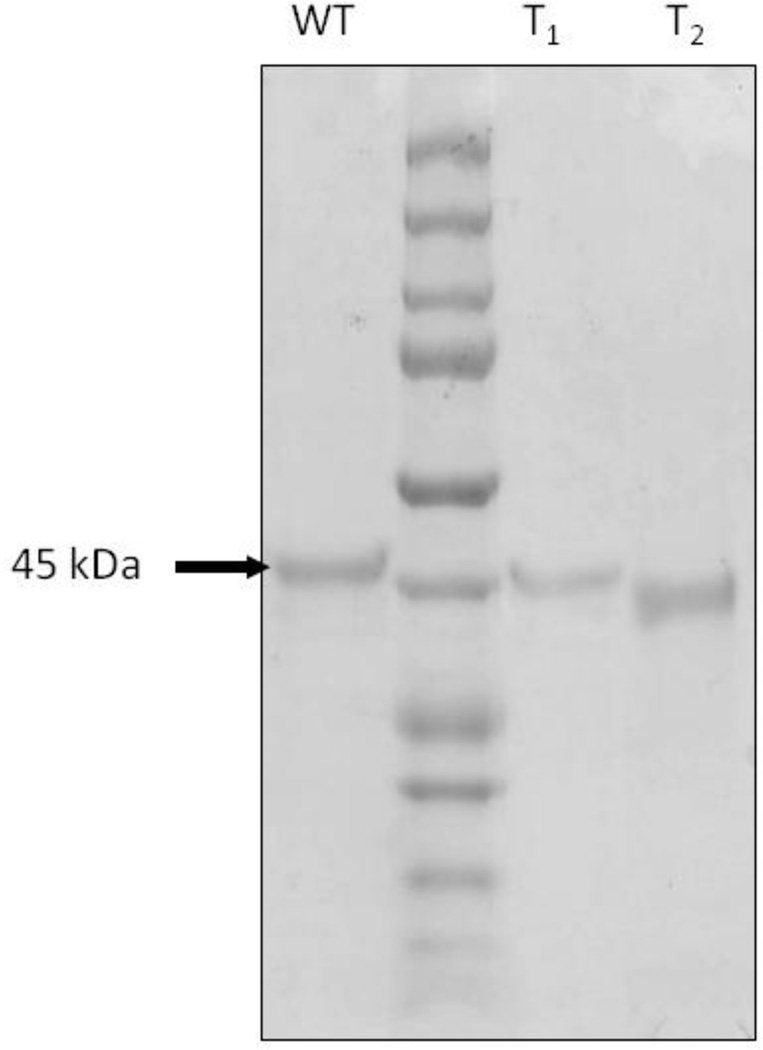
Recently Marcia et al [13] proposed a new classification of SQRs into six different groups. Analysis of the amino acid sequence of the SQR from C. maquilingensis (Figure 1) reveals that this enzyme belongs to Type III, or SqrC according to the nomenclature of Gregersen et al [5]. The His-tagged protein was heterologously expressed in E. coli, solubilized from membranes using the detergent DDM, and purified by affinity chromatography. The protein yield was 2 mg from 1 liter of cell culture. SDS-PAGE analysis shows a single band with apparent molecular weight of 45 kDa (Figure 2), similar to other SQRs [13, 20, 21, 31]. The UV-visible spectrum shows the presence of FAD in the protein, and a shoulder at 465 nm indicating the cofactor is in an apolar environment [32] (Figure 3A). Precipitation of the protein with 5% tricholoracetic acid leaves the FAD cofactor in solution, demonstrating that the FAD is not covalently bound to the protein (Figure 3B). Quantitation of the extracted flavin shows about 50–60% of the expected value based on one equivalent of FAD per SQR polypeptide. A low flavin content has been observed with other heterologously expressed flavoproteins and is not unusual [23, 33]. Several methods were attempted to increase the FAD content without success. Among the methods used were 1) addition of 0.5 mg/L riboflavin to the growth medium; 2) decrease the growth rate after induction with IPTG, by reducing both the temperature (to 30° C) and agitation rate (to 100 rpm); 3) reduction of the rate of protein expression by decreasing the IPTG concentration to 0.5 mM; 4) addition of 20 µM FAD to all of the the purification buffers; and 5) flavin reconstitution by a cycle of deflavination-flavination of the purified protein [34]. These were all unsuccessful at increasing the incorporation of FAD into the protein.
Figure 2. Expression and purification of SQR, SQRT1 and SQRT2.
SDS-PAGE gel showing the purified C. maquilingensis SQR (WT), along with the water-soluble truncated forms of the enzyme. T1 has the last 21 amino acids removed, including the C-terminal amphiphilic helix, and T2 lacks 45 amino acids and both amphiphilic helices.
Figure 3. UV-visible and fluorescence spectra of the purified C. maquilingensis SQR.
(A) The pure protein exhibits a typical UV-visible absorption spectrum of a flavoprotein, with characteristic peaks at 375 and 440 nm for the oxidized FAD. (B) After a brief incubation with 5% trichloroacetic acid at room temperature, the fluorescence emission spectra of supernatant (dashed line) and resuspended protein (straight line) were recorded between 470 nm and 650 nm (excitation at 365 nm). The data show that the FAD is not covalently bound to the protein.
