Abstract
Aldehyde dehydrogenase (ALDH) enzymes catalyze the NAD(P)+-dependent oxidation of a wide variety of endogenous and exogenous aldehydes to their corresponding acids. Some members of the ALDH superfamily of enzymes are abundantly expressed in the mammalian cornea and lens in a taxon-specific manner. Considered to be corneal and lens crystallins, they confer protective and transparent properties upon these ocular tissues. ALDH3A1 is highly expressed in the cornea of most mammals, with the exception of rabbit that expresses exclusively ALDH1A1 in the cornea. ALDH1A1 is present in both the cornea and lens of several animal species. As a result of their catalytic and non-catalytic functions, ALDH3A1 and ALDH1A1 proteins protect inner ocular tissues from ultraviolet radiation and reactive oxygen-induced damage. In addition, these corneal crystallins contribute to cellular transparency in corneal stromal keratocytes, supporting a structural role of these ALDH proteins. A putative regulatory function of ALDH3A1 on corneal cell proliferation has also been proposed. Finally, the three retinaldehye dehydrogenases cooperatively mediate retinoic acid signaling during the eye development.
Keywords: ALDH, crystalline, cornea, lens, ultraviolet damage, cataract, transparency
1. Introduction
The human eye is routinely exposed to sunlight and artificial light. While transmission of incident light through the eye is fundamental for vision, this radiation can pose a hazard to ocular tissues, potentially leading to impaired vision. A plethora of studies have established an association between ultraviolet radiation (UVR) exposure and numerous ocular disease states, such as cataracts and macular degeneration (Roberts, 2011).
The aldehyde dehydrogenase (ALDH) superfamily of enzymes plays an important role in the metabolism of endogenous and exogenous aldehydes (Marchitti et al., 2008). Through their catalytic functions, ALDH enzymes detoxify reactive aldehydes and modulate some important cellular processes, such as embryogenesis and neurotransmission. Through involvement in retinoic acid (RA) biosynthesis, members of the ALDH family are also implicated in vertebrate eye development (Duester, 2009). Additionally, ALDHs can exhibit biological functions unrelated to catalytic activity, an example of which is physicochemical binding to hormones and small molecules. Specific ALDH isozymes have been found to be abundantly expressed in the cornea and lens in a taxon-specific manner (Cooper et al., 1993). Studies using cell cultures and transgenic animal models have identified these ALDHs to be corneal and lens crystallins (Estey et al., 2007a; Estey et al., 2007b; Lassen et al., 2007). Like other crystallin proteins, the ALDH molecules have a structural role and contribute to cellular transparency (Jester, 2008). They also function as important components of cellular defense mechanisms against UVR and reactive oxygen species-induced ocular damage (Lassen et al., 2008). This review summarizes the current state of knowledge about the properties and functions of ocular ALDHs, with a focus on the unique roles of ALDH1A1 and ALDH3A1 as corneal and lens crystallins. The discussion covers the following topics: (i) an overview of the ALDH superfamily of enzymes, (ii) taxon-specific expression of ALDHs in the cornea and lens, (iii) protective properties of ALDH1A1/3A1 against UV exposure, (iv) structural and regulatory roles of ALDH1A1/3A1 in the cornea, and (v) ALDH enzymes in the developing eye.
2. ALDH superfamily of enzymes
2.1 Structure and catalytic sites of ALDH enzymes
The ALDH superfamily comprises nicotinamide-adenine dinucleotide phosphate (NAD(P)+)-dependent enzymes that irreversibly catalyze the oxidation of aldehyde substrates to their respective carboxylic acids (Jackson et al., 2011). Isozymes are given the name based on their peptide sequence identity such that families within the superfamily share > 40% identity and members of the same subfamily share > 60% identity. ALDH proteins are widely expressed in mammalian tissues, albeit different isozymes exhibit distinct tissue distributions. Although these enzymes share some common physiological functions and substrates, each of the isozymes exhibits distinct substrate specificities (Fig. 1) (Marchitti et al., 2008). Most ALDH isozymes share the same catalytic mechanism for the dehydrogenase activity (Cobessi et al., 2000; D'Ambrosio et al., 2006). This may be explained by highly conserved residues necessary for catalytic activity (i.e., Cys302 and Glu268; numbering based on human ALDH1A1) and for co-factor binding (i.e., Lys192, Gly-245, Gly-250, Glu-399, and Phe-401).
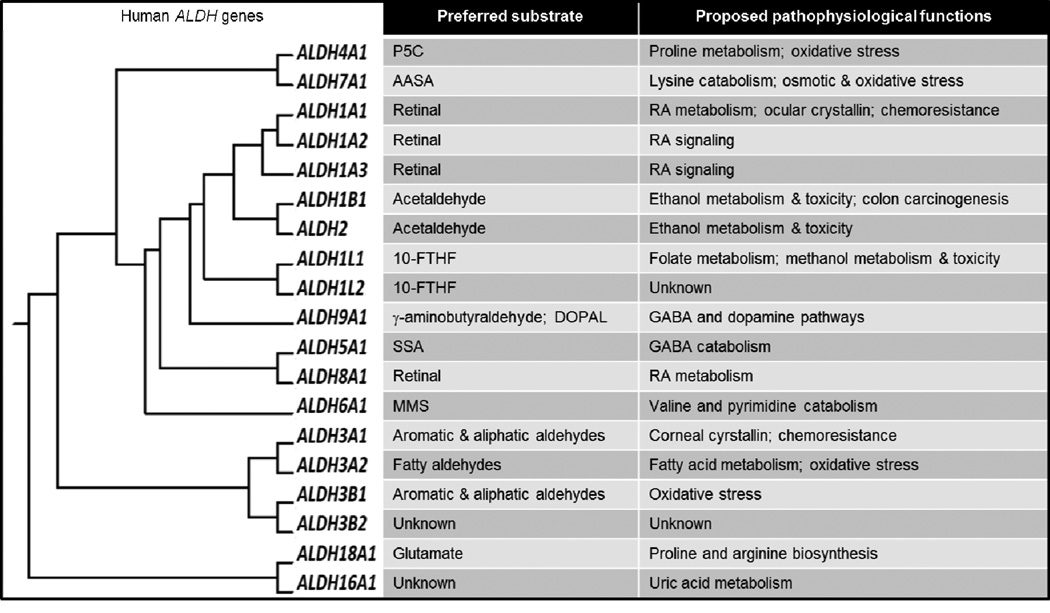
Fig. 1. Human ALDH superfamily.
Clustering dendrogram of the nineteen human ALDH genes shows the evolution of human ALDH superfamily from a single gene. For each isozyme, the preferred substrates and proposed pathophysiological function are provided (Marchitti et al., 2008). 10-FTHF, 10-formyltetrahydrofolate; AASA, α-aminoadipic semialdehyde; DOPAL, 3,4-dihydroxyphenylacetaldehyde; GABA, γ-aminobutyric acid; MMS, methylmalonate semialdehyde; P5C, pyrroline-5-carboxylate; RA, retinoic acid; SSA, succinic semialdehyde.
Mammalian ALDHs function as either homotetramers or homodimers, with a catalytic active site present within each monomer (Cobessi et al., 2000; Colby et al., 1998; D'Ambrosio et al., 2006; Liu et al., 1997). In general, each ALDH monomer comprises 500 to 600 amino acids. A few exceptions that have up to 800 or 900 amino acids exist and include ALDH16A1, ALDH18A1, ALDH1L1 and ALDH1L2. X-ray crystallographic studies indicate that the ALDH monomers across all isozyme classes share high homology in three distinct functional regions: a βαβ N-terminal co-factor (NAD(P)+) binding domain, a βαβ catalytic domain and a C-terminal small β-sheet oligomerization domain (Figs. 2A&C). In the dimeric (Fig. 2B) or tetrameric holoenzyme (Fig. 2D), the active sites reside at the base of a hydrophobic tunnel projecting inward from the surface of the enzyme. The active sites are close to the tetrameric interface and opposite to the cofactor binding sites. Residues lining the hydrophobic tunnel are thought to confer the substrate specificity of each isozyme. The rate-limiting step of the ALDH-mediated dehydrogenation reaction differs between the isozymes and depends upon the effect of co-factor binding, the presence of divalent ions and the pH of microenvironment (Perez-Miller and Hurley, 2003). For example, reduced NAD+ release is the rate-limiting step for ALDH1A1, whereas the hydride transfer to the pyridine ring of NADP+ is rate-limiting for ALDH3A1. In addition to the dehydrogenase activity, some ALDH enzymes possess esterase activity, the catalytic mechanism of which has been discussed elsewhere (Kazmi et al., 1992). In some ALDH isozymes (like human ALDH1A1 and ALDH16A1), coiled-coil motifs are predicted to reside in the nucleotide-binding-domain (Chen et al., 2011a). Such coiled-coil domains are highly stable oligomerization motifs found in proteins functioning in gene regulation, cell communication, membrane fusion and drug extrusion. It is worth noting that ALDH16A1 is a novel and unique member of the ALDH superfamily (Vasiliou et al 2012, submitted manuscript). Mammalian ALDH16A1 proteins contain two ALDH domains and lack the catalytically important cysteine residue. The recent report on the association between gout and a rare missense single nucleotide polymporphism of human ALDH16A1 gene (Hanna and Blackstone, 2009) is suggestive of a potential role of ALDH16A1 in uric acid metabolism.
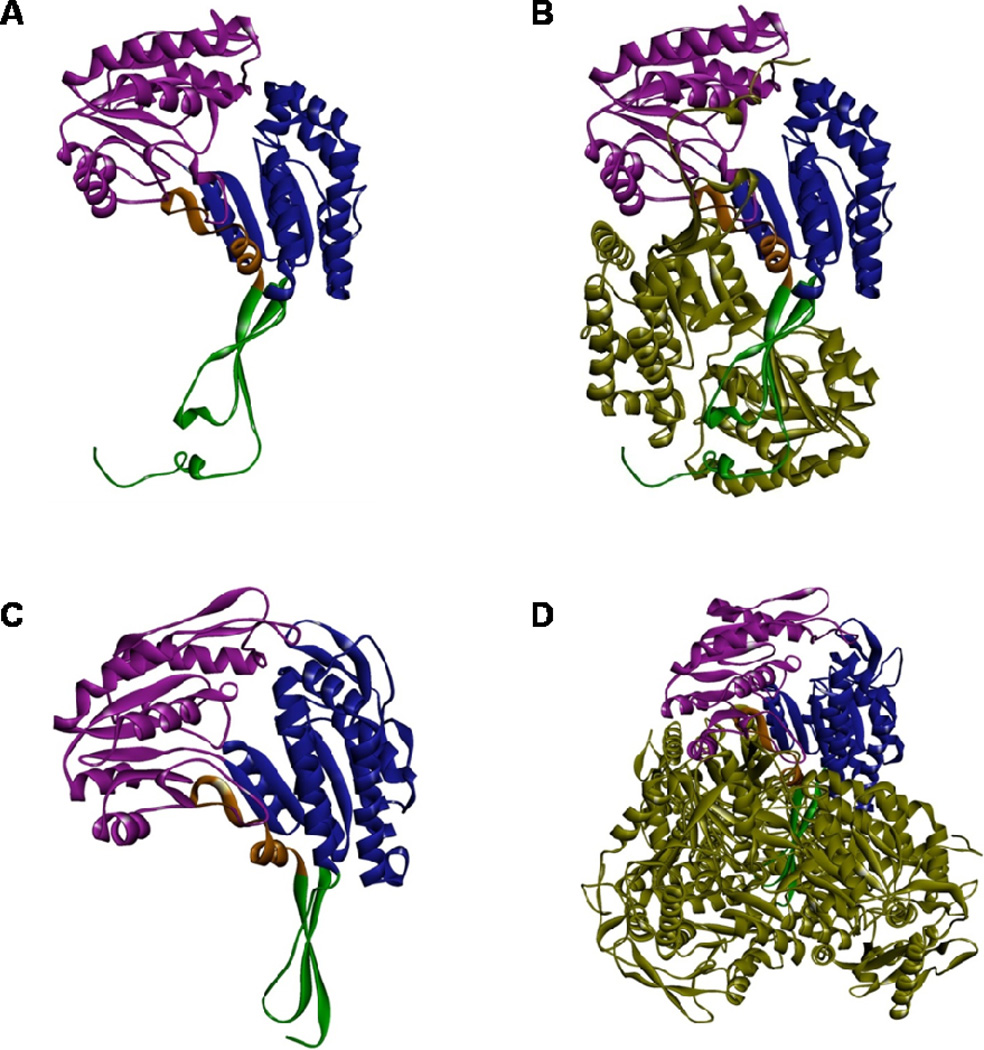
Fig. 2. Protein structure of ALDH3A1 and ALDH1A1.
