Abstract
Data from eight breast cancer genome sequencing projects identified 25 patients with HER2 somatic mutations in cancers lacking HER2 gene amplification. To determine the phenotype of these mutations, we functionally characterized thirteen HER2 mutations using in vitro kinase assays, protein structure analysis, cell culture and xenograft experiments. Seven of these mutations are activating mutations, including G309A, D769H, D769Y, V777L, P780ins, V842I, and R896C. HER2 in-frame deletion 755-759, which is homologous to EGFR exon 19 in-frame deletions, had a neomorphic phenotype with increased phosphorylation of EGFR or HER3. L755S produced lapatinib resistance, but was not an activating mutation in our experimental systems. All of these mutations were sensitive to the irreversible kinase inhibitor, neratinib. These findings demonstrate that HER2 somatic mutation is an alternative mechanism to activate HER2 in breast cancer and they validate HER2 somatic mutations as drug targets for breast cancer treatment.
Keywords: Genomics, Breast Cancer, Receptor Tyrosine Kinase, Oncogene
INTRODUCTION
Functional characterization of cancer-associated genetic alterations has led to new therapeutic approaches that have dramatically improved patient outcomes. Examples of this include trastuzumab and lapatinib for HER2-gene amplification positive breast cancer, EGFR tyrosine kinase inhibitors for EGFR mutant lung cancers, and imatinib, nilotinib, and dasatinib for BCR-ABL gene fusion in chronic myelogenous leukemias (1). Cancer genome sequencing studies are identifying novel cancer-associated genetic alterations at an unprecedented rate, but the functional consequences of these genetic alterations are known only in a few cases (2).
HER2 gene amplification is a major therapeutic target in breast cancer and it is the clinical criteria for the use of FDA-approved, HER2-targeted drugs, which currently are trastuzumab, pertuzumab, and lapatinib (3). Additional HER2-targeted drugs undergoing clinical trials include neratinib, an irreversible HER2/EGFR tyrosine kinase inhibitor, and trastuzumab-DM1, an antibody-drug conjugate (4). We and others have recently identified HER2 somatic mutations in HER2 gene-amplification negative breast cancer patients through cancer genome sequencing (5-12). If these HER2 somatic mutations are driver events in breast cancer, then these patients may benefit from existing HER2 targeted drugs. In this study, we characterize thirteen HER2 somatic mutations using multiple experimental methods. The sensitivity of these mutations to several HER2 targeted drugs was tested to provide critical preclinical data for HER2 sequencing-directed breast cancer clinical trials.
RESULTS
Identification of HER2 Somatic Mutations in Patients without HER2 Gene Amplification
We identified two breast cancer patients with HER2 somatic mutations by performing next-generation, massively parallel sequencing on DNA samples accrued in the American College of Surgeons Oncology Group (ACOSOG) Z1031 clinical trial (Fig. 1A) (5). These two cancers, code numbers 15687 and 16757, were classified as HER2 negative by the Z1031 trial based on standard clinical tests for HER2 gene amplification, which are HER2 fluorescence in situ hybridization (FISH) and HER2 immunohistochemistry (IHC). To find more breast cancer cases with HER2 somatic mutations, The Cancer Genome Atlas (TCGA) breast cancer project sequencing data was searched and seven additional patients with HER2 somatic mutations were identified (Fig. 1A) (6). Six of these patients were classified as HER2 negative by TCGA and one patient, TCGA-C8-A135, was HER2 positive on the basis of 3+ HER2 IHC score. We searched further for breast cancer patients with HER2 mutations by carefully reviewing six other breast cancer sequencing studies (Supplementary table 1) (7-12). These studies contained 16 more cases of HER2 somatic mutations, thus a total of 25 breast cancer patients with HER2 somatic mutations have been identified to date. Several recurrent mutations were found, including G309A/E, S310F, L755S, in frame deletion of residues 755-759 (abbreviated del.755-759), D769H/Y, and V777L (Fig. 1B). By examining the location of these mutations in the HER2 domain structure (Fig. 1B), we found that these mutations clustered in two major areas, with 20% of patients (5 out of 25) having extracellular domain (ECD) mutations at residues 309-310 and 68% of patients (17 out of 25) having kinase domain mutations between residues 755-781.