The sulfide oxidase activity of the purified SQR was examined with several different quinones as oxidants: menadione, duroquinone, ubiquinone-1 and decylubiquinone. The highest steady state activity was observed with decylubiquinone (Table 1), and all subsequent characterizations were performed with decylubiquinone as the electron acceptor. The steady state activity increased as a function of temperature between room temperature and 60°C, the highest temperature examined, to a value of 0.60 nmol quinone reduced s−1 nmol FAD−1 (kcat = 0.6 s−1). The optimal growth temperature for C. maquilingensis is 85°C [24], but measurements above 60°C were not feasible. The activity was not increased by the addition of FAD (20 µM) to the assay buffer. At 60°C, the Km values for both sulfide and decylubiquinone are in the micromolar range (Table 2). The enzyme activity is inhibited completely in the presence of 250 nM aurachin C, a quinone analogue [35, 36], and by iodoacetamide (300 µM), a sulfhydryl blocking agent, which is consistent with reports of SQRs from other organisms [2].
Table 1.
Relative sulfide:quinone oxidoreductase activities of the C. maquilingensis SQR with different short chain quinones.
| Electron acceptor | Activity (%) |
|---|---|
| Decylubiquinone | 100a |
| Menadione | 25 |
| Duroquinone | 23 |
| Ubiquinol-1 | 15 |
Activity measured with decylubiquinone is 0.60 ± 0.07 nmol quinone reduced s−1 nmol FAD−1 (100%), and all activities are expressed as the percent relative to this value. Measurements were performed at 60°C as described in the text.
Table 2.
Steady state kinetics parameters for wild type and mutant C. maquilingensis SQRs
| Mutant SQR | Localization (E. coli) |
kcata (s−1) |
Km(Na2S) (µM) |
Km (decylubiquinone) (µM) |
|---|---|---|---|---|
| WT | membrane | 0.60 ± 0.07 | 77 ± 8 | 30 ± 1 |
| SQRT1 | cytoplasm | 0 | ----- | ----- |
| SQRT2 | cytoplasm | 0 | ----- | ----- |
| Y383Q/F384K | membrane (50%) |
0.82 ± 0.04 | 46 ± 2 | 33 ± 3 |
| cytoplasm (50%) |
1.20 ± 0.08 | 77 ± 5 | 36 ± 2 | |
| L379D/M380N | membrane (50%) |
0 | ---- | ---- |
| cytoplasm (50%) |
0 | ---- | ---- | |
| L379D | membrane | 0 | ---- | ---- |
| L379N | membrane | 0 | ---- | ---- |
| M380N | membrane | 0.62 ± 0.06 | 73 ± 6 | 32 ± 3 |
| Y383Q/F389K L379D/M380N |
cytoplasm | 0 | ---- | ---- |
Activity is expressed as nanomol of decylubiquinone reduced per second per nanomol of FAD, measured as described in the text at 60 °C. Data are expressed as average ± SD of three independent experiments.
3.2. Conversion of C. maquilingensis SQR from a membrane protein to a soluble protein