X-ray crystal structures of human ALDH3A1 monomer (A) and sheep liver ALDH1A1 monomer (C) showing the catalytic domain (purple), NAD(P)+ binding domain (blue), and oligomerization domain (green). Crystal structures were obtained from RCSB database; PDBIDs: 3SZA (ALDH3A1) and 1BXS (ALDH1A1). Computational modeling of dimeric human ALDH3A1 holoenzyme (B) and tetrameric sheep ALDH1A1 holoenzyme (D). The 2nd ALDH3A1 subunit and three other ALDH1A1 subunits are shown in forest green.
2.2 Functions of ALDH isozymes
ALDH isozymes play a key role in the metabolism of aldehydes of both endogenous and exogenous derivation. Some aldehydes participate in vital physiological functions, including vision, embryonic development and neurotransmission (Marchitti et al., 2008). Others, such as 4-hydroxy-2-nonenal (4-HNE) and malondialdehyde (MDA), the major products of lipid peroxidation (LPO), and acetaldehyde, an ethanol metabolite, are cytotoxic and carcinogenic. As such, ALDHs exhibit a broad spectrum of biological activities of both physiological and toxicological significance (Marchitti et al., 2008). Genetic polymorphisms of and mutations in human ALDH genes are associated with several disease states (Marchitti et al., 2008), underscoring the significant pathophysiological roles played by the ALDH isozymes.
The ALDH1 subfamily (i.e., ALDH1A1/1A2/1A3) catalyzes the synthesis of RA from retinal (retinaldehyde), a retinol (vitamin A) metabolite. In so doing, these isozymes play a central role in biological functions involving RA signaling, such as vertebrate development. The ALDH1 isozymes have a high affinity for both all-trans- and 9-cis-retinal, exhibiting a Km in the lower µM range (Bhat and Samaha, 1999; Yoshida et al., 1992). RA acts as a ligand for the heterdimer of the nuclear RA receptor and the retinoic X receptor, which transcriptionally regulates many downstream developmental genes (Matt et al., 2003). During vertebrate embryogenesis, the ALDH1 isozymes are expressed in tissues in a spatial-temporal fashion (Downes et al., 1992; Duester, 2009), an observation that substantiates a developmental role for these enzymes. Some stem cells, such as those associated with hematopoiesis, exhibit high ALDH activity. This feature has been exploited for the isolation of primitive stem cell populations (Storms et al., 1999). In a recent study, an important role for ALDH1A1 and ALDH3A1 in hematopoietic stem cell biology has been reported (Gasparetto et al., 2011). Their effects seem due, at least in part, to metabolism of reactive oxygen species (ROS) and reactive aldehydes.
The catalytic functions of ALDHs support cellular homeostasis through the detoxification of reactive aldehydes. The ALDH2 isozyme is primarily responsible for acetaldehyde detoxification in the second step of alcohol metabolism (Klyosov et al., 1996). ALDH1A1 and ALDH1B1 also contribute to this process, albeit with less affinity for acetaldehyde than the ALDH2 isozyme (Stagos et al., 2010a). ALDH7A1 has been shown to protect cells against hyperosmotic stress by generating osmolytes and metabolizing toxic aldehydes derived from LPO (Brocker et al., 2010). Some ALDHs appear to participate in neurotransmission and neuroprotection. For example, both ALDH1A1 and ALDH2 have been implicated in dopamine metabolism. Inhibition of these isozymes leads to the accumulation of neurotoxic dopamine metabolites that are thought to contribute to the pathogenesis of Parkinson’s disease (Allen et al., 2010; Burke et al., 2003; Marchitti et al., 2008). In recent years, several ALDH isozymes have been discovered to have both regulatory as well as metabolic roles in cancer. Cancer stem cells exhibit elevated ALDH catalytic activity. Metabolic inactivation by such activity is believed to underlie resistance to selected anti-cancer agents and consequent cancer relapse (Ma and Allan, 2010). Several ALDH isozymes, including ALDH1A1 (Deng et al., 2010), ALDH3A1 (Ucar et al., 2009), ALDH1B1 (Chen et al., 2011b), and ALDH7A1 (van den Hoogen et al., 2011), are induced in some human cancers. The pathophysiological consequences of such induction remain to be established. Finally, some ALDH isozymes, especially ALDH3A1 and ALDH1A1, are highly expressed in the cornea and lens of various species. These isozymes serve important structural and protective roles in these ocular tissues. Their properties, functions and mechanisms of action in ocular tissues are addressed in detail below.
3. ALDH enzymes are corneal and lens crystallins
3.1 Taxon-specific expression of ALDHs in the cornea and lens
Based on the “Refracton Hypothesis” first proposed by Piatigorsky (Piatigorsky, 2001), the cornea and lens in the anterior segment of the eye form a single refractive unit (Fig. 3) that permits light entry and focusing onto the retina. The cornea, the outermost layer of the eye, is composed of three cellular layers (Fig. 3) including: (i) a stratified squamous epithelium, (ii) a thick stroma containing collagen fibers, proteoglycans, glycosaminoglycans and keratocytes, and (iii) a posterior monolayer of endothelium. The cornea is the first cellular defense against environmental insults. Accordingly, it plays a pivotal role in protecting the inner ocular tissues against damage and provides most of the refractive power of the eye. The lens, through accommodative mechanisms changes in its biconvex shape and functions sharpen the focus of light reflected from an object back onto the retina. The lens comprises three main parts (Fig. 3): (i) an elastic capsule surrounding the lens, (ii) the lamina composed of lens fibers that form the bulk of the lens and (iii) a simple cuboidal epithelial layer that lines the anterior portion of the lens between the capsule and the fibers. Lens fibers are formed by firmly packed, long, thin, transparent cells that stretch lengthwise from the posterior to the anterior poles. The lens epithelium is a major site of transport, metabolism and detoxification and is crucial for the homeostatic functions of the lens, while also serving as the progenitors for new lens fibers.
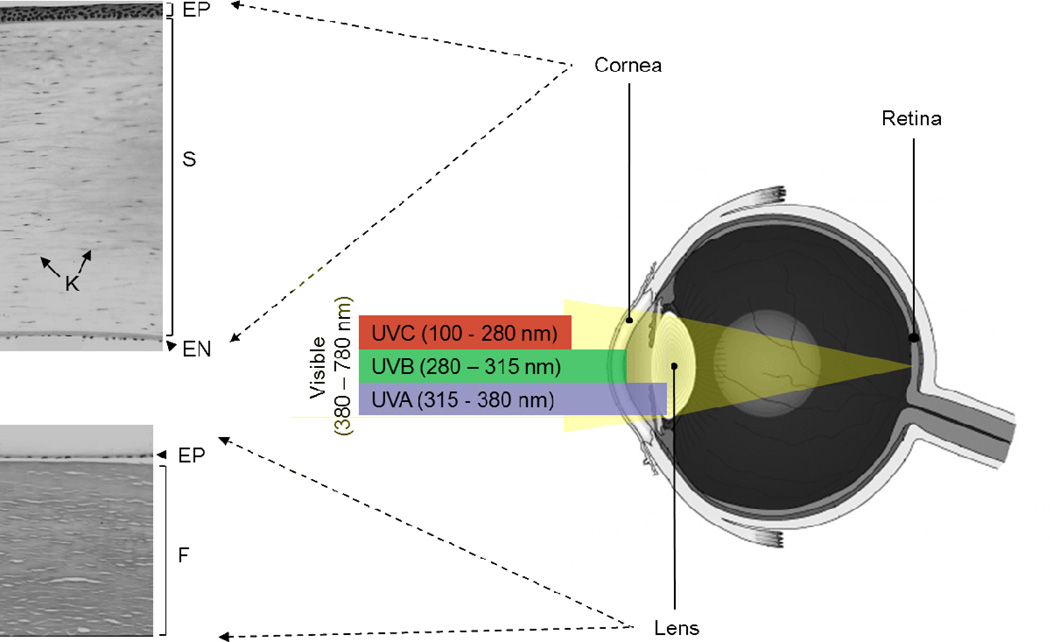
Fig. 3. The refractive unit of and ultraviolet radiation absorption by the human eye.
The cornea and lens constitute the refractive unit, permitting light entry and focusing onto the retina. The cornea is composed of three cellular layers: a stratified squamous epithelium (EP), a thick stroma (S) containing collagen fibers and keratocytes (K), and a posterior monolayer of endothelium (EN). The lens comprises three main parts: an elastic capsule surrounding the lens, the lamina composed of lens fibers (F) that form the bulk of the lens, and a simple cuboidal epithelial layer (EP) between the anterior capsule and the fibers. The incident light consists of a spectrum of wavelengths ranging from ultraviolet radiation (UVR) to visible and infrared (IR). UVR comprises UVC, UVB and UVA. UVC is precluded from reaching the Earth’s surface by the upper atmosphere. Most UVB is absorbed by the cornea, whereas UVA is primarily absorbed by the lens. Visible light passes through the cornea and lens and reaches the retina to form images.
The term “crystallins” was originally given to lens proteins that compose up to 90% of the total water-soluble proteins within the lens and account for the transparency of the lens (Eguchi, 1966). It is now clear that some proteins function as corneal crystallins due to their similarities to the lens crystallins (Piatigorsky, 1998a) in that they: (i) are, in most cases, diverse cytoplasmic proteins with metabolic functions, (ii) display taxon-specificity (i.e. differ between species) and (iii) accumulate to high levels in transparent tissues, while being present at lower levels in other tissues. It has been proposed that, as a consequence of evolutionary events, metabolic proteins were recruited to tissues to serve a second and distinct function, a phenomenon termed “gene sharing” (Piatigorsky, 1998a). It is becoming increasingly recognized that, in addition to serving a structural role, crystallins may have other metabolic and regulatory functions, both within the eye and in other parts of the body.
Members of the ALDH superfamily have been identified as corneal and lens crystallins in both vertebrates and invertebrates (Table 1). In vertebrates, the bovine corneal protein 54 (BCP54) was first reported to comprise 20–40% of total soluble proteins of the bovine cornea; in latter studies, BCP54 was identified to be ALDH3A1 (Verhagen et al., 1991). It is now well established that, in most mammalian species including human, ALDH3A1 is a major soluble protein in the cornea; in contrast, rabbits express ALDH1A1 (rather than ALDH3A1) in the cornea (Holmes et al., 1989). Other vertebrates, such as chicken, frog and fish, express ALDH1A1 and, in some cases, ALDH2 rather than ALDH3A1 in the cornea (Pappa et al., 2001). Other members of ALDH class 1 proteins are lens crystallins in the elephant shrew (ALDH1A8/η-crystallin), scallops (ALDH1A9/Ω-crystallin) and cephalopods (ALDH1C1/2/Ω-crystallins) (Graham et al., 1996). Although expressed in a taxon-specific fashion, members of ALDH class 1 and ALDH3A1 are well conserved in mammals, showing ~90% homology in amino acid sequence among human, rabbit, cow, sheep, mouse and rat (Manzer et al., 2003).
Table 1.