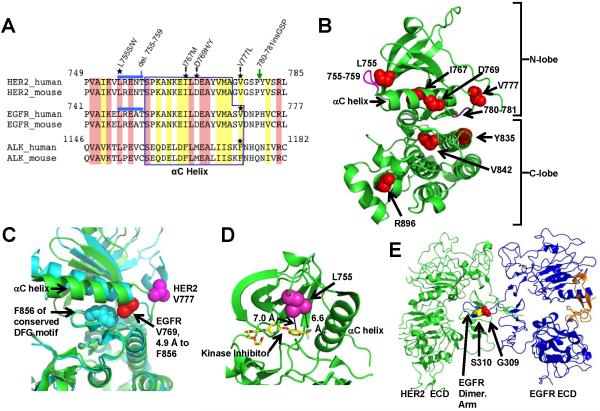
Figure 1.
HER2 somatic mutations in breast cancer. A, Clinical information on patients with HER2 somatic mutations. ER = estrogen receptor, PR = progesterone receptor. B, HER2 somatic mutations observed in 25 patients are shown. Blue circles represent each case of the indicated mutation. Two patients had 2 HER2 somatic mutations each, resulting in a total of 27 mutations in 25 patients. del.755-759* indicates that 2 patients had del.755-759 and one patient had del.755-759 with a S760A change. ECD = Extracellular domain, TM = transmembrane region, JM = juxtamembrane region. C, HER2 gene copy number in patient 15687, who has V777L mutation. X-axis values represent the genomic position on chromosome 17, with numbering based on the March 2006 human reference sequence (NCBI Build 36.1/hg18). D, The exome sequencing-based copy number, as determined by VarScan 2 (45), for the ERBB2 gene and the level 3 gene expression values from the Agilent 244K Custom Gene Expression 4502A-07-03 platform are plotted. The red data points and the arrows to them indicate the patients with HER2 somatic mutations. E, Expression of mutant allele in tumors harboring HER2 mutations by Sanger sequencing of patient mRNA samples.
In order to confirm that these cancers did not have HER2 gene amplification, we checked the read counts from the next-generation DNA sequencing (Fig. 1C-D). Patient 15687 had diploid HER2 gene copy number in her tumor and normal DNA (Fig. 1C). Patient 16757 had focused sequencing of only 25 genes and therefore, the read counts cannot be used to estimate gene copy number here. Read counts for all 507 TCGA patients (Fig. 1D) demonstrated that the majority of cancers clustered at two HER2 gene copies, and a subset of patients had increased HER2 gene copy number along with increased HER2 expression, as measured by the TCGA microarray data (data points in upper right hand corner of Fig. 1D). Of the seven TCGA patients with HER2 somatic mutations, only one patient, TCGA-C8-A135, had evidence for HER2 gene amplification by the sequencing read counts and this correlates well with the 3+ HER2 IHC score obtained in this patient.
In order to determine whether these HER2 mutations were expressed, we searched the next-generation RNA sequencing data (RNA-seq) on TCGA patients and performed RT-PCR and Sanger sequencing on tumor RNA from patients 15687 and 16757 (Fig. 1A and 1E). RNA-seq data was available on six of the TCGA patients and in all six cases, the mutant allele was detected. In 5 cases, the mutant HER2 allele expression accounted for 51% to 76 % of the read counts, demonstrating that this is likely a heterozygous mutation, while in patient TCGA-A8-A06Z, the mutant HER2 allele was only 16% of the total read counts (Fig. 1A). For patients 15687 and 16757, RNA samples from their tumor were obtained and RT-PCR was performed to amplify the region of the HER2 mRNA containing the predicted mutation. These PCR products were subjected to Sanger sequencing and for both cases, expression of both the mutant and WT allele was clearly detectable (Fig 1E). These data unequivocally demonstrate that the HER2 somatic mutations are being expressed at the RNA level.