The amino acid sequence analysis of C. maquilingensis SQR shows no predicted transmembrane helices. Therefore, like other SQRs [17], the C. maquilingensis SQR is a monotopic membrane protein, using the classification of Blobel [37]. One experimental manifestation of this is that treating the E. coli membranes containing C. maquilingensis SQR with 1 M NaCl (or higher) results in quantitatively removing the protein from the membrane. Although the salt-extracted enzyme has no sulfide:decylubiquione oxidoreductase activity, subsequent dialysis to remove the salt in the presence of 0.05% DDM fully restores the activity (not shown). This indicates no irreversible damage has been done to the protein by this treatment.
Monotypic membrane proteins have one or more segments that enter and leave the membrane bilayer from the same side. A computational study [22] of 11 different monotopic membrane proteins showed that the interactions with the membrane were mediated primarily through hydrophobic amino acid residues (particularly F, L, I and V, but also Y and W) which insert into the non-polar portion of the bilayer, as well as basic residues (R and K, but also H to a lesser extent). These residues may be distributed over the protein interface in contact with the membrane, or may be localized within one or more amphiphilic helices.
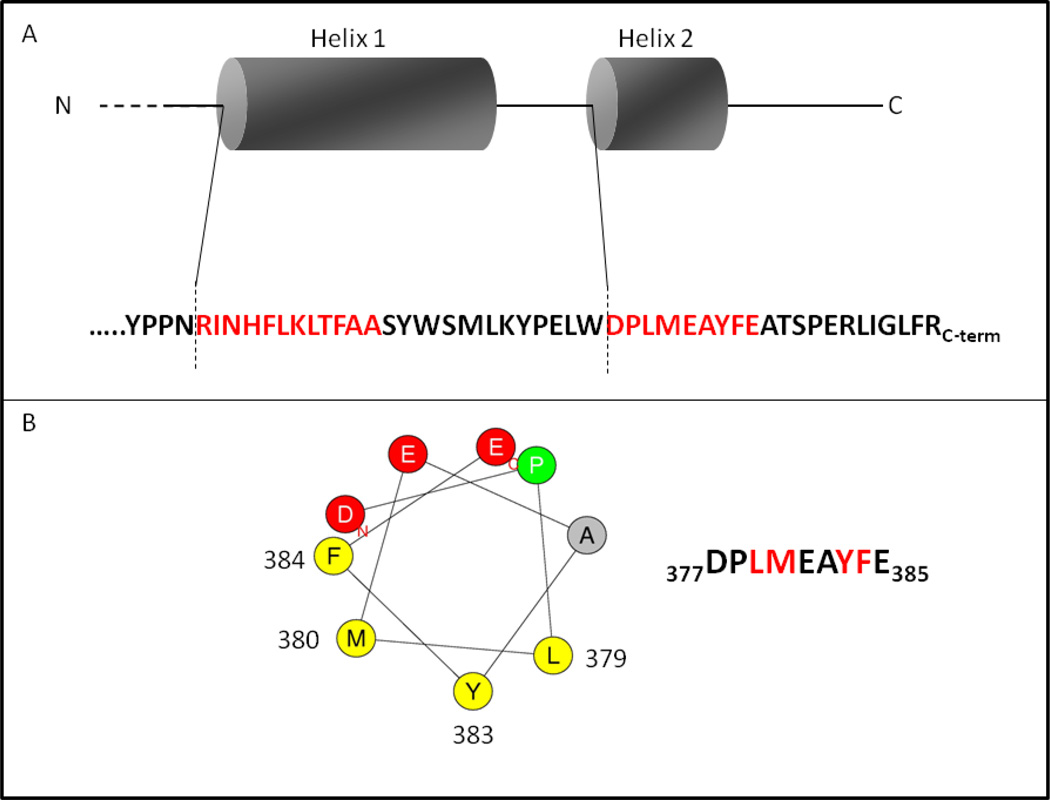
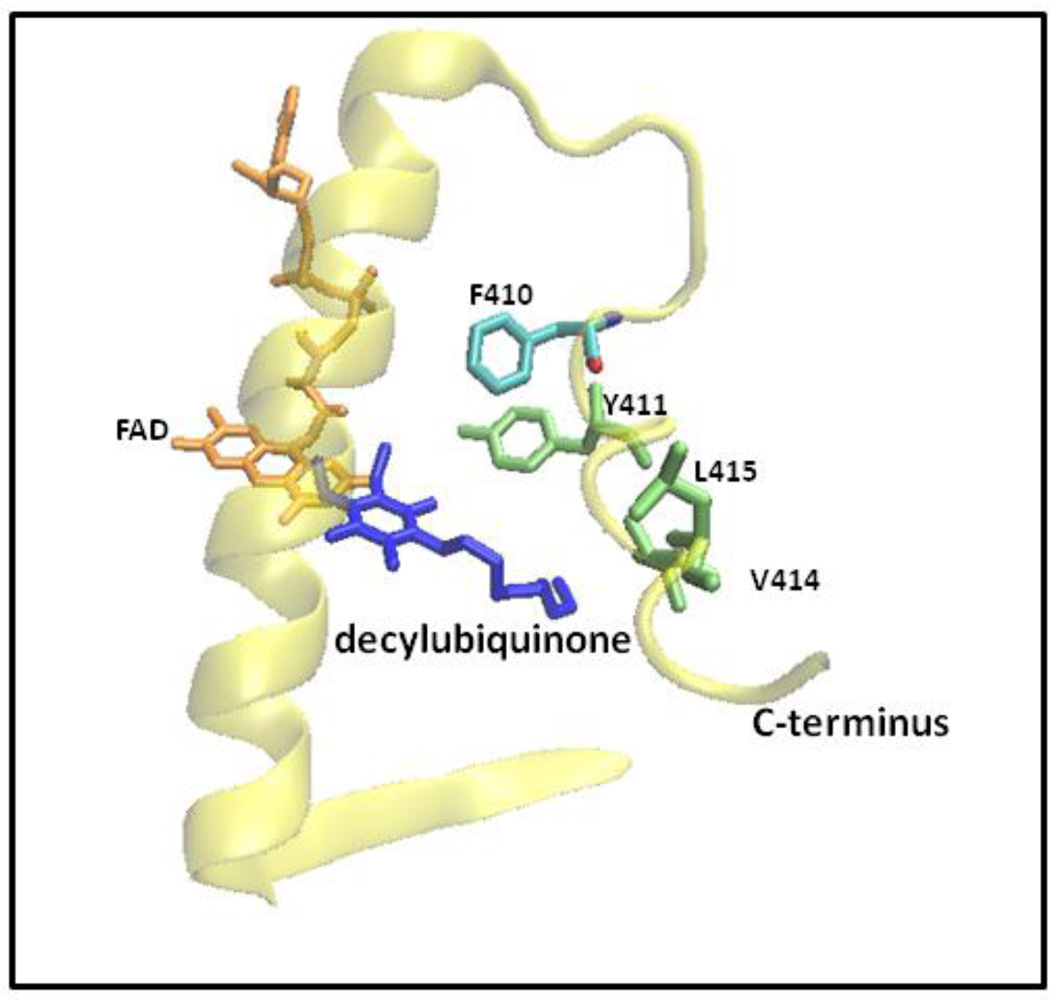
The C. maquilingensis SQR has two amphiphilic helices near the C-terminus (Figure 4). The hypothesis that these mediate membrane binding was tested by site-directed mutagenesis both by removal of the helices and by point mutations (Table 2). The mutated residues were chosen based on Heliquest software predictions [38]. The amino acids selected for mutation were hydrophobic and proposed to be located on the hydrophobic side of the helix (Figure 4). Figure 5 shows the X-ray structure of the C-terminal region of the SQR from A. ferrooxidans [19, 20] corresponding to that which was modified by mutagenesis in the C. maquilingensis SQR. One evident feature is that residues in the C-terminal amphipathic helices are also at or near the binding site for the substrate decylubiquinone (Figure 5).
Truncations: Two constructs were made. In SQRT1 a stop codon was introduced to eliminate the last 21 amino acids from the C-terminus, removing one putative amphiphilic helix. Figure 4B shows a helical wheel diagram of this helix. In construct SQRT2, the last 45 amino acids are removed, thus eliminating both of the amphiphilic helices (Figure 4). Both SQRT1 and SQRT2 when expressed in E. coli resulted in water-soluble proteins (Table 2). In each case the yield of protein was nearly 5-fold higher than the wild type construct, in which the recombinant protein is bound to the membrane. The FAD content of each of the truncated proteins, as well as the characteristics of the absorption spectra, were identical to those of the detergent-solubilized, wild type SQR. However, no sulfide:decylubiquinone oxidoreductase activity was observed in either case. These results show that minimally, the last C-terminal amphiphilic helix is both necessary and sufficient to mediate the binding of C. maquilingensis SQR to the membrane bilayer.