ALDH isozymes identified as corneal and lens crystallins
3.2 ALDH3A1, a corneal crystallin
The first recognized corneal crystallin was ALDH3A1, which shows a taxon-specific pattern of expression. Among mammals, constitutive expression of ALDH3A1 has been found in the cornea of human, cow, pig, mouse, rat, baboon, opossum and kangaroo (Piatigorsky, 1998b). As noted, rabbits are exceptional in that no detectable ALDH3A1 protein is observed in their corneas (Jester et al., 1999; Pappa et al., 2001). In human and mouse corneas, ALDH3A1 protein is strongly expressed in epithelial cells and stromal keratocytes, but not in endothelial cells (Pappa et al., 2003b). Depending on the species, ALDH3A1 accounts for 5–50% of the total water-soluble proteins of the corneal epithelium (Abedinia et al., 1990). In contrast to most mammals, ALDH3A1 enzymatic activity is absent from birds, such as the chicken and turkey (Pappa et al., 2001). Similarly, neither ALDH3A1 protein nor ALDH3A1 catalytic activity is detected in corneal extracts from frogs or a variety of fish species, including trout, zebrafish, chain pickerel and redhorse (Cooper et al., 1993; Pappa et al., 2001). These results suggest that corneal expression of ALDH3A1 is specific to mammals. Interestingly, the SWR/J inbred mouse strain exists as a ‘natural Aldh3a1 gene knockout’ model by showing in the cornea a lack of detectable catalytic activity of ALDH3A1, low levels of ALDH3A1 protein, and reduced soluble protein levels (Downes et al., 1997). This strain also exhibits greater levels of corneal clouding after UVR exposure (Downes et al., 1997). The molecular basis of this defect has been investigated and found to be 13 nucleotide changes in the Aldh3a1c allele carried by these mice, four of which cause amino acid substitutions that may affect the catalytic activity and the stability of the protein (Shiao et al., 1999).
ALDH3A1 is a homodimer with the capacity to oxidize medium-chain aliphatic and aromatic aldehydes (Pappa et al., 2003b). It was considered to be a cytosolic protein until the recent documentation of its nuclear localization in transfected human corneal epithelial cells (Pappa et al., 2005) and transfected rabbit corneal keratocytes (Stagos et al., 2010b). In human corneal epithelial cells, ALDH3A1 expression inversely correlates with cellular proliferation rate (Pappa et al., 2005). In mouse, the expression of corneal ALDH3A1 is temporally regulated beginning at birth and increasing substantially by six weeks of age (Davis et al., 2008), a process likely controlled at the transcriptional level. The promoter activity of the mouse Aldh3a1 gene is downstream of numerous transcription factors and co-activators, including Paired Box 6 (PAX6), Octamer-Binding transcription factor 1 (OCT1), Krüppel-like factor 4 (KLF4) and P300 (Davis et al., 2008; Swamynathan et al., 2008). PAX6 is an important transcription factor regulating gene expression of crystallins in the development of the lens (Cvekl and Piatigorsky, 1996; Gehring and Ikeo, 1999). The differential expression of ALDH3A1 in mouse cornea is regulated by PAX6. Both OCT1 and P300 regulate Aldh3a1 expression through a PAX6-dependent mechanism (Davis et al., 2008). KLF4, one of the most highly expressed transcription factors in the mouse cornea, plays an important role in maturation and maintenance of the ocular surface (Swamynathan et al., 2008). Thus, expression of Aldh3a1 in the mouse appears to be controlled through the synergistic effects of transcription factors and co-factors that are also important in regulating expression of other lens crystallins. These features of corneal ALDH3A1, including nuclear localization, effect on cell proliferation and transcriptional controlled by similar mechanisms regulating lens crystallins, may have important implications for the physiological roles of ALDH3A1 in the cornea (see below).
3.3 ALDH1A1, a lens and corneal crystallin
ALDH1A1 is a homotetramer and is well noted for its capacity to oxidize retinaldehyde to form retinoic acid (Bhat and Samaha, 1999). ALDH1A1 also metabolizes highly reactive aldehydes, including the ethanol metabolite acetaldehyde and major products of LPO (Manzer et al., 2003; Xiao et al., 2009). In addition to its metabolic functions, ALDH1A1 is capable of non-catalytic interactions with both endobiotics (e.g., androgen, thyroid hormone and cholesterol) and xenobiotics (e.g., flavopiridol, daunorubicin and quinolone) (Marchitti et al., 2008). The pathophysiological role(s) of such interactions remain to be identified. In humans, ALDH1A1 constitutes 3% and 2% of the soluble proteins in the cornea and lens epithelium, respectively (King and Holmes, 1993). Mouse ALDH1A1 is found mainly in the lens and to a lesser extent (<5%) in the corneal epithelium (Lassen et al., 2007). Rabbits primarily express ALDH1A1 in the cornea, with no detectable amount of ALDH3A1 (Jester et al., 2005). In this species, ALDH1A1 represents 3%, 16%, and 11% of the total soluble proteins of the epithelium, stromal keratocytes and endothelium, respectively (Jester et al., 2005). It has been proposed that the presence of ALDH1A1 in the corneal epithelium counterbalances the lack of expression of ALDH3A1 and thereby provides the necessary transparent, refractive and protective properties of the cornea. Indeed, our preliminary analysis of the rabbit genome shows that the Aldh3a1 gene does not exist in this species (unpublished data), although the particular genome may not be complete.
The molecular basis of this characteristic expression pattern of ALDH1A1 in the rabbit cornea has been explored using transfected cells and transgenic mice (Hough and Piatigorsky, 2004). The promoter region of the rabbit ALDH1A1 gene contains several response elements that are also present in the promoter region of other mammalian genes that are highly expressed in the cornea (Hough and Piatigorsky, 2004). Given that the cornea is constantly exposed to environmental stresses and high oxygen tension, it is believed that environmental induction plays a key role in the regulation of corneal crystallins; this is in contrast to the notion that developmental regulation plays a major role in the regulation of lens crystallins (Cvekl et al., 1995; Cvekl and Piatigorsky, 1996). At least two xenobiotic response elements and an E-box have been identified as being involved in the promoter activity of rabbit Aldh1a1 gene. In addition, several transcription factors, potentially interacting with these cis-elements to mediate responses to hypoxia and xenobiotics, are also expressed in rabbit cornea (Hough and Piatigorsky, 2004). It should be noted that the 5’ flanking sequence of human ALDH1A1 gene has 60.2% homology with that of rabbit Aldh1a1 gene and around 50% with that of rodents (Hough and Piatigorsky, 2004), indicating a closer phylogenetic relationship and expression pattern of ocular ALDH1A1 between humans and rabbits than humans and rodents.
4. ALDH enzymes protect against UVR-induced damage to the eye
4.1 UVR-induced damage to the eye
Primary determinants of UVR-induced injury to the human eye include the intensity of light, the wavelength absorbed and the duration of exposure. UVR consists of three wavebands: UVA (315–400 nm), UVB (280–315 nm) and UVC (100–280 nm) (Fig. 3). The most genotoxic and damaging is UVC, due to its high energy and short wavelength. However, human exposure to UVC is minimized due to its failure to penetrate the earth’s upper atmosphere. The cornea absorbs most UVB, whereas UVA is primarily absorbed by the lens. Accordingly, no UVB and very little UVA (<1%) reaches the retina (Bergmanson and Soderberg, 1995). Acute exposure to high levels of UVR, such as may occur from sunlight reflected from snow, leads to thermal damage (or sunburn) in the eye. The resultant inflammatory response, characterized by the release of interleukin-1, inflammatory infiltration at the site of irritation and subsequent formation of ROS, serves to eventually damage the ocular tissues (Klein et al., 2009; Sliney, 1997). Chronic low level UVR exposure, particularly that associated with the shorter and higher energy wavelengths of UVB, induces oxidative stress in the eye through the generation of ROS (Bergmanson and Soderberg, 1995).
ROS are produced as by-products during cellular metabolism (Fig. 4). The addition of one electron to dioxygen forms the superoxide anion radical (O2−•), which occurs mostly during the process of mitochondrial respiration (Cadenas and Sies, 1998). The dismutation of this molecule by superoxide dismutase forms hydrogen peroxide (H2O2) (Johnson and Giulivi, 2005) which can be efficiently scavenged by glutathione peroxidase in the mitochondria (Brigelius-Flohe, 2006) or by catalase in peroxisomes (Schrader and Fahimi, 2006). Although H2O2 is less reactive than superoxide, the breakdown of this molecule by various transition metals (e.g., Fe2+ or Cu+) via the Fenton reaction can generate the highly-reactive hydroxyl radical (OH•) (Leonard et al., 2004). The reaction of OH• or singlet oxygen (1O2; formed from molecular oxygen) with polyunsaturated fatty acids (PUFAs) in the cell membrane bilayer initiates LPO (Fig. 4) and generates peroxyl radical (LOO•) and lipid hydroperoxides (LOOH) (Aikens and Dix, 1991). Degradation of the lipid hydroperoxides yields a variety of products including more than 200 species of aldehydes (Esterbauer, 1993). When compared with the ROS that initiated the LPO, the aldehydes are relatively stable allowing them to diffuse intracellularly and intercellularly from the site of their generation. Several of these aldehydic breakdown products are electrophilic in nature, including acrolein, MDA and 4-HNE.
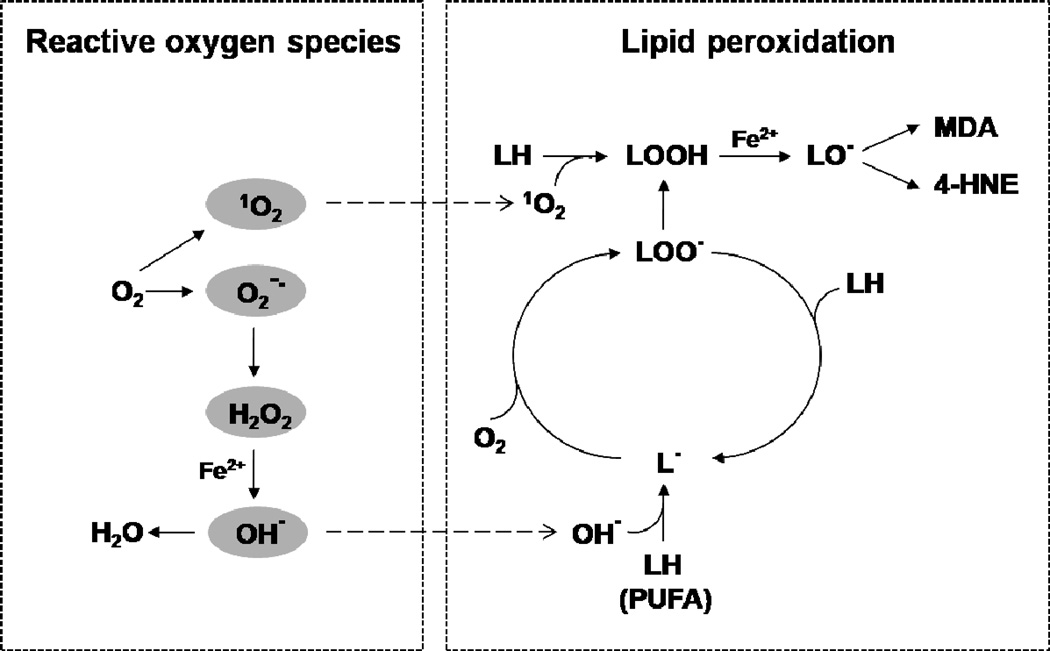
Fig. 4. Formation of reactive oxygen species and initiation and propagation of lipid peroxidation.
Reactive oxygen species (ROS) are derived from diatomic oxygen (O2), including singlet oxygen (1O2), superoxide anion radical (O2−•), hydrogen peroxide (H2O2), hydroxyl radical (OH•). ROS can initiate lipid peroxidation (LPO) by reacting with polyunsaturated fatty acids (PUFAs) in the cell membrane bilayer and thereby lead to the formation of lipid radicals (LOO•, LO• and L•) that propagate the reaction, and lipid hydroperoxide (LOOH) breakdown products (4-HNE and MDA).
ROS have the potential to attack important biomolecules. Excessive ROS can cause damage to DNA, cellular lipids and proteins and thereby inhibit their biological functions (Bergendi et al., 1999). As a very reactive molecule, OH• can react with each component of the DNA molecule, attacking the purine and pyrimidine bases and the deoxyribose backbone (Halliwell, 1998). Such modification of DNA has the potential to initiate mutagenesis and carcinogenesis. The amino acids of proteins, especially cysteine and methionine residues, are prone to oxidation by ROS (Dean et al., 1997). While irreversible oxidation of structurally or functionally important sites result in protein inactivation or misfolding, reversible oxidation of redox-responsive proteins functions as an important regulatory mechanism by which cells respond to oxidative stress (Finkel, 2011). 4-HNE has been identified as the most cytotoxic breakdown product generated from LPO. Its high reactivity is attributed to three main functional groups that can react synergistically with biomolecules containing amino and thiol groups (Poli and Schaur, 2000; Schaur, 2003). The cellular implications of 4-HNE reactivity include growth inhibition, a decrease in levels of the antioxidant glutathione (GSH), inhibition of sulfhydryl- and thiol-containing proteins, inhibition of calcium sequestration by microsomes, inhibition of protein synthesis and degradation, and alterations in signal transduction and gene expression profiles (Dianzani, 1998; Esterbauer et al., 1991).