HER2 Protein Structure Visualization and Comparison of Breast Cancer HER2 Mutations to Previous Described Kinase Mutations
We will first consider the mutations clustering at amino acid (aa) residues 755-781, which is in the tyrosine kinase domain of HER2. These mutations either flank or are within the αC helix of the kinase domain (Fig. 2A-B). The HER2 780-781 insertion mutation has been previously described in non-small cell lung cancer (NSCLC), but the rest of the breast cancer HER2 somatic mutations were distinct from HER2 mutations found in NSCLC (13-15). The HER2 in frame deletion of aa 755-759 (del.755-759) matches the EGFR exon 19 deletions found in NSCLC, which are known activating mutations that confer sensitivity to the EGFR tyrosine kinase inhibitor, gefitinib (16-18).
Figure 2.
Protein structure visualizations of the HER2 somatic mutations. A, Multi-sequence alignment of HER2, EGFR, and ALK tyrosine kinases. Location of somatic mutations is marked with stars. Blue line and green arrow indicate del.755-759 and P780ins, respectively. Identical residues are shaded pink and similar residues yellow. B, HER2 kinase domain structure (PDB: 3PP0) showing the locations of point mutations (red spheres) and deletions/insertions (magenta). C, HER2 V777 and EGFR V769 are visualized in the active conformation of the EGFR kinase domain (cyan, PDB: 2GS2) and the inhibitor bound conformation of HER2 (green, PDB: 3PP0). D, Proximity of the HER2 L755 to the tyrosine kinase inhibitor SYR127063 is shown. SYR127063 has the same core chemical structure as lapatinib (23). E, Alignment of HER2-EGFR extracellular domain structures PDB: 1N8Y (HER2) and PDB: 1IVO (EGFR).
The HER2 V777L mutation is homologous to ALK F1174L and EGFR V769L mutations (Fig 2A, mutations indicated by a star on the multi-sequence alignment). ALK F1174L is a known activating mutation, found in neuroblastoma (19-21). EGFR V769L is a rare NSCLC-associated mutation, but to our knowledge, no functional studies on it have yet been reported (22). HER2 V777 and EGFR V769 residues were visualized by superimposing crystal structures of the active conformation of EGFR kinase domain with an inhibitor bound conformation of HER2 kinase domain (Fig. 2C). There are only two crystal structures of HER2 kinase domain available, and both contain kinase inhibitors and represent an identical, inhibitor-bound conformation (23, 24). In contrast, at least 40 crystal structures on the EGFR kinase domain are available and these represent the active, inactive and inhibitor-bound conformations (25). In order to understand the various conformations that HER2 kinase domain is likely to adopt in vivo, we utilized EGFR kinase domain active conformation crystal structure and the HER2 inhibitor-bound crystal structure (Fig. 2C). In the EGFR active conformation, the V769 side chain abuts the F856 side chain (distance 4.9 Å), which is part of the conserved DFG motif. Similarly, ALK F1174 is known to pack against F1271 of ALK’s DFG motif, matching the EGFR crystal structure (26). Since the Asp residue in the DFG motif is part of the kinase catalytic site and the Phe residue in DFG forms part of the regulatory spine of the EGFR kinase domain (25), we infer that EGFR V769L and HER2 V777L mutations will likely affect tyrosine kinase activity.