- Point mutations: Several combinations of multiple mutations as well as single mutations in the C-terminal amphiphilic helix were examined.
- Y383Q/F384K/L379D/M380N: Guided by the amphiphilic pattern of the putative C-terminal helix shown in Figure 4, four point mutations were introduced into the SQR, replacing hydrophobic residues by polar or ionizable amino acids. The resulting protein is found entirely in the cytoplasmic fraction but there is no catalytic activity (Table 2). This shows that the complete deletion of the C-terminus is not required to free the protein from the membrane.
- Y383Q/F384K: This double mutant corresponds to two of the four mutations shown above to eliminate membrane binding. This protein was expressed in a yield similar to the wild type SQR, and was found equally in the cytoplasmic and membrane fractions after cell disruption (Table 2), i.e., half of the protein was found in a water-soluble form. The SQR was isolated from each fraction, using DDM to solubilize the membrane-bound fraction as with the wild type protein. The isolated proteins from each fraction contained FAD to the same extent as the wild type SQR. Remarkably, both the soluble and membrane-bound versions of this double-mutant are catalytically active (sulfide:decylubiquinone oxidoreductase activity). The comparison of the steady state kinetic parameters of the enzymes isolated from the membrane and cytoplasmic fractions is shown in Table 2. The membrane-bound mutant enzyme has a specific activity about 30% higher than the wild type SQR and the Km for sulfide is about half of the value found for the wild type (46 µM vs. 77 µM). The water-soluble version of this mutant SQR is twice as active as the wild type SQR (1.20 vs. 0.60 nmol quinone reduced s−1 nmol FAD−1) and the Km values for both sulfide and decylubiquinone are about the same as the wild type, membrane-bound form. This double mutant does not eliminate, but weakens the membrane association of the protein. The soluble form is, however, functional with decylubiquinone as substrate.
- L379D/M380N: This double mutant corresponds to the second pair of mutations in the quadruple mutant, Y383Q/F384K/L379D/M380N, which eliminates membrane binding. As with the Y383Q/F384K double mutant (above), the expressed protein was found in both the cytoplasmic and membrane fractions in equal proportions after disruption of the E. coli cells (Table 2), and each fraction had the same FAD content as the membrane-bound wild type SQR (about 50%). The total yield of this recombinant protein was comparable to the wild type. However, both the membrane-bound and soluble forms of this protein were inactive (Table 2). Hence, this double mutant, like the Y383Q/F384K mutant described above, weakens the membrane affinity but does not eliminate membrane binding of the protein. Furthermore, one or both of the mutations in the L379D/M380N double mutant must be responsible for the loss of catalytic activity.
- M380N: This is one of the two mutations in the L379D/M380N double mutant. The M380N mutation by itself results in protein that is entirely membrane-bound, but which has the same activity as wild type SQR. Hence, this substitution by itself is not responsible for the loss of activity of the L379D/M380N double mutant but is insufficient to result in any of the recombinant protein being free of the membrane.
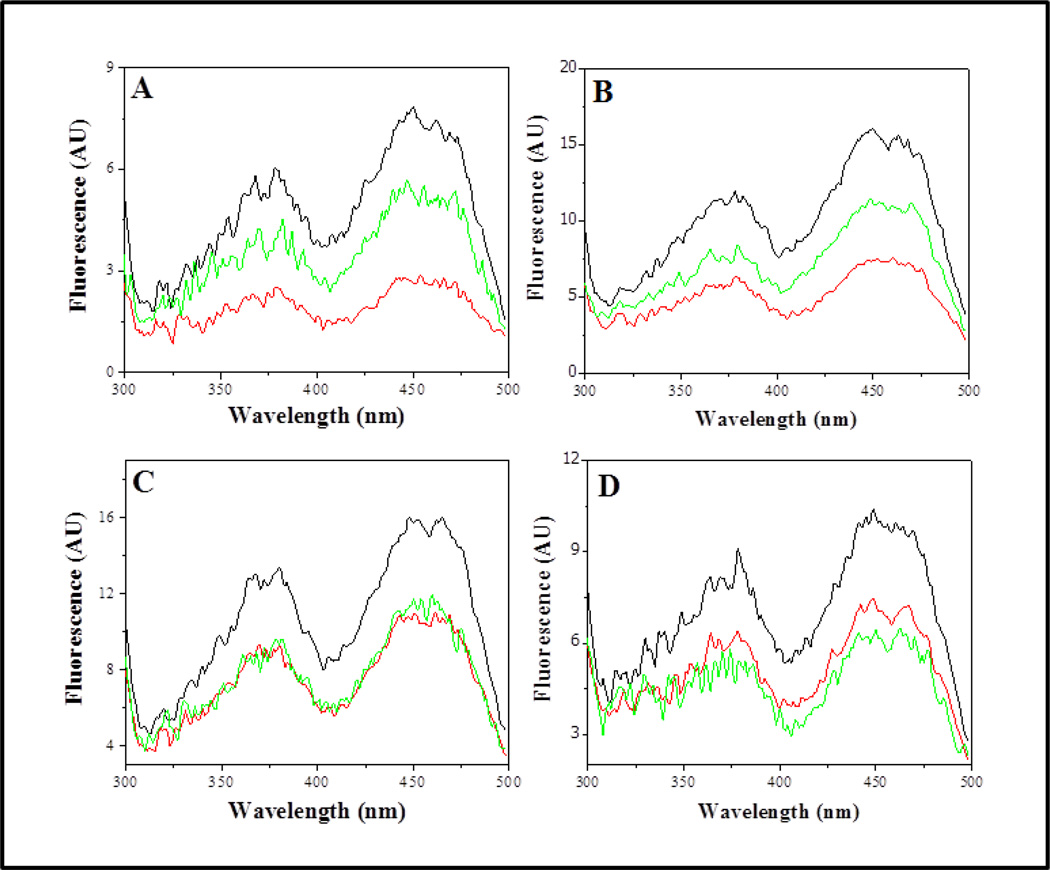
- L379D: This single mutation is responsible for the loss of catalytic activity of the Y383Q/F384K/L379D/M380N quadruple mutant of the C. maquilingensis SQR. All of the expressed protein is membrane-bound. Because of the likely proximity of this region of the protein to the binding site for decylubiquinone (Figure 5), the function of this site was investigated further. Figure 6 shows the fluorescence excitation spectrum of the protein-bound flavin for the wild type as well as several mutant SQRs. In each case the spectrum is shown: 1) in the absence of substrates; 2) after the addition of sulfide; and 3) after the subsequent addition of decylubiquinone. For the wild type SQR as well as for each mutant examined, the addition of sulfide results in reduction of the bound FAD, and the lower intensity of the fluorescence. Subsequent addition of decylubiquinone to the wild type SQR restores the FAD to the oxidized state, with concomitant increase in the FAD fluorescence intensity. The same result is obtained with the fully active M380N mutant. However, the addition of decylubiquinone to the L379D mutant fails to reoxidize the protein-bound FAD (Table 2 and Figure 6). It is concluded that this mutant is inactive due to the selective perturbation of the interaction with decylubiquinone.