4.2 Ocular diseases associated with UVR exposure
UVR-induced molecular modifications in the eye have been implicated in the pathogenesis of numerous forms of eye disease including photokeratitis (Roberts, 2001), pingueculae and pterygia (Tenkate, 1999), cataracts (Andley et al., 1994; Balasubramanian, 2000; Zigman, 2000) and macular degeneration (Taylor et al., 1990). Any intense exposure to UVR can lead to photokeratitis, a painful eye condition that is seen as "snow blindness" in skiers or "arc welder's flash" in people operating metal welding equipment without sufficient eye protection. In photokeratitis, only the superficial layers of the eye (the cornea and the lining of the eyelids) are affected. No damage to the lens or retina occurs and it does not cause permanent blindness. Pinguecula and pterygium are conditions that involve non-malignant, slow-growing proliferation of conjunctival connective tissue in the eye; it extends over the cornea in pterygium but not in pingueculae. While the exact causes of these disorders are unknown, UVR exposure has been proposed as a major etiologic factor. This hypothesis is supported by: (i) epidemiological data that show a high prevalence of these conditions in people who live in sunny climates, have jobs that expose them to UVR or are elderly (Mackenzie et al., 1992; Moran and Hollows, 1984; Wong et al., 2001), and (ii) histopathological changes in lesions from pingueculae and pterygia that share the common features with UVR-damaged skin (Perra et al., 2006).
A cataract occurs when lens opacity forms and impairs vision. It is the leading cause of blindness, accounting for 16 million people worldwide. Cataracts are usually age-related, appearing in more than 50% of those over 65 years of age (Roberts, 2001). They can also manifest in young people in the case of congenital cataracts and in patients with diabetes. While numerous studies in humans and experimental animals have shown a strong correlation between cataract formation and UVR exposure (Balasubramanian, 2000), the molecular mechanisms of UVR-induced cataractogenesis are not completely understood. It is believed that UVR-induced production of ROS and reactive metabolites (e.g., 4-HNE) are critically involved in these processes and function to damage lenticular DNA and proteins and destroy antioxidant nutrients (Eaton, 1994). In addition, UVR-induced modifications to proteins can promote enzyme inactivation, partial unfolding and non-native aggregation of proteins (Estey et al., 2007a; Estey et al., 2010), all of which may contribute to the accumulation of aggregated proteins in the cataract lens. Age-related macular degeneration (AMD) represents another leading cause of blindness in the western world. It is characterized by the accumulation of intracellular lipofuscin, extracellular drusen deposits and Bruch membrane thickening, changes that lead to progressive degeneration and loss of retinal pigment epithelium (RPE) and photoreceptor cells in the macular region (Bergmanson and Soderberg, 1995). Based upon existing epidemiologic evidence, the relationship between UVR exposure and AMD is inconclusive, although, in experimental settings, retinal ganglion and RPE cells are highly sensitive to UVR-induced phototoxicity mediated by ROS production (Chalam et al., 2011).
Absorption of UVR by ocular tissues leads to photochemically-generated ROS and oxidative tissue damage which contribute to the pathological eye conditions noted above. Due to its ability to absorb UVR, the cornea plays a pivotal role in protecting the internal ocular tissues (e.g., lens and retina) from UVR-induced damage. To withstand this challenge, the cornea is equipped with robust antioxidant systems, including low-molecular weight antioxidants and antioxidant enzymes (Chen et al., 2009). Non-enzymatic antioxidants in the cornea include ascorbate, glutathione, α-tocopherol and NAD(P)H. Enzymatic antioxidants that have been detected in the cornea include superoxide dismutases (SODs), catalase (CAT), glutathione peroxidases (GPXs) and reductase (GR), glucose-6-phospate dehydrogenase (G6PDH) and ALDH enzymes.
4.3 Mechanisms of ALDH-mediated protection from UVR damage
The expression of corneal crystallins, ALDH3A1 and ALDH1A1, at levels exceeding those needed for metabolism in the cornea has led to the proposal that these proteins may have additional roles in this ocular structure. An important role for ALDH3A1 in the cornea is suggested by the observation that diseased corneas are associated with decreased ALDH3A1 catalytic activity (Gondhowiardjo et al., 1993). More direct evidence comes from mouse models in which ablation of the Aldh3a1 gene (and to a lesser extent Aldh1a1 gene) renders mice highly susceptible to cataract development (Lassen et al., 2007). These observations are in agreement with a protective function for ALDH3A1 and ALDH1A1 against UVR-induced damage. Accumulating lines of evidence suggest that ALDH3A1 and ALDH1A1 mediate their effects by the following mechanisms (Fig. 5): (i) direct absorption of UVR, (ii) metabolism of toxic aldehydes produced by UVR-induced lipid peroxidation, and (iii) acting as antioxidants by scavenging directly UVR-induced free radicals or (iv) by producing the antioxidant NAD(P)H. These are considered in more detail below.
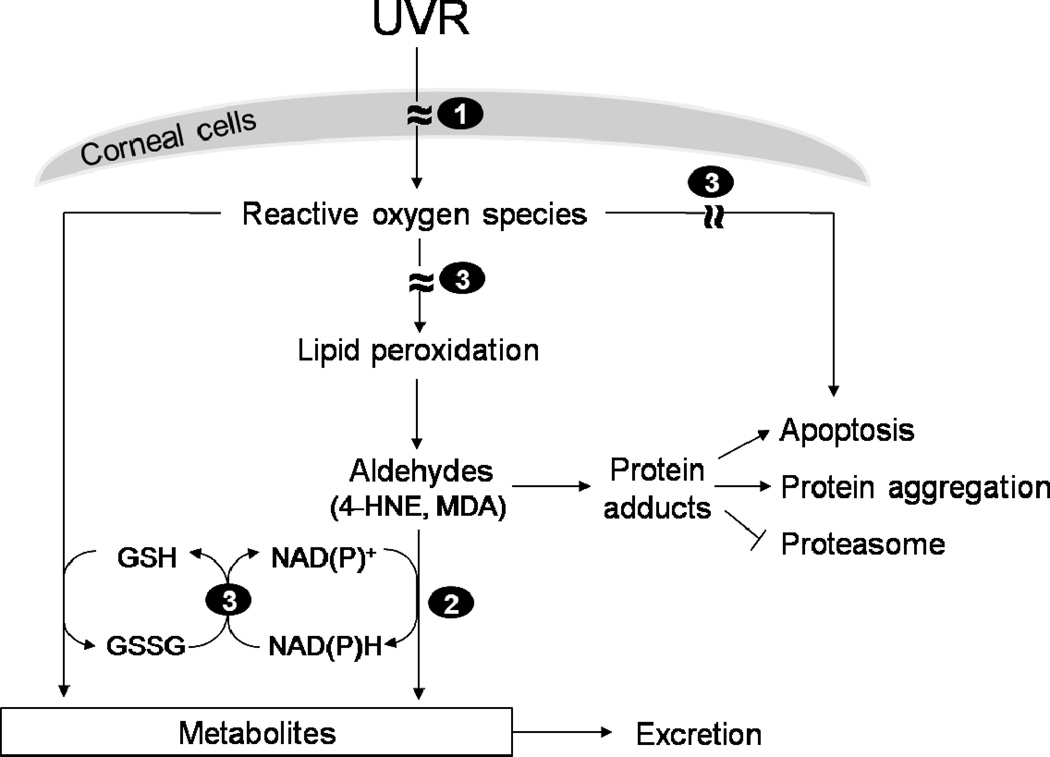
Fig. 5. Proposed mechanisms underlying the protective role of corneal ALDH3A1/1A1 against UVR damage.
ALDH3A1 and/or ALDH1A1 may directly absorb UVR (1), detoxify reactive aldehydes produced by lipid peroxidation to non-toxic metabolites (2) and act as antioxidants by scavenging reactive oxygen species (ROS) (3) or by generating NAD(P)H (3), which can be used in the regeneration of reduced glutathione (GSH) from its oxidized form (GSSG). GSH and NAD(P)H may also eliminate ROS and thereby contribute to the cellular defense against UVR-induced oxidative damage.
ALDH3A1 directly absorbs UVR energy, thereby reducing damage to inner ocular tissues. It was first proposed that ALDH3A1/BCP54 in the bovine cornea could directly absorb UVR (Abedinia et al., 1990). This was based upon the observation that the water-soluble protein fraction of bovine cornea accounted for only 17% of the total cellular protein but was responsible for almost 50% of the total absorption of UVB. Additional evidence supporting such an “absorbin” role for ALDH3A1 include the observation that the majority of UVR is absorbed by the corneal epithelial layer, a site of abundant ALDH3A1 expression (Kolozsvari et al., 2002). It has been proposed that NADPH bound to ALDH3A1 may facilitate the UVR-absorbing capacity of ALDH3A1 (Atherton et al., 1999). A recent study elucidated the molecular details of structural and functional modifications to ALDH3A1 caused by UVR using recombinant proteins (Estey et al., 2010). UVR exposure caused non-native aggregation and associated structural changes of ALDH3A1, but spared the conserved catalytic active site Cys residue. These UVR-induced conformational changes in ALDH3A1 rendered the enzyme completely catalytically-inactive. Such “suicidal” protection by ALDH3A1 may serve to protect other corneal proteins from UVR-induced inactivation (Downes et al., 1993).
ALDH3A1 and ALDH1A1 detoxify reactive aldehydes generated upon UVR exposure. Elevated levels of 4-HNE and/or MDA have been observed in cataracts (Bhuyan et al., 1992), corneal pathologies (Buddi et al., 2002) and retinal disorders (Verdejo et al., 1999), suggesting potential roles for these aldehydes in the pathogenesis of these diseases. As noted, human ALDH3A1 has a high affinity for 4-HNE (Pappa et al., 2003b) and ALDH1A1 metabolizes both 4-HNE and MDA (Manzer et al., 2003) (Table 2). Thus, the metabolic detoxification of these lipid-derived aldehydes represents one mechanism by which ALDH3A1 and ALDH1A1 could protect ocular tissues from UVR-induced damage. It should be noted that the presence of ALDH1A1 in the cornea may complement ALDH3A1 by oxidizing MDA (which is a poor substrate for ALDH3A1) (Pappa et al., 2003b). In human corneal epithelial cells and transfected rabbit corneal keratocytes, ALDH3A1 expression inhibits the formation of 4-HNE-adducted proteins and protects against 4-HNE-induced cytotoxicity and apoptosis (Pappa et al., 2003a). Suppression of ALDH1A1 expression in human lens epithelial cells results in reduced oxidation of 4-HNE and increased susceptibility of cells to oxidant-induced apoptosis (Choudhary et al., 2005). An in vivo role for ocular ALDH3A1 and ALDH1A1 in detoxifying 4-HNE and MDA has been demonstrated in intact animals. In these studies, elevated levels of 4-HNE- and MDAadducted proteins were detected in lens extracts from mice rendered incapable of expressing ALDH1A1 and ALDH3A1, i.e., Aldh3a1−/−/Aldh1a1−/− null mice (Lassen et al., 2007).
Table 2.