The HER2 L755S mutation is a known lapatinib resistance mutation and it was initially identified from an in vitro mutagenesis screen (27, 28). The HER2 L755 side chain is close proximity to the binding site for small molecule kinase inhibitors (Fig. 2D), thus explaining how mutations at this residue could produce drug resistance. The remaining mutations in the kinase domain (Y835F, V842I, and R896C) occur in the C-lobe (Fig. 2B) and each of these mutations was found only in single patient to date. A striking finding is that no activation loop (A-loop) mutations are found in these breast cancer patients. A-loop mutations are a major group of kinase domain mutations found in cancer, with prominent examples including EGFR L858R, BRAF V600E, and ALK R1275Q (16-18, 26, 29). The absence of HER2 A-loop mutations in breast cancer underscores the differences between the mutation spectrum seen here versus in other cancer types.
Mutations in the extracellular domain (ECD) of HER2 are found at aa 309 and 310 (Fig 1B and 2E). By modeling the ECD’s of a HER2-EGFR heterodimer, it is clear that HER2 G309 and S310 contact the dimerization arm of EGFR. HER2 G309 abuts the EGFR M253 side chain with a distance of 4.1 Å from the G309 α-carbon to the terminal carbon of the M253 side chain (not shown). Mutations at these residues are also found in NSCLC and a recent publication demonstrated that HER2 G309E and S310F are activating mutations when introduced into NIH 3T3 fibroblasts, AALE human airway epithelial cells, and Ba/F3 murine lymphoid cell line (30).
Effect on Tyrosine Kinase Activity and Cellular Signaling
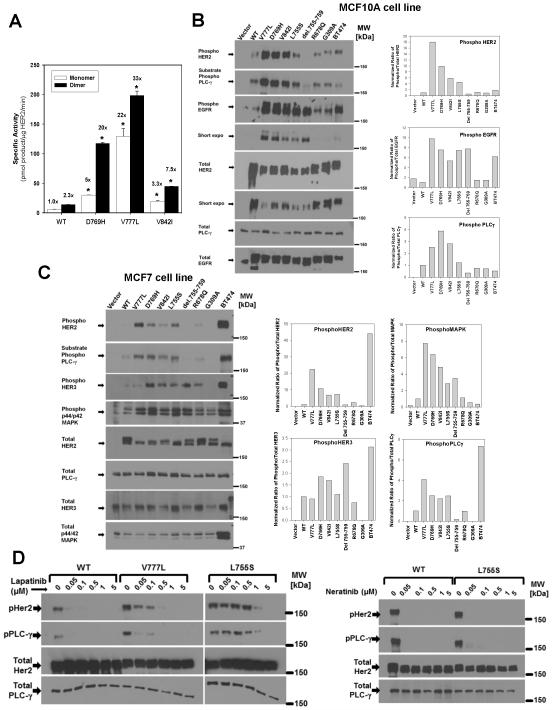
In vitro kinase assays were performed on the HER2 kinase domain mutations using recombinant expression of the isolated HER2 kinase domain (31). In this system, the activity of monomeric or dimeric HER2 can be compared (31, 32). WT HER2 showed a 2.3-fold increase in kinase activity upon in vitro dimerization (Fig. 3A). All three mutations tested here, HER2 V777L, D769H, and V842I, had greater tyrosine kinase specific activity than WT, with further increases seen with in vitro dimer formation. While the V842I had the lowest activity of these three mutations, it did show a statistically significant increase over WT HER2. The L755S and del.755-759 mutations were poorly expressed and could not be assayed in this system.
Figure 3.
HER2 mutations differentially activate HER2 signaling. A, HER2 WT or mutant kinase domain constructs were recombinantly expressed and assayed in vitro. Fold change relative to HER2 WT monomer is indicated above each bar. * = p<0.01. B, MCF10A cells were retrovirally transduced with HER2 WT or the respective mutants and lysates were probed with the indicated antibodies. Bar graphs represents quantification of western blots bands, which was performed with Bio-Rad ChemiDoc XRS system or ImageJ software. C, MCF7 cells were retrovirally transduced with HER2 WT or the respective mutants and lysates were probed and quantified as in B. D, MCF10A cells expressing HER2 WT, V777L, or L755S were treated with lapatinib (left) or neratinib (right) at the indicated concentrations for 4 hours, and analyzed by western blot as above.