- L379N: This mutation places a polar but non-ionizable residue in place of L379. The mutant behaves identically to L379D, described above, and is inactive due to a perturbation of the decylubiquinone binding site (Figure 6). Hence, placing a potential negative charge at this site is not required to inactivate the enzyme.
Figure 4. The amphiphilic helices at the C-terminal region of the C. maquilingensis SQR.
(A) Schematic showing the sequences and locations of the two amphipathic helices. The sequence in red corresponds to the predicted amphiphilic helices, and the dashed lines indicate the locations of the C-termini of the two truncations that result in water-soluble protein. (B) A helical wheel depiction of the amphiphilic helix closest to the C-terminus (Helix 2 in Panel A). The sequence is shown, with the two pairs of residues in red, which were altered by mutagenesis.
Figure 5. A portion of the C-terminal structure of the Acidithiobacillus ferrooxidans SQR (PDB ID: 3T31) [19, 20] highlighting the homologous amino acids mutated in C. maquilingensis.
F410 in the A. ferroxidans sequence corresponds to L379 residue in the C. maquilingensis sequence.
Figure 6. Fluorescence excitation spectra of purified wild type and mutant C. maquilingensis SQR.
Excitation spectra are shown for (A) wild type, (B) M380N, (C) L379D and (D) L379N SQR proteins. Spectra were recorded prior to substrate addition, (black lines); 20 min after addition of 150 µM sulfide (red lines); 20 min after addition of 100 µM decylubiquinone (green lines). All experiments were performed at room temperature, resulting in slow oxidation and reduction of the FAD. Reoxidation of the FAD upon addition of decylubiquinone is not observed for the L379D and L379N mutants. The protein concentration was 1 mg ml−1 (22 µM protein; ~11µM FAD). The emission was monitored at 520 nm. Data are representative of results of at least three separate experiments. AU, arbitrary units.
In summary, these data demonstrate that the affinity of recombinant C. maquilingensis SQR for the E. coli membrane is substantially reduced by the substitution of two amino acids in the C-terminal amphiphilic helix, resulting in half of the isolated protein being found in the cytoplasm. Substitution of four non-polar residues in the amphiphilic helix with more polar amino acids is sufficient to convert the monotypic membrane protein to a water-soluble form. The role of the C-terminal amphiphilic helix in the binding of the quinone substrate is also demonstrated by the L379 mutations. This is not surprising since in vivo the quinone must access the enzyme-bound flavin through the membrane bilayer.
4. Discussion
4.1. Biochemical characteristics of the enzyme
The first significant aspect of this work is the demonstration that the gene from C. maquilingensis predicted to encode a Type III/SqrC SQR [5, 13] does, indeed, encode an enzyme with SQR activity. This is the first enzyme in this classification to be characterized and shown to be an SQR. Furthermore, the C. maquilingensis SQR is only the second representative of SQR from an archaeal source, the first being from A. ambivalens [21]. The activity of C. maquilingensis SQR is comparable to the activity reported for A. ambivalens SQR (at 50 °C) [21]. This is 10-fold slower than the SQR from R. capsulatus [31], 60-fold slower than the SQR from A. ferrooxidans [39] and 100-fold slower than the SQR from A. aeolicus (at 80 °C) [17].
To date, all SQRs appear to contain FAD as a cofactor. The SQRs from A. ambivalens [21] and from A. aeolicus [18] each have covalently-bound FAD linked to a cysteine residue. Although the homologous cysteine is present in the SQR from A. ferrooxidans [20], the FAD is non-covalently bound to this enzyme. The homologous cysteine is not present in the sequence of C. maquilingensis SQR (Type III) or in Type II or bacterial Type V SQRs [13]. Also, it has been demonstrated in R. capsulatus SQR that mutation of this cysteine (C127S) does not alter the FAD content of the mutated enzyme compared to the wild type, although the sulfide-dependent reduction of the FAD is inhibited [23]. Certainly, when present, the cysteine at this sequence position in SQRs is at the FAD binding site, but this cysteine is not essential for all SQRs for either flavin binding or function.