Kinetic properties of ALDH1A1/3A1 towards lipid derived aldehydes
ALDH3A1 and ALDH1A1 are both antioxidants. The high concentration of ALDH3A1 in the cornea may contribute to the antioxidant defense mechanisms of the cornea by serving as a direct target for ROS and reactive aldehydes (Estey et al., 2007a; Uma et al., 1996). Indeed, the level of ALDH3A1 carbonylation, a marker of oxidative damage to ALDH3A1, increases 2 to 3-fold in rabbit corneal fibroblasts treated with oxidative agents (Lassen et al., 2006). ALDH3A1 may also protect against UVR-induced oxidative damage by modifying proteasome activity, the major proteolytic system responsible for removing damaged proteins (Davies, 2001). This is supported by two observations: overexpression of ALDH3A1 inhibits UVR-induced loss of proteasome activity in rabbit corneal fibroblasts (Lassen et al., 2006) and decreased lens proteasome activity is detected in Aldh3a1−/− and Aldh3a1−/−/Aldh1a1−/− null mice following UVR exposure (Lassen et al., 2007). ALDH3A1 and ALDH1A1 may also exhibit indirect antioxidant activity by producing NAD(P)H. NAD(P)H functions as an antioxidant in the cornea via multiple mechanisms including (i) serving as a reducing agent for the regeneration of GSH from its oxidized form GSSG (Dickinson and Forman, 2002), (ii) acting as a direct antioxidant by reducing glutathiyl and tyrosyl radicals generated during oxidative stress (Forni and Willson, 1986; Kirsch and De, 2001), (iii) directly absorbing UVR (Atherton et al., 1999; Davies and Truscott, 2001), and (iv) maintaining a reducing potential for pyridine nucleotide-dependent redox-active enzymes (Pappa et al., 2003a).
5. ALDH enzymes contribute to corneal cellular transparency
5.1 Cellular transparency in the cornea
Three-dimensional reconstruction of confocal images through the corneas of living rabbits (Moller-Pedersen, 2004) show that the major sources of light scattering are the superficial anterior corneal epithelial cell layer and the posterior corneal endothelium, the two interfaces between aqueous solutions. Within the cornea, however, little light scattering is detected from the stratified corneal epithelial layers and light scattering within the stroma is limited to the keratocyte nuclei, not from the broad cell bodies (Jester, 2008). According to the proposed “refracton hypothesis” (Piatigorsky, 2001), the lack of light scattering in the corneal epithelium and keratocyte cell bodies indicates that fluctuations in refractive index within these cells is minimized.
The lens has evolved a mechanism to maximize transparency and minimize light scattering through high concentrations of crystallin proteins that are organized in a short-range order within the cytoplasm of the lens fibers (Benedek, 1971; Delaye and Tardieu, 1983; Tardieu, 1998). In the corneas from a wide range of species, a few water-soluble crystallin enzymes/proteins have been found to be identical to lens crystallins, including ALDH1/3 (Fig. 6). Such similarities between corneal and lens crystallins, viz. abundant expression and taxon-specificity, have led to the proposal that the cornea and lens co-evolved through gene-sharing to form a refractive unit that allows maximal light transmission and refraction to the retina (Piatigorsky, 2007). Recent experimental evidence has confirmed an association between expression of the corneal crystallin ALDH1A1 and cellular transparency, particularly involving corneal stromal keratocytes (Jester, 2008).
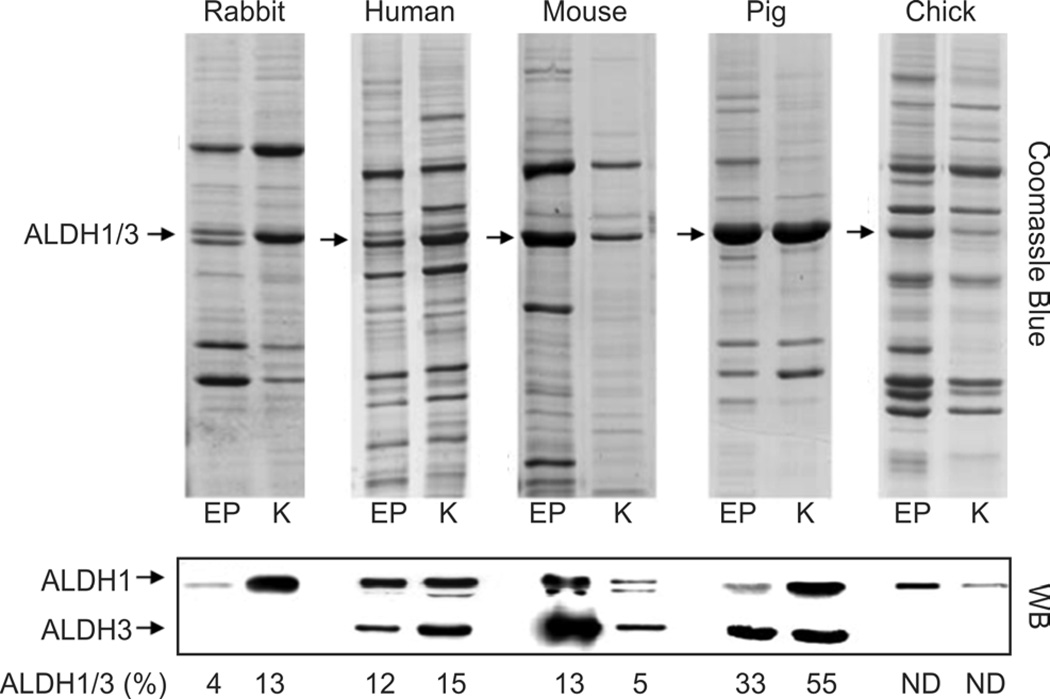
Fig. 6. Abundant and taxon-specific expression of ALDH1A1/3A1 in the cornea.
Watersoluble proteins extracted from corneal epithelium (EP) and keratocytes (K) of different species were resolved by SDS-PAGE and visualized by Coomassie blue or analyzed by Western Blot (WB) for ALDH1A1 (ALDH1) and ALDH3A1 (ALDH3). The contribution of ALDH1A1 and ALDH3A1 together (ALDH1/3) is expressed as a percentage of the total proteins in each sample, i.e., ALDH1/3 (%). ND, not determined. [Adapted from (Jester et al., 2005)].
5.2 ALDH1A1 and ALDH3A1 may serve a structural role in the cornea
Abundant expression of a few water-soluble proteins in keratocytes was first identified in the rabbit (Jester et al., 1999). In this species, transketolase (TKT) and ALDH1A1 comprise, respectively, 14% and 12.7% of total water-soluble protein in corneal keratotyes. Rabbit corneal epithelial cells predominantly express lactate dehydrogenase (14.7%) and, to a lesser extent, TKT (7.5%), G3PDH (6.9%) and α-enolase (4.5%). In other mammalian species including the mouse, human, cow and pig, keratocytes consistently show abundant expression of ALDH3A1 and ALDH1A1, as does the corneal epithelium (Beales et al., 1999; Jester et al., 2005).
Several lines of evidence suggest that ALDH1A1 and ALDH3A1 may function as the structural elements of the cornea. ALDH1A1 appears to contribute substantially to the transparent and refractive aspects of the cornea in the rabbit, consistent with a structural role as a corneal crystallin (Jester et al., 2005). Loss of ALDH1A1 expression in the corneal stroma is associated with injury-induced corneal haze. Further, the development of postnatal corneal transparency is associated with stromal keratocyte quiescence and increased ALDH1A1 expression. By contrast, ALDH3A1 is undetectable in mouse embryo eyes until 9 and 14 days after birth at which time its levels increase dramatically in the cornea. This time is coincident with eye opening (Davis et al., 2008). Based upon these observations, it is possible that reduced expression of ALDH3A1 contributes to the loss of corneal transparency in humans after injury (Pei et al., 2006). The finding of increased light scattering and corneal haze in ALDH3A1 knockout mice (manuscript submitted) support this proposal, as do in vitro studies showing an association between decreased corneal crystallin levels and increased cellular light scattering in rabbit keratocytes (Jester et al., 2005) and that transfection with ALDH3A1 leads to reduced light scattering (Jester et al., 2012).
6. ALDH enzymes may play regulatory roles in the cornea
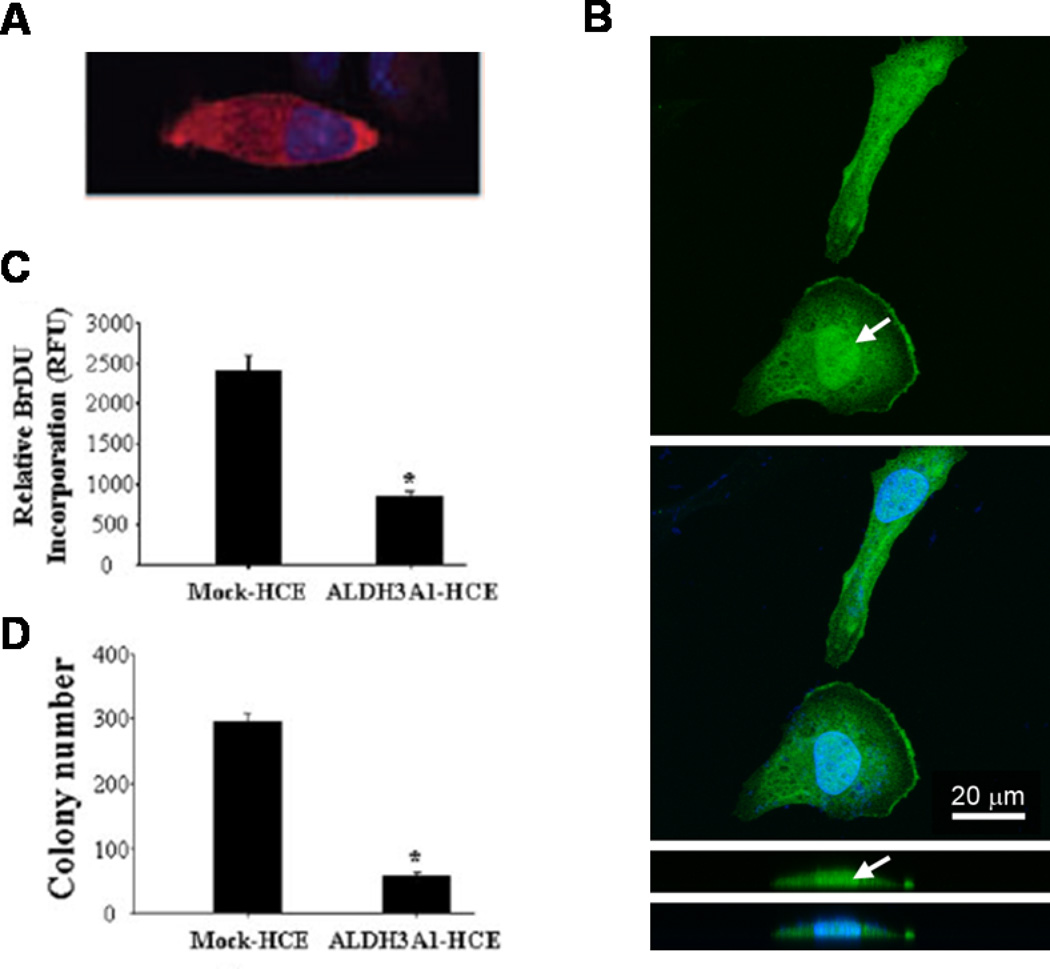
The observation of nuclear localization of ALDH3A1 and ALDH1A1 proteins in transfected human corneal epithelial cells (Pappa et al., 2005) and in transfected and normal rabbit corneal keratocytes (Jester et al., 2012; Stagos et al., 2010b) introduces the possibility for novel physiological roles for these proteins in the cornea, potentially in relation to modulation of cell proliferation (Fig. 7). In this context, an inverse relationship between ALDH3A1 expression and cell proliferation rate has been reported (Pappa et al., 2005). It was found that in actively proliferating primary human corneal epithelial cells, there was a progressive loss of ALDH3A1 gene expression. On the other hand, human corneal epithelial and human skin keratinocyte cells, when stably transfected with human ALDH3A1, showed attenuated growth accompanied by a prolonged cell cycle. Such growth inhibition was associated with suppressed DNA synthesis, reduced activity of cyclin-dependent kinases, dephosphorylation of retinoblastoma protein and down-regulation of several cell cycle regulators including cyclins A, B and E, E2F and p21. This inhibitory effect of ALDH3A1 on cell cycle progression may have important implications for a potential regulatory role of ALDH3A1 in the cornea. First, it may serve as an additional mechanism by which this protein protects corneal cells from damage, i.e., by facilitating DNA repair and cellular survival. Second, ALDH3A1 may participate in controlling the mitotic phenotypes of corneal epithelial cells and fibroblasts in response to physiological or stress signals. The mechanisms underlying this effect on cellular proliferation are currently unknown. One possible mechanism could involve ALDH3A1 metabolizing 4-HNE, an aldehyde known to modulate cell proliferation and differentiation at low concentrations (Barrera et al., 2004). Alternatively, ALDH3A1 may regulate cellular proliferation by physically interacting with (and thereby modulating the actions of) other nuclear mitotic regulators. It is conceivable that ALDH1A1 could function similarly to ALDH3A1, although there is no direct evidence suggesting a role for this protein in cellular growth regulation.