To assess the effect of HER2 mutations on signal transduction pathways, retroviral vectors were used to transduce mutant or WT HER2 into MCF10A (Fig. 3B), MCF7 (Fig. 3C), and NIH 3T3 (Supplementary Fig.1) cells. MCF10A cells are a non-transformed breast epithelial cell line which express endogenous EGFR and require supplementary EGF in their media (33, 34). MCF10A cells are widely used as a model for oncogenic transformation and tumorigenesis in breast cancer (33, 35). MCF7 cells are an estrogen receptor positive breast cancer cell line and constitute a model for hormone receptor positive breast cancer. MCF7 cells have high endogenous HER3, but low levels of endogenous EGFR and HER2 (34). NIH 3T3 cells are a fibroblast cell line that has been extensively used to study oncogenes and they lack or have very low levels of endogenous EGFR, HER2, and HER3 (36, 37). Using these cell lines, we measured HER2 autophosphorylation, and phosphorylation of HER2’s dimerization partners (EGFR or HER3) and downstream signaling proteins (phospholipase γ C1, PLCγ and MAP kinase). PLCγ is a well-characterized HER2 and EGFR substrate protein and provides a direct measurement of HER2 and EGFR substrate phosphorylation (37).
The V777L, D769H, and V842I mutations strongly increased phosphorylation of all signaling proteins in all three cell lines, indicating that these are activating mutations (Fig. 3B-C and Supplementary Fig. 1). D769H had a particularly strong effect on HER3 phosphorylation in MCF7 cells (Fig. 3C). HER2 L755S increased HER2 and EGFR phosphorylation in MCF10A cells (Fig. 3B) and HER2, PLCγ and MAP kinase in MCF7 cells (Fig. 3C), but it did not increase HER2 or PLCγ phosphorylation in NIH 3T3 cells (Supplementary Fig. 1). The del.755-759 mutation showed a marked increase in EGFR phosphorylation in MCF10A cells and HER3 and MAP kinase in MCF7 cells, but did not increase HER2 and PLCγ phosphorylation in any of the three cell lines (Fig. 3B-C and Supplementary Fig. 1). This suggests that HER2 del.755-759 may interact differently with its dimerization partners, EGFR and HER3, and have a greater dependence on them than the other HER2 mutants do. In all three cell lines, R678Q and G309A showed little effect on protein phosphorylation. To ensure that the mutant HER2 proteins are being expressed on the cell surface, we performed flow cytometry on the retrovirally transduced MCF10A cells and observed that the cell surface HER2 expression correlated well with the total HER2 levels measured by western blot (Supplementary Fig. 2 and Fig. 3B). Consistent with prior reports on a NSCLC-associated HER2 mutation (30), the HER2 mutations that strongly activate signaling showed lower amount of total HER2 as compared to WT (Fig 3B, total HER2 blot with shorter exposure).
The HER2 L755S was previously shown to produce lapatinib resistance in mammalian cells (27, 28), and we therefore tested the lapatinib sensitivity of MCF10A-HER2 V777L and L755S cells. MCF10A-HER2 V777L cells were sensitive to lapatinib with reduced HER2 autophosphorylation seen at 50nM and complete inhibition observed with 500nM lapatinib (Fig. 3D). In contrast, L755S required doses of >1μm lapatinib to inhibit HER2 signaling, but it was sensitive to the irreversible tyrosine kinase inhibitors, neratinib (Fig. 3D) and canertinib (CI-1033, Supplementary Fig. 3). Complete inhibition of signaling by HER2 L755S was observed even at the lowest dose of neratinib used here (50nM).