4.2. Attachment of the SQR protein to the membrane
SQRs have no transmembrane segments and are classified as monotopic proteins [37]. Other monotopic proteins include prostaglandin H2 synthase-1 [40], prostaglandin H2 synthase-2 [41], the electron transfer flavoprotein:ubiquinone oxidoreductase [42, 43], glycerol-3-phosphate dehydrogenase [44] and oxidosqualene cyclase [45]. These proteins interact at the surface of one side of the membrane bilayer, but the depth of penetration and extent of perturbation of the membrane may differ substantially for different monotopic proteins [22]. There is no signature folding motif or sequence to identify the membrane-interacting domain(s). In some cases, the primary mode of attachment is through an amphiphilic helix which lies more-or-less parallel to the plane of the bilayer with hydrophobic residues penetrating into the hydrophobic region of the membrane. Examples are fatty acid amide hydrolase [46] and the electron transfer flavoprotein:ubiquinol oxidoreductase [42, 43]. Other monotopic membrane proteins interact through a hydrophobic patch consisting of residues from different parts of the protein sequence, exemplified by apocarotenoid cleavage oxygenase [47]. Computational [22] as well as bioinformatics surveys [48] indicate that the stabilizing interactions are through hydrophobic residues and through basic residues which interact with the phospholipid headgroups. It is noted that the membrane of C. maquilingensis, contains large amounts of tetraether core lipids [24], which are very different from the E. coli membrane lipids. Nevertheless, it is likely that the pattern of affinities of mutant SQRs for the E. coli membrane will also apply qualitatively for the natural membrane under physiological conditions.
The C. maquilingensis SQR has two predicted amphiphilic helices near the C-terminus. The fact that the protein can be readily removed from the E. coli membrane by 1 M NaCl suggests that the membrane-protein interaction is fairly weak, consistent with the main interaction being through amphiphilic helices that do not penetrate deeply into the bilayer. The essential functional requirement is that the membrane-bound quinone must be able to access the FAD to facilitate electron transfer. The X-ray structures of the SQRs from A. aeolicus [18] and from A. ferrooxidans [20] each contain a quinone, and each of these proteins has been modeled to interact with the membrane through two C-terminal amphiphilic helices, with an opening facing the hydrophobic core of the bilayer which leads to an hydrophobic channel through which the quinone can access the reduced FADH2. This is likely to also be true for the C. maquilingensis SQR.
In attempting to define residues critical for membrane association by site-directed mutagenesis, the overlap between regions of the protein important for quinone interaction and for membrane binding is a complication. This was encountered when the C-terminal helix was removed by truncation of the final 21 amino acid residues. The resulting protein is water-soluble but has no catalytic function, presumably because the access of the decylubiquinone substrate was perturbed in the mutant. The same is also true for the L379D/M380N mutant SQR, half of which is isolated in a soluble form, but which has no catalytic activity. The lack of activity was shown to be entirely due to mutation of L379, and the lack of enzyme activity is due to the inability of the protein-bound FADH2 to be oxidized by decylubiquinone (Table 2 and Figure 6). L379 corresponds to F410 in the structure of A. ferrooxidans SQR (Figure 6), which interacts with the aliphatic tail of decylubiquinone. Hence, this mutation likely either restricts access of the quinone to the active site flavin, or destabilizes quinone binding.
Mutation of the pair of residues Y383Q/F384K also results in 50% of the protein being water-soluble. However, in this case the soluble version of the C. maquilingensis SQR has twice the sulfide:decylubiquinone activity as the detergent-solubilized wild type enzyme. The decreased sulfide Km of the detergent-solubilized version of this mutant SQR could be related to the introduction of a positive charge close to the flavin, possibly altering the redox potential of the cofactor [49]. Alternatively, the positive charge of the lysine might increase the attraction of the anionic substrate (sulfide).
There are relatively few experimental data on the interactions of monotopic membrane proteins with the lipid bilayer [22]. The protein-membrane interaction of C. maquilingensis is relatively weak and disrupted by just a pair of mutations in one amphiphilic helix. In order to convert 100% of the C. maquilingensis SQR to a water-soluble form, we substituted 4 hydrophobic amino acids within the C-terminal amphiphilic helix with polar residues. The data do not rule out favorable interactions between the membrane and the second putative amphiphilic helix located near the C-terminus (Figure 4). However, the data do show that membrane interactions with the C-terminal helix are necessary to maintain the C. maquilingensis SQR bound to the membrane, and are also important for the access of the quinone substrate to the protein-bound flavin at the enzyme active site.
Highlights.
The sulfide:quinone oxidoreductase (SQR) from C. maquilingensis was characterized.
This is the first representative of a “Class III” SQR to be charcterized.
Membrane binding is mediated by residues in the C-terminal amphiphilic helix.
Mutations in this portion of the protein converts the enzyme to a soluble form.
Acknowledgments
We thank Dr C. House for providing the genomic DNA from Caldivirga maquilingensis. This work was supported by grants from the National Institutes of Health: HL16101 (RBG).