Fig. 7. Inhibitory effect of ALDH3A1 on corneal cell proliferation.
Dual (cytoplasmic and nuclear) localization of human ALDH3A1 in ALDH3A1-transfected human corneal epithelial cells (HCE) (A) and rabbit corneal keratocytes (TRK 43) (B). Expression of human ALDH3A1 inhibits cellular proliferation of HCE cells, as reflected by a decrease in BrdU incorporation (C) and colony formation efficiency (D) in ALDH3A1-expressing HCE cells. [Adapted from (Pappa et al., 2005)].
7. ALDH enzymes in the developing eye
Retinol metabolism plays a crucial role in vertebrate eye development and function (Duester, 2008; Duester, 2009). Gestational vitamin A deficiency manifests itself in a xerophthalmia, a worsened condition of night blindness prevalent in pediatric populations of developing countries. Retinitis pigmentosa is an inherited eye disorder in which progressive vision loss manifests as a result of abnormalities in the retinal pigment epithelium. The genetic heterogeneity associated with this disease challenges our understanding of its etiology. Nevertheless, those genes linked to retinol metabolism appear to play a major role in the disease mechanism.
As noted above, the conversion of retinal to RA is carried out by three retinaldehye dehydrogenases (RALDHs), namely ALDH1A1, ALDH1A2 and ALDH1A3 (Duester, 2000; Duester et al., 2003). Three mouse genes encoding these enzymes have unique spatio-temporal expression patterns (Duester, 2009). ALDH1A2 alone accounts for the initial RA synthesis from embryonic days 7.5–9.5 (E7.5–9.5), but RA synthesis from E9.5–10.5 involves participation of all three ALDH1 enzymes. This expression is critical for development of the neural components of the eye. During the process of optic cup formation, ALDH1A2 expression starts at E9.0 in the peri-optic mesenchyme and terminates at E10.0. ALDH1A3 expression begins in the surface ectoderm at E8.75 and in the dorsal retinal pigment epithelium at E9.5 and continues into the dorsoventral patterning of the retina (Suzuki et al., 2000). ALDH1A1 expression begins in the dorsal neutral retina at E10.5 and continues to be expressed along with ALDH1A3 in the choroid fissure at E13.5.
Studies using transgenic mouse models have elucidated the mechanism of RA signaling in the developing eye. ALDH1A1-deficent mice survive into adulthood and display no eye defects (Fan et al., 2003). However, mice deficient in both ALDH1A1 and ALDH1A2 exhibit defective apoptosis in perioptic mesenchyme (POM) (Mic et al., 2004). This finding suggests that these two proteins may have redundant functions in the optic nerve development. ALDH1A3-deficient mice die at birth due to blockage of nasal passages (choanal atresia) and show incomplete ventral optic cup formation as embryos (Dupe et al., 2003). On the other hand, fetuses deficient in both ALDH1A1 and ALDH1A3 exhibit multiple and severe ocular malformations affecting the eyelids, the cornea, the lens and the retina (Matt et al., 2005). ALDH1A2-deficient mice also display under-developed optic cup formation (Mic et al., 2004), but these mice can be rescued by maternal RA administration; this treatment seems to result in ALDH1A3 levels being nearly normal in these mice, restoring all RA signaling in these tissues (Fan et al., 2003). These results are consistent with ALDH1A3 being a very important contributor to RA signaling. Collectively, studies using genetic approaches in the mouse demonstrate that RA, produced by ALDH1A isozymes in the neural retina, the retinal pigmented epithelium and the corneal ectoderm, acts in a paracrine fashion on POM which controls the development of anterior eye segment and the growth of the ventral retina (Matt et al., 2005).
In addition to a developmental role, ALDHs expressed in the retina may have an important function in protecting the retina from oxidative damage. The retina is exposed to a range of oxidative stress-associated insults, from environmental pollutants to UVR. The high levels of PUFAs in the photoreceptor membranes of the retina represent a formidable threat from photo-oxidation and subsequent ROS-mediated LPO (Cai and McGinnis, 2012). Anti-oxidant defenses in the retina are critical for maintaining the integrity and function of this tissue. These defenses can be compromised in disease states, such as diabetic retinopathy, cataracts, glaucoma and AMD, where reactive aldehydes comprise the bulk of the oxidative assault (Madsen-Bouterse and Kowluru, 2008). Immunohistochemical studies have revealed alcohol dehydrogenase 3 (ADH3) and ALDH2 to be co-localized with peroxydic aldehydes in rat photoreceptors and inner retinal layers (Galbis-Estrada et al., 2012; Pinazo-Duran et al., 2000), consistent with a possible role for these enzymes in cellular antioxidant defenses against ROS.
8. Conclusions and future directions
ALDH enzymes are a group of structurally-related proteins that facilitate the metabolism of aldehyde substrates of physiological and pathophysiological importance. In so doing, ALDH isozymes have diverse but distinct functions in various tissues. In the cornea and lens, with the characteristics of abundant expression and active metabolism of toxic aldehydes, ALDH3A1 and ALDH1A1 are being recognized as multifunctional crystallin proteins that play important structural, metabolic and regulatory roles. Further elucidation of the molecular mechanisms governing the expression of these ALDHs, as well as the definition of their cellular functions, will provide valuable insights into the pathophysiological mechanisms of ocular diseases, particularly those associated with UVR exposure. Such information may permit the development of novel treatment interventions for these diseases in the future.
Acknowledgement
This work was supported in part by the National Institutes of Health, National Eye Institute Grants EY017963, EY011490, EY016663, and EY07348.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Abedinia M, Pain T, Algar EM, Holmes RS. Bovine corneal aldehyde dehydrogenase: the major soluble corneal protein with a possible dual protective role for the eye. Exp. Eye Res. 1990;51:419–426. doi: 10.1016/0014-4835(90)90154-m. [DOI] [PubMed] [Google Scholar]
- Aikens J, Dix TA. Perhydroxyl radical (HOO.) initiated lipid peroxidation. The role of fatty acid hydroperoxides. J. Biol. Chem. 1991;266:15091–15098. [PubMed] [Google Scholar]
- Allen EM, Anderson DG, Florang VR, Khanna M, Hurley TD, Doorn JA. Relative inhibitory potency of molinate and metabolites with aldehyde dehydrogenase 2: implications for the mechanism of enzyme inhibition. Chem. Res. Toxicol. 2010;23:1843–1850. doi: 10.1021/tx100317q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andley UP, Rhim JS, Chylack LT, Jr, Fleming TP. Propagation and immortalization of human lens epithelial cells in culture. Invest Ophthalmol. Vis. Sci. 1994;35:3094–3102. [PubMed] [Google Scholar]
- Atherton SJ, Lambert C, Schultz J, Williams N, Zigman S. Fluorescence studies of lens epithelial cells and their constituents. Photochem. Photobiol. 1999;70:823–828. [PubMed] [Google Scholar]
- Balasubramanian D. Ultraviolet radiation and cataract. J. Ocul. Pharmacol. Ther. 2000;16:285–297. doi: 10.1089/jop.2000.16.285. [DOI] [PubMed] [Google Scholar]
- Barrera G, Pizzimenti S, Dianzani MU. 4-hydroxynonenal and regulation of cell cycle: effects on the pRb/E2F pathway. Free Radic. Biol. Med. 2004;37:597–606. doi: 10.1016/j.freeradbiomed.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Beales MP, Funderburgh JL, Jester JV, Hassell JR. Proteoglycan synthesis by bovine keratocytes and corneal fibroblasts: maintenance of the keratocyte phenotype in culture. Invest Ophthalmol. Vis. Sci. 1999;40:1658–1663. [PubMed] [Google Scholar]
- Benedek GB. Theory of transparency of the eye. Appl. Opt. 1971;10:459–473. doi: 10.1364/AO.10.000459. [DOI] [PubMed] [Google Scholar]
- Bergendi L, Benes L, Durackova Z, Ferencik M. Chemistry, physiology and pathology of free radicals. Life Sci. 1999;65:1865–1874. doi: 10.1016/s0024-3205(99)00439-7. [DOI] [PubMed] [Google Scholar]
- Bergmanson JP, Soderberg PG. The significance of ultraviolet radiation for eye diseases. A review with comments on the efficacy of UV-blocking contact lenses. Ophthalmic Physiol Opt. 1995;15:83–91. [PubMed] [Google Scholar]
- Bhat PV, Samaha H. Kinetic properties of the human liver cytosolic aldehyde dehydrogenase for retinal isomers. Biochem. Pharmacol. 1999;57:195–197. doi: 10.1016/s0006-2952(98)00261-5. [DOI] [PubMed] [Google Scholar]
- Bhuyan KC, Bhuyan DK, Santos O, Podos SM. Antioxidant and anticataractogenic effects of topical captopril in diquat-induced cataract in rabbits. Free Radic. Biol. Med. 1992;12:251–261. doi: 10.1016/0891-5849(92)90112-t. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R. Glutathione peroxidases and redox-regulated transcription factors. Biol. Chem. 2006;387:1329–1335. doi: 10.1515/BC.2006.166. [DOI] [PubMed] [Google Scholar]
- Brocker C, Lassen N, Estey T, Pappa A, Cantore M, Orlova VV, Chavakis T, Kavanagh KL, Oppermann U, Vasiliou V. Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J. Biol. Chem. 2010;285:18452–18463. doi: 10.1074/jbc.M109.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddi R, Lin B, Atilano SR, Zorapapel NC, Kenney MC, Brown DJ. Evidence of oxidative stress in human corneal diseases. J. Histochem. Cytochem. 2002;50:341–351. doi: 10.1177/002215540205000306. [DOI] [PubMed] [Google Scholar]
- Burke WJ, Li SW, Williams EA, Nonneman R, Zahm DS. 3,4-Dihydroxyphenylacetaldehyde is the toxic dopamine metabolite in vivo: implications for Parkinson's disease pathogenesis. Brain Res. 2003;989:205–213. doi: 10.1016/s0006-8993(03)03354-7. [DOI] [PubMed] [Google Scholar]
- Cadenas E, Sies H. The lag phase. Free Radic. Res. 1998;28:601–609. doi: 10.3109/10715769809065816. [DOI] [PubMed] [Google Scholar]
- Cai X, McGinnis JF. Oxidative stress: the achilles' heel of neurodegenerative diseases of the retina. Front Biosci. 2012;17:1976–1995. doi: 10.2741/4033. [DOI] [PubMed] [Google Scholar]
- Chalam KV, Khetpal V, Rusovici R, Balaiya S. A review: role of ultraviolet radiation in agerelated macular degeneration. Eye Contact Lens. 2011;37:225–232. doi: 10.1097/ICL.0b013e31821fbd3e. [DOI] [PubMed] [Google Scholar]
- Chen Y, Koppaka V, Thompson DC, Vasiliou V. Focus on Molecules: ALDH1A1: From lens and corneal crystallin to stem cell marker. Exp. Eye Res. 2011a doi: 10.1016/j.exer.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Mehta G, Vasiliou V. Antioxidant defenses in the ocular surface. Ocul. Surf. 2009;7:176–185. doi: 10.1016/s1542-0124(12)70185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem. Biophys. Res. Commun. 2011b;405:173–179. doi: 10.1016/j.bbrc.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S, Xiao T, Vergara LA, Srivastava S, Nees D, Piatigorsky J, Ansari NH. Role of aldehyde dehydrogenase isozymes in the defense of rat lens and human lens epithelial cells against oxidative stress. Invest Ophthalmol. Vis. Sci. 2005;46:259–267. doi: 10.1167/iovs.04-0120. [DOI] [PubMed] [Google Scholar]