Abbreviations
- DDM
dodecyl-β-D-maltoside
- NTA
nitrilotriacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.YaH Shahak G. Sulfide Oxidation from Cyanobacteria to Humans: Sulfide–Quinone Oxidoreductase (SQR) In: Hell R, Dahl S, Knaff DB, Leustek T, editors. Advances in Photosynthesis and Respiration vol. 27. Germany: Springer, Heidelberg; 2008. pp. 319–335. [Google Scholar]
- 2.Griesbeck C, Hauska G, Schütz M. Biological Sulfide Oxidation: Sulfide-Quinone Reductase (SQR), the Primary Reaction. In: Pandalai SG, editor. Recent Research Developments in Microbiology vol. 4. Trivandrum, India: Research Signpost; 2000. pp. 179–203. [Google Scholar]
- 3.Kletzin A, Urich T, Muller F, Bandeiras TM, Gomes CM. Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J Bioenerg Biomembr. 2004;36:77–91. doi: 10.1023/b:jobb.0000019600.36757.8c. [DOI] [PubMed] [Google Scholar]
- 4.Guiral M, Prunetti L, Lignon S, Lebrun Rg, Moinier D, Giudici-Orticoni M-Trs. New Insights into the Respiratory Chains of the Chemolithoautotrophic and Hyperthermophilic Bacterium Aquifex aeolicus. Journal of Proteome Research. 2009;8:1717–1730. doi: 10.1021/pr8007946. [DOI] [PubMed] [Google Scholar]
- 5.Gregersen LH, Bryant DA, Frigaard NU. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Frontiers in microbiology. 2011;2:116. doi: 10.3389/fmicb.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mimoun S, Andriamihaja M, Chaumontet C, Atanasiu C, Benamouzig R, Blouin JM, Tome D, Bouillaud F, Blachier F. Detoxification of h(2)s by differentiated colonic epithelial cells: implication of the sulfide oxidizing unit and of the cell respiratory capacity. Antioxidants & redox signaling. 2012;17:1–10. doi: 10.1089/ars.2011.4186. [DOI] [PubMed] [Google Scholar]
- 7.Bouillaud F, Blachier F. Mitochondria and sulfide: a very old story of poisoning, feeding, and signaling? Antioxidants & redox signaling. 2011;15:379–391. doi: 10.1089/ars.2010.3678. [DOI] [PubMed] [Google Scholar]
- 8.Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 9.Wang R. The gasotransmitter role of hydrogen sulfide. Antioxidants & redox signaling. 2003;5:493–501. doi: 10.1089/152308603768295249. [DOI] [PubMed] [Google Scholar]
- 10.Kabil O, Banerjee R. Redox Biochemistry of Hydrogen Sulfide. Journal of Biological Chemistry. 2010;285:21903–21907. doi: 10.1074/jbc.R110.128363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura H. Hydrogen sulfide as a neuromodulator. Molecular neurobiology. 2002;26:13–19. doi: 10.1385/MN:26:1:013. [DOI] [PubMed] [Google Scholar]
- 12.Guiral M, Tron P, Aubert C, Gloter A, Iobbi-Nivol C, Giudici-Orticoni MT. A membrane-bound multienzyme, hydrogen-oxidizing, and sulfur-reducing complex from the hyperthermophilic bacterium Aquifex aeolicus. J Biol Chem. 2005;280:42004–42015. doi: 10.1074/jbc.M508034200. [DOI] [PubMed] [Google Scholar]
- 13.Marcia M, Ermler U, Peng G, Michel H. A new structure-based classification of sulfide:quinone oxidoreductases. Proteins: Structure, Function, and Bioinformatics. 2010;78:1073–1083. doi: 10.1002/prot.22665. [DOI] [PubMed] [Google Scholar]
- 14.Chan LK, Morgan-Kiss RM, Hanson TE. Functional analysis of three sulfide:quinone oxidoreductase homologs in Chlorobaculum tepidum. J Bacteriol. 2009;191:1026–1034. doi: 10.1128/JB.01154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holkenbrink C, Barbas SO, Mellerup A, Otaki H, Frigaard NU. Sulfur globule oxidation in green sulfur bacteria is dependent on the dissimilatory sulfite reductase system. Microbiology. 2011;157:1229–1239. doi: 10.1099/mic.0.044669-0. [DOI] [PubMed] [Google Scholar]
- 16.Theissen U, Hoffmeister M, Grieshaber M, Martin W. Single eubacterial origin of eukaryotic sulfide:quinone oxidoreductase, a mitochondrial enzyme conserved from the early evolution of eukaryotes during anoxic and sulfidic times. Mol Biol Evol. 2003;20:1564–1574. doi: 10.1093/molbev/msg174. [DOI] [PubMed] [Google Scholar]
- 17.Marcia M, Langer JD, Parcej D, Vogel V, Peng G, Michel H. Characterizing a monotopic membrane enzyme. Biochemical, enzymatic and crystallization studies on Aquifex aeolicus sulfide:quinone oxidoreductase. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2010;1798:2114–2123. doi: 10.1016/j.bbamem.2010.07.033. [DOI] [PubMed] [Google Scholar]
- 18.Marcia M, Ermler U, Peng G, Michel H. The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc Natl Acad Sci U S A. 2009;106:9625–9630. doi: 10.1073/pnas.0904165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherney MM, Zhang Y, James MN, Weiner JH. Structure-activity characterization of sulfide:quinone oxidoreductase variants. Journal of structural biology. 2012 doi: 10.1016/j.jsb.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Cherney MM, Zhang Y, Solomonson M, Weiner JH, James MN. Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification. J Mol Biol. 2010;398:292–305. doi: 10.1016/j.jmb.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Brito JA, Sousa FL, Stelter M, Bandeiras TM, Vonrhein C, Teixeira M, Pereira MM, Archer M. Structural and Functional Insights into Sulfide:Quinone Oxidoreductase. Biochemistry. 2009;48:5613–5622. doi: 10.1021/bi9003827. [DOI] [PubMed] [Google Scholar]
- 22.Balali-Mood K, Bond PJ, Sansom MS. Interaction of monotopic membrane enzymes with a lipid bilayer: a coarse-grained MD simulation study. Biochemistry. 2009;48:2135–2145. doi: 10.1021/bi8017398. [DOI] [PubMed] [Google Scholar]
- 23.Griesbeck C, Schutz M, Schodl T, Bathe S, Nausch L, Mederer N, Vielreicher M, Hauska G. Mechanism of sulfide-quinone reductase investigated using sitedirected mutagenesis and sulfur analysis. Biochemistry. 2002;41:11552–11565. doi: 10.1021/bi026032b. [DOI] [PubMed] [Google Scholar]
- 24.Itoh T, Suzuki K, Sanchez PC, Nakase T. Caldivirga maquilingensis gen. nov., sp. nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines. International journal of systematic bacteriology. 1999;49(Pt 3):1157–1163. doi: 10.1099/00207713-49-3-1157. [DOI] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 27.Weyler W, Salach JI. Purification and properties of mitochondrial monoamine oxidase type A from human placenta. J Biol Chem. 1985;260:13199–13207. [PubMed] [Google Scholar]
- 28.Susin S, Abian J, Sanchez-Baeza F, Peleato ML, Abadia A, Gelpi E, Abadia J. Riboflavin 3'- and 5'-sulfate, two novel flavins accumulating in the roots of iron-deficient sugar beet (Beta vulgaris) J Biol Chem. 1993;268:20958–20965. [PubMed] [Google Scholar]
- 29.Shahak Y, Klughammer C, Schreiber U, Padan E, Herrman I, Hauska G. Sulfide-quinone and sulfide-cytochrome reduction in Rhodobacter capsulatus. Photosynth Res. 1994;39:175–181. doi: 10.1007/BF00029384. [DOI] [PubMed] [Google Scholar]
- 30.Morton RA. Quinones as a Biological Catalysts. Endeavour. 1965;24:81–86. doi: 10.1016/0160-9327(65)90005-0. [DOI] [PubMed] [Google Scholar]
- 31.Schutz M, Shahak Y, Padan E, Hauska G. Sulfide-quinone reductase from Rhodobacter capsulatus. Purification, cloning, and expression. J Biol Chem. 1997;272:9890–9894. doi: 10.1074/jbc.272.15.9890. [DOI] [PubMed] [Google Scholar]
- 32.Ghisla S, Massey V. New flavins for old: artificial flavins as active site probes of flavoproteins. Biochem J. 1986;239:1–12. doi: 10.1042/bj2390001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vande Weghe JG, Ow DW. A fission yeast gene for mitochondrial sulfide oxidation. J Biol Chem. 1999;274:13250–13257. doi: 10.1074/jbc.274.19.13250. [DOI] [PubMed] [Google Scholar]
- 34.Hefti MH, Vervoort J, van Berkel WJ. Deflavination and reconstitution of flavoproteins. Eur J Biochem. 2003;270:4227–4242. doi: 10.1046/j.1432-1033.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- 35.Kunze B, Hofle G, Reichenbach H. The aurachins, new quinoline antibiotics from myxobacteria: production, physico-chemical and biological properties. J Antibiot (Tokyo) 1987;40:258–265. doi: 10.7164/antibiotics.40.258. [DOI] [PubMed] [Google Scholar]
- 36.Romagnoli S, Oettmeier W, Zannoni D. The effects of decyl aurachins C and D on the respiratory electron flow of facultative phototrophic bacteria. Biochem Mol Biol Int. 1996;39:671–678. doi: 10.1080/15216549600201741. [DOI] [PubMed] [Google Scholar]
- 37.Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci U S A. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 39.Wakai S, Tsujita M, Kikumoto M, Manchur MA, Kanao T, Kamimura K. Purification and characterization of sulfide:quinone oxidoreductase from an acidophilic iron-oxidizing bacterium. Acidithiobacillus ferrooxidans, Bioscience, biotechnology, and biochemistry. 2007;71:2735–2742. doi: 10.1271/bbb.70332. [DOI] [PubMed] [Google Scholar]
- 40.Gupta K, Selinsky BS, Kaub CJ, Katz AK, Loll PJ. The 2.0 A resolution crystal structure of prostaglandin H2 synthase-1: structural insights into an unusual peroxidase. J Mol Biol. 2004;335:503–518. doi: 10.1016/j.jmb.2003.10.073. [DOI] [PubMed] [Google Scholar]
- 41.Kiefer JR, Pawlitz JL, Moreland KT, Stegeman RA, Hood WF, Gierse JK, Stevens AM, Goodwin DC, Rowlinson SW, Marnett LJ, Stallings WC, Kurumbail RG. Structural insights into the stereochemistry of the cyclooxygenase reaction. Nature. 2000;405:97–101. doi: 10.1038/35011103. [DOI] [PubMed] [Google Scholar]
- 42.Watmough NJ, Frerman FE. The electron transfer flavoprotein: Ubiquinone oxidoreductases. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2010;1797:1910–1916. doi: 10.1016/j.bbabio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Frerman FE, Kim JJ. Structure of electron transfer flavoproteinubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. Proc Natl Acad Sci U S A. 2006;103:16212–16217. doi: 10.1073/pnas.0604567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeh JI, Chinte U, Du S. Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism. Proc Natl Acad Sci U S A. 2008;105:3280–3285. doi: 10.1073/pnas.0712331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thoma R, Schulz-Gasch T, D'Arcy B, Benz J, Aebi J, Dehmlow H, Hennig M, Stihle M, Ruf A. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature. 2004;432:118–122. doi: 10.1038/nature02993. [DOI] [PubMed] [Google Scholar]
- 46.Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002;298:1793–1796. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]
- 47.Kloer DP, Ruch S, Al-Babili S, Beyer P, Schulz GE. The structure of a retinal-forming carotenoid oxygenase. Science. 2005;308:267–269. doi: 10.1126/science.1108965. [DOI] [PubMed] [Google Scholar]
- 48.Granseth E, von Heijne G, Elofsson A. A Study of the Membrane–Water Interface Region of Membrane Proteins. Journal of Molecular Biology. 2005;346:377–385. doi: 10.1016/j.jmb.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 49.Ghisla S, Massey V. Mechanisms of flavoprotein-catalyzed reactions. Eur J Biochem. 1989;181:1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]