- Cobessi D, Tete-Favier F, Marchal S, Branlant G, Aubry A. Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. J. Mol. Biol. 2000;300:141–152. doi: 10.1006/jmbi.2000.3824. [DOI] [PubMed] [Google Scholar]
- Colby TD, Bahnson BJ, Chin JK, Klinman JP, Goldstein BM. Active site modifications in a double mutant of liver alcohol dehydrogenase: structural studies of two enzyme-ligand complexes. Biochemistry. 1998;37:9295–9304. doi: 10.1021/bi973184b. [DOI] [PubMed] [Google Scholar]
- Cooper DL, Isola NR, Stevenson K, Baptist EW. Members of the ALDH gene family are lens and corneal crystallins. Adv. Exp. Med. Biol. 1993;328:169–179. doi: 10.1007/978-1-4615-2904-0_19. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Piatigorsky J. Lens development and crystallin gene expression: many roles for Pax-6. Bioessays. 1996;18:621–630. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Sax CM, Li X, McDermott JB, Piatigorsky J. Pax-6 and lens-specific transcription of the chicken delta 1-crystallin gene. Proc. Natl. Acad. Sci. U. S. A. 1995;92:4681–4685. doi: 10.1073/pnas.92.10.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrosio K, Pailot A, Talfournier F, Didierjean C, Benedetti E, Aubry A, Branlant G, Corbier C. The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis. Biochemistry. 2006;45:2978–2986. doi: 10.1021/bi0515117. [DOI] [PubMed] [Google Scholar]
- Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- Davies MJ, Truscott RJ. Photo-oxidation of proteins and its role in cataractogenesis. J. Photochem Photobiol. B. 2001;63:114–125. doi: 10.1016/s1011-1344(01)00208-1. [DOI] [PubMed] [Google Scholar]
- Davis J, Davis D, Norman B, Piatigorsky J. Gene expression of the mouse corneal crystallin Aldh3a1: activation by Pax6, Oct1, and p300. Invest Ophthalmol. Vis. Sci. 2008;49:1814–1826. doi: 10.1167/iovs.07-1057. [DOI] [PubMed] [Google Scholar]
- Dean RT, Fu S, Stocker R, Davies MJ. Biochemistry and pathology of radical-mediated protein oxidation. Biochem. J. 1997;324(Pt 1):1–18. doi: 10.1042/bj3240001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaye M, Tardieu A. Short-range order of crystallin proteins accounts for eye lens transparency. Nature. 1983;302:415–417. doi: 10.1038/302415a0. [DOI] [PubMed] [Google Scholar]
- Deng S, Yang X, Lassus H, Liang S, Kaur S, Ye Q, Li C, Wang LP, Roby KF, Orsulic S, Connolly DC, Zhang Y, Montone K, Butzow R, Coukos G, Zhang L. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS. One. 2010;5:e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianzani MU. 4-Hydroxynonenal and cell signalling. Free Radic. Res. 1998;28:553–560. doi: 10.3109/10715769809065811. [DOI] [PubMed] [Google Scholar]
- Dickinson DA, Forman HJ. Glutathione in defense and signaling: lessons from a small thiol. Ann. N. Y. Acad. Sci. 2002;973:488–504. doi: 10.1111/j.1749-6632.2002.tb04690.x. [DOI] [PubMed] [Google Scholar]
- Downes JE, Swann PG, Holmes RS. Ultraviolet light-induced pathology in the eye: associated changes in ocular aldehyde dehydrogenase and alcohol dehydrogenase activities. Cornea. 1993;12:241–248. doi: 10.1097/00003226-199305000-00010. [DOI] [PubMed] [Google Scholar]
- Downes JE, Swann PG, Holmes RS. A genetic basis for corneal sensitivity to ultraviolet light among recombinant SWXJ inbred strains of mice. Curr. Eye Res. 1997;16:539–546. doi: 10.1076/ceyr.16.6.539.5075. [DOI] [PubMed] [Google Scholar]
- Downes JE, VandeBerg JL, Hubbard GB, Holmes RS. Regional distribution of mammalian corneal aldehyde dehydrogenase and alcohol dehydrogenase. Cornea. 1992;11:560–566. doi: 10.1097/00003226-199211000-00013. [DOI] [PubMed] [Google Scholar]
- Duester G. Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur. J. Biochem. 2000;267:4315–4324. doi: 10.1046/j.1432-1327.2000.01497.x. [DOI] [PubMed] [Google Scholar]
- Duester G. Keeping an eye on retinoic acid signaling during eye development. Chem. Biol. Interact. 2008 doi: 10.1016/j.cbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. Keeping an eye on retinoic acid signaling during eye development. Chem. Biol. Interact. 2009;178:178–181. doi: 10.1016/j.cbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G, Mic FA, Molotkov A. Cytosolic retinoid dehydrogenases govern ubiquitous metabolism of retinol to retinaldehyde followed by tissue-specific metabolism to retinoic acid. Chem. Biol. Interact. 2003;143–144:201–210. doi: 10.1016/s0009-2797(02)00204-1. [DOI] [PubMed] [Google Scholar]
- Dupe V, Matt N, Garnier JM, Chambon P, Mark M, Ghyselinck NB. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton JW. UV-mediated cataractogenesis: a radical perspective. Doc. Ophthalmol. 1994;88:233–242. doi: 10.1007/BF01203677. [DOI] [PubMed] [Google Scholar]
- Eguchi G. [Crystalline lens] Tanpakushitsu Kakusan Koso. 1966;11:1083–1084. [PubMed] [Google Scholar]
- Esterbauer H. Cytotoxicity and genotoxicity of lipid-oxidation products. Am. J. Clin. Nutr. 1993;57:779S–785S. doi: 10.1093/ajcn/57.5.779S. [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- Estey T, Cantore M, Weston PA, Carpenter JF, Petrash JM, Vasiliou V. Mechanisms involved in the protection of UV-induced protein inactivation by the corneal crystallin ALDH3A1. J Biol. Chem. 2007a;282:4382–4392. doi: 10.1074/jbc.M607546200. [DOI] [PubMed] [Google Scholar]
- Estey T, Chen Y, Carpenter JF, Vasiliou V. Structural and functional modifications of corneal crystallin ALDH3A1 by UVB light. PLoS. One. 2010;5:e15218. doi: 10.1371/journal.pone.0015218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estey T, Piatigorsky J, Lassen N, Vasiliou V. ALDH3A1: a corneal crystallin with diverse functions. Exp. Eye Res. 2007b;84:3–12. doi: 10.1016/j.exer.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol. Cell Biol. 2003;23:4637–4648. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni LG, Willson RL. Thiyl free radicals and the oxidation of ferrocytochrome c. Direct observation of coupled hydrogen-atom- and electron-transfer reactions. Biochem. J. 1986;240:905–907. doi: 10.1042/bj2400905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbis-Estrada C, Pons-Vazquez S, Gallego-Pinazo R, Lleo-Perez A, Garcia-Medina JJ, Bou VV, Sanz-Solana P, Pinazo-Duran MD. Glutathione-dependent formaldehyde dehydrogenase (ADH3) and low km mitochondrial aldehyde dehydrogenase (ALDH2). New evidence for differential expression in the rat retina in response to oxidative stress. Free Radic. Res. 2012;46:77–84. doi: 10.3109/10715762.2011.640324. [DOI] [PubMed] [Google Scholar]
- Gasparetto M, Sekulovic S, Brocker C, Tang P, Zakaryan A, Xiang P, Kuchenbauer F, Wen M, Kasaian K, Witty MF, Rosten P, Chen Y, Imren S, Duester G, Thompson DC, Humphries RK, Vasiliou V, Smith C. Aldehyde dehydrogenases are regulators of hematopoietic stem cell numbers and B-cell development. Exp. Hematol. 2011 doi: 10.1016/j.exphem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Gondhowiardjo TD, van Haeringen NJ, Volker-Dieben HJ, Beekhuis HW, Kok JH, van RG, Pels L, Kijlstra A. Analysis of corneal aldehyde dehydrogenase patterns in pathologic corneas. Cornea. 1993;12:146–154. doi: 10.1097/00003226-199303000-00010. [DOI] [PubMed] [Google Scholar]
- Graham C, Hodin J, Wistow G. A retinaldehyde dehydrogenase as a structural protein in a mammalian eye lens. Gene recruitment of eta-crystallin. J Biol. Chem. 1996;271:15623–15628. doi: 10.1074/jbc.271.26.15623. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Can oxidative DNA damage be used as a biomarker of cancer risk in humans? Problems, resolutions and preliminary results from nutritional supplementation studies. Free Radic. Res. 1998;29:469–486. doi: 10.1080/10715769800300531. [DOI] [PubMed] [Google Scholar]
- Hanna MC, Blackstone C. Interaction of the SPG21 protein ACP33/maspardin with the aldehyde dehydrogenase ALDH16A1. Neurogenetics. 2009;10:217–228. doi: 10.1007/s10048-009-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RS, Cheung B, VandeBerg JL. Isoelectric focusing studies of aldehyde dehydrogenases, alcohol dehydrogenases and oxidases from mammalian anterior eye tissues. Comp Biochem. Physiol B. 1989;93:271–277. doi: 10.1016/0305-0491(89)90081-3. [DOI] [PubMed] [Google Scholar]
- Hough RB, Piatigorsky J. Preferential transcription of rabbit Aldh1a1 in the cornea: implication of hypoxia-related pathways. Mol. Cell Biol. 2004;24:1324–1340. doi: 10.1128/MCB.24.3.1324-1340.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genomics. 2011;5:283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jester JV. Corneal crystallins and the development of cellular transparency. Semin. Cell Dev. Biol. 2008;19:82–93. doi: 10.1016/j.semcdb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jester JV, Brown D, Pappa A, Vasiliou V. Myofibroblast differentiation modulates keratocyte crystallin protein expression, concentration, and cellular light scattering. Invest Ophthalmol. Vis. Sci. 2012;53:770–778. doi: 10.1167/iovs.11-9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jester JV, Budge A, Fisher S, Huang J. Corneal keratocytes: phenotypic and species differences in abundant protein expression and in vitro light-scattering. Invest Ophthalmol. Vis. Sci. 2005;46:2369–2378. doi: 10.1167/iovs.04-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jester JV, Moller-Pedersen T, Huang J, Sax CM, Kays WT, Cavangh HD, Petroll WM, Piatigorsky J. The cellular basis of corneal transparency: evidence for 'corneal crystallins. J. Cell Sci. 1999;112(Pt 5):613–622. doi: 10.1242/jcs.112.5.613. [DOI] [PubMed] [Google Scholar]
- Johnson F, Giulivi C. Superoxide dismutases and their impact upon human health. Mol. Aspects Med. 2005;26:340–352. doi: 10.1016/j.mam.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Kazmi SM, Li D, Koop K, Conant J, Lau CY. Role of aldehyde dehydrogenase in the biological activity of spermine dialdehyde, a novel immunosuppressive/purging agent. Pharmacol. Res. 1992;25:383–392. doi: 10.1016/1043-6618(92)90675-2. [DOI] [PubMed] [Google Scholar]
- King G, Holmes RS. Human corneal aldehyde dehydrogenase: purification, kinetic characterisation and phenotypic variation. Biochem. Mol. Biol. Int. 1993;31:49–63. [PubMed] [Google Scholar]
- Kirsch M, De GH. NAD(P)H, a directly operating antioxidant? FASEB J. 2001;15:1569–1574. doi: 10.1096/fj.00-0823hyp. [DOI] [PubMed] [Google Scholar]
- Klein RS, Werth VP, Dowdy JC, Sayre RM. Analysis of compact fluorescent lights for use by patients with photosensitive conditions. Photochem. Photobiol. 2009;85:1004–1010. doi: 10.1111/j.1751-1097.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyosov AA, Rashkovetsky LG, Tahir MK, Keung WM. Possible role of liver cytosolic and mitochondrial aldehyde dehydrogenases in acetaldehyde metabolism. Biochemistry. 1996;35:4445–4456. doi: 10.1021/bi9521093. [DOI] [PubMed] [Google Scholar]
- Kolozsvari L, Nogradi A, Hopp B, Bor Z. UV absorbance of the human cornea in the 240- to 400-nm range. Invest Ophthalmol. Vis. Sci. 2002;43:2165–2168. [PubMed] [Google Scholar]
- Lassen N, Bateman JB, Estey T, Kuszak JR, Nees DW, Piatigorsky J, Duester G, Day BJ, Huang J, Hines LM, Vasiliou V. Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(-/-)/Aldh1a1(-/-) knock-out mice. J. Biol. Chem. 2007;282:25668–25676. doi: 10.1074/jbc.M702076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen N, Black WJ, Estey T, Vasiliou V. The role of corneal crystallins in the cellular defense mechanisms against oxidative stress. Semin. Cell Dev. Biol. 2008;19:100–112. doi: 10.1016/j.semcdb.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Lassen N, Pappa A, Black WJ, Jester JV, Day BJ, Min E, Vasiliou V. Antioxidant function of corneal ALDH3A1 in cultured stromal fibroblasts. Free Radic. Biol. Med. 2006;41:1459–1469. doi: 10.1016/j.freeradbiomed.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Leonard SS, Harris GK, Shi X. Metal-induced oxidative stress and signal transduction. Free Radic. Biol. Med. 2004;37:1921–1942. doi: 10.1016/j.freeradbiomed.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Hempel J, Sun J, Rose J, Hsiao D, Chang WR, Chung YJ, Kuo I, Lindahl R, Wang BC. Crystal structure of a class 3 aldehyde dehydrogenase at 2.6 A resolution. Adv. Exp. Med. Biol. 1997;414:1–7. doi: 10.1007/978-1-4615-5871-2_1. [DOI] [PubMed] [Google Scholar]
- Ma I, Allan AL. The Role of Human Aldehyde Dehydrogenase in Normal and Cancer Stem Cells. Stem Cell Rev. 2010 doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- Mackenzie FD, Hirst LW, Battistutta D, Green A. Risk analysis in the development of pterygia. Ophthalmology. 1992;99:1056–1061. doi: 10.1016/s0161-6420(92)31850-0. [DOI] [PubMed] [Google Scholar]
- Madsen-Bouterse SA, Kowluru RA. Oxidative stress and diabetic retinopathy: pathophysiological mechanisms and treatment perspectives. Rev. Endocr. Metab Disord. 2008;9:315–327. doi: 10.1007/s11154-008-9090-4. [DOI] [PubMed] [Google Scholar]
- Manzer R, Qamar L, Estey T, Pappa A, Petersen DR, Vasiliou V. Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA. DNA Cell Biol. 2003;22:329–338. doi: 10.1089/104454903322216671. [DOI] [PubMed] [Google Scholar]
- Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert. Opin. Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt N, Dupe V, Garnier JM, Dennefeld C, Chambon P, Mark M, Ghyselinck NB. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development. 2005;132:4789–4800. doi: 10.1242/dev.02031. [DOI] [PubMed] [Google Scholar]
- Matt N, Ghyselinck NB, Wendling O, Chambon P, Mark M. Retinoic acid-induced developmental defects are mediated by RARbeta/RXR heterodimers in the pharyngeal endoderm. Development. 2003;130:2083–2093. doi: 10.1242/dev.00428. [DOI] [PubMed] [Google Scholar]
- Mic FA, Molotkov A, Molotkova N, Duester G. Raldh2 expression in optic vesicle generates a retinoic acid signal needed for invagination of retina during optic cup formation. Dev. Dyn. 2004;231:270–277. doi: 10.1002/dvdy.20128. [DOI] [PubMed] [Google Scholar]
- Moller-Pedersen T. Keratocyte reflectivity and corneal haze. Exp. Eye Res. 2004;78:553–560. doi: 10.1016/s0014-4835(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Moran DJ, Hollows FC. Pterygium and ultraviolet radiation: a positive correlation. Br. J. Ophthalmol. 1984;68:343–346. doi: 10.1136/bjo.68.5.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappa A, Brown D, Koutalos Y, DeGregori J, White C, Vasiliou V. Human aldehyde dehydrogenase 3A1 inhibits proliferation and promotes survival of human corneal epithelial cells. J. Biol. Chem. 2005;280:27998–28006. doi: 10.1074/jbc.M503698200. [DOI] [PubMed] [Google Scholar]
- Pappa A, Chen C, Koutalos Y, Townsend AJ, Vasiliou V. Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radic. Biol. Med. 2003a;34:1178–1189. doi: 10.1016/s0891-5849(03)00070-4. [DOI] [PubMed] [Google Scholar]
- Pappa A, Estey T, Manzer R, Brown D, Vasiliou V. Human aldehyde dehydrogenase 3A1 (ALDH3A1): biochemical characterization and immunohistochemical localization in the cornea. Biochem. J. 2003b;376:615–623. doi: 10.1042/BJ20030810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappa A, Sophos NA, Vasiliou V. Corneal and stomach expression of aldehyde dehydrogenases: from fish to mammals. Chem. Biol. Interact. 2001;130–132:181–191. doi: 10.1016/s0009-2797(00)00233-7. [DOI] [PubMed] [Google Scholar]
- Pei Y, Reins RY, McDermott AM. Aldehyde dehydrogenase (ALDH) 3A1 expression by the human keratocyte and its repair phenotypes. Exp. Eye Res. 2006;83:1063–1073. doi: 10.1016/j.exer.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Perez-Miller SJ, Hurley TD. Coenzyme isomerization is integral to catalysis in aldehyde dehydrogenase. Biochemistry. 2003;42:7100–7109. doi: 10.1021/bi034182w. [DOI] [PubMed] [Google Scholar]
- Perra MT, Maxia C, Corbu A, Minerba L, Demurtas P, Colombari R, Murtas D, Bravo S, Piras F, Sirigu P. Oxidative stress in pterygium: relationship between p53 and 8-hydroxydeoxyguanosine. Mol. Vis. 2006;12:1136–1142. [PubMed] [Google Scholar]
- Piatigorsky J. Gene sharing in lens and cornea: facts and implications. Prog. Retin. Eye Res. 1998a;17:145–174. doi: 10.1016/s1350-9462(97)00004-9. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. Multifunctional lens crystallins and corneal enzymes. More than meets the eye. Ann. N. Y. Acad. Sci. 1998b;842:7–15. doi: 10.1111/j.1749-6632.1998.tb09626.x. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. Enigma of the abundant water-soluble cytoplasmic proteins of the cornea: the "refracton" hypothesis. Cornea. 2001;20:853–858. doi: 10.1097/00003226-200111000-00015. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. Gene Sharing and Evolution: The Diversity of Protein Functions. Cambridge, MA: Harvard University Press; 2007. [Google Scholar]
- Pinazo-Duran MD, Verdejo C, Azorin I, Renau-Piqueras J, Iborra FJ. Colocalization of aldehyde dehydrogenases and Fe/NADPH-induced lipid peroxidation in tissue sections of rat retina. Ophthalmic Res. 2000;32:61–68. doi: 10.1159/000055591. [DOI] [PubMed] [Google Scholar]
- Poli G, Schaur RJ. 4-Hydroxynonenal in the pathomechanisms of oxidative stress. IUBMB. Life. 2000;50:315–321. doi: 10.1080/713803726. [DOI] [PubMed] [Google Scholar]
- Roberts JE. Ocular phototoxicity. J. Photochem. Photobiol. B. 2001;64:136–143. doi: 10.1016/s1011-1344(01)00196-8. [DOI] [PubMed] [Google Scholar]
- Roberts JE. Ultraviolet radiation as a risk factor for cataract and macular degeneration. Eye Contact Lens. 2011;37:246–249. doi: 10.1097/ICL.0b013e31821cbcc9. [DOI] [PubMed] [Google Scholar]
- Schaur RJ. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol. Aspects Med. 2003;24:149–159. doi: 10.1016/s0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim. Biophys. Acta. 2006;1763:1755–1766. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Shiao T, Tran P, Siegel D, Lee J, Vasiliou V. Four amino acid changes are associated with the Aldh3a1 locus polymorphism in mice which may be responsible for corneal sensitivity to ultraviolet light. Pharmacogenetics. 1999;9:145–153. [PubMed] [Google Scholar]
- Sliney DH. Optical radiation safety of medical light sources. Phys. Med. Biol. 1997;42:981–996. doi: 10.1088/0031-9155/42/5/016. [DOI] [PubMed] [Google Scholar]
- Stagos D, Chen Y, Brocker C, Donald E, Jackson BC, Orlicky DJ, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1: molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab Dispos. 2010a;38:1679–1687. doi: 10.1124/dmd.110.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagos D, Chen Y, Cantore M, Jester JV, Vasiliou V. Corneal aldehyde dehydrogenases: multiple functions and novel nuclear localization. Brain Res. Bull. 2010b;81:211–218. doi: 10.1016/j.brainresbull.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storms RW, Trujillo AP, Springer JB, Shah L, Colvin OM, Ludeman SM, Smith C. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc. Natl. Acad. Sci. U. S.A. 1999;96:9118–9123. doi: 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shintani T, Sakuta H, Kato A, Ohkawara T, Osumi N, Noda M. Identification of RALDH-3, a novel retinaldehyde dehydrogenase, expressed in the ventral region of the retina. Mech. Dev. 2000;98:37–50. doi: 10.1016/s0925-4773(00)00450-0. [DOI] [PubMed] [Google Scholar]
- Swamynathan SK, Davis J, Piatigorsky J. Identification of candidate Klf4 target genes reveals the molecular basis of the diverse regulatory roles of Klf4 in the mouse cornea. Invest Ophthalmol. Vis. Sci. 2008;49:3360–3370. doi: 10.1167/iovs.08-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu A. alpha-Crystallin quaternary structure and interactive properties control eye lens transparency. Int. J. Biol. Macromol. 1998;22:211–217. doi: 10.1016/s0141-8130(98)00018-x. [DOI] [PubMed] [Google Scholar]
- Taylor HR, Munoz B, West S, Bressler NM, Bressler SB, Rosenthal FS. Visible light and risk of age-related macular degeneration. Trans. Am. Ophthalmol. Soc. 1990;88:163–173. [PMC free article] [PubMed] [Google Scholar]
- Tenkate TD. Occupational exposure to ultraviolet radiation: a health risk assessment. Rev. Environ. Health. 1999;14:187–209. doi: 10.1515/reveh.1999.14.4.187. [DOI] [PubMed] [Google Scholar]
- Ucar D, Cogle CR, Zucali JR, Ostmark B, Scott EW, Zori R, Gray BA, Moreb JS. Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chem. Biol. Interact. 2009;178:48–55. doi: 10.1016/j.cbi.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uma L, Hariharan J, Sharma Y, Balasubramanian D. Corneal aldehyde dehydrogenase displays antioxidant properties. Exp. Eye Res. 1996;63:117–120. doi: 10.1006/exer.1996.0098. [DOI] [PubMed] [Google Scholar]
- van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Pelger RC, van der Pluijm G. The aldehyde dehydrogenase enzyme 7A1 is functionally involved in prostate cancer bone metastasis. Clin. Exp. Metastasis. 2011;28:615–625. doi: 10.1007/s10585-011-9395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdejo C, Marco P, Renau-Piqueras J, Pinazo-Duran MD. Lipid peroxidation in proliferative vitreoretinopathies. Eye. 1999;13(Pt 2):183–188. doi: 10.1038/eye.1999.48. [DOI] [PubMed] [Google Scholar]
- Verhagen C, Hoekzema R, Verjans GM, Kijlstra A. Identification of bovine corneal protein 54 (BCP 54) as an aldehyde dehydrogenase. Exp. Eye Res. 1991;53:283–284. doi: 10.1016/0014-4835(91)90085-s. [DOI] [PubMed] [Google Scholar]
- Wong TY, Foster PJ, Johnson GJ, Seah SK, Tan DT. The prevalence and risk factors for pterygium in an adult Chinese population in Singapore: the Tanjong Pagar survey. Am. J. Ophthalmol. 2001;131:176–183. doi: 10.1016/s0002-9394(00)00703-0. [DOI] [PubMed] [Google Scholar]
- Xiao T, Shoeb M, Siddiqui MS, Zhang M, Ramana KV, Srivastava SK, Vasiliou V, Ansari NH. Molecular cloning and oxidative modification of human lens ALDH1A1: implication in impaired detoxification of lipid aldehydes. J. Toxicol. Environ. Health A. 2009;72:577–584. doi: 10.1080/15287390802706371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Hsu LC, Dave V. Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase. Enzyme. 1992;46:239–244. doi: 10.1159/000468794. [DOI] [PubMed] [Google Scholar]
- Zigman S. Lens UVA photobiology. J. Ocul. Pharmacol. Ther. 2000;16:161–165. doi: 10.1089/jop.2000.16.161. [DOI] [PubMed] [Google Scholar]