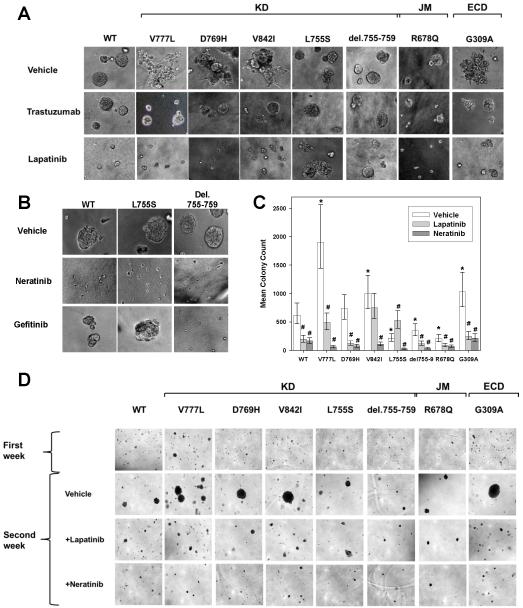
Effect on Cell Growth in Matrigel and Soft Agar
To assess growth effects in three dimensional cultures, MCF10A cells transduced with mutant or WT HER2 were seeded into Matrigel. As previously reported (35), MCF10A-HER2 WT cells formed spherical structures (Fig. 4A, upper left). MCF10A cells bearing HER2 V777L, D769H, V842I, or G309A mutations formed irregular structures with spiculated, invading protrusions (Fig. 4A, top row). Addition of the HER2 antibody trastuzumab restored the spherical morphology to these mutations (Fig. 4A, middle row). HER2 L755S, del.755-759, and R678Q formed spherical structures, similar to WT HER2. MCF10A cells are an EGF-dependent cell line and addition of lapatinib, a dual HER2/EGFR inhibitor, blocked the growth of MCF10A-HER2 WT cells (Fig. 4A, lower row). Most of the HER2 mutant cells were also growth inhibited by lapatinib, but L755S and del.755-759 were resistant to lapatinib and continued to form large spherical structures. L755S and del.755-759 were sensitive to the irreversible inhibitor, neratinib (Fig. 4B). The del.755-759 mutation was also sensitive to gefitinib, consistent with the idea that this mutation is more dependent upon a heterodimerization partner, such as EGFR, for its functional effects (Fig. 4B, 3B, and 3C).
Figure 4.
HER2 mutations V777L, D769H, V842I, G309A induce gain of function over HER2 WT in MCF10A mammary epithelial cells. A, HER2 WT or mutants were seeded on 3D Matrigel culture in the presence or absence of DMSO vehicle (0.5%), trastuzumab (100 μg/ml) or lapatinib (0.5 μM). Phase contrast images of acini or invasive structures were obtained at 200x magnification on day 8. KD = kinase domain, JM = juxtamembrane region, and ECD = extracellular domain. B, HER2 WT, L755S, and del.755-759 cells were grown in Matrigel in the presence of DMSO vehicle (0.5%), neratinib (0.5 μM) or gefitinib (0.5 μM). Phase contrast images were obtained as in A. C, MCF10A-HER2 WT or mutants were seeded in soft agar. After 7 days of growth, they were treated with DMSO vehicle (0.5%), lapatinib (0.5μM) or neratinib (0.5μM) for an additional week. Error bars represent 95% highest posterior density intervals. * indicates a significant difference between the HER2 mutant and HER2 WT and # indicates the effect of inhibitor treatment was significant (95% highest posterior density interval did not contain 0 for both). D, Photomicrographs of the colonies in soft agar on day 12, magnification 40x.
The above experiment (Fig. 4A-B) was performed by seeding the cells in the presence or absence of drugs. In order to mimic the situation with established tumors, we also performed drug treatment of MCF10A-HER2 cells after allowing them to form invasive colonies (Supplementary Fig. 4). Treatment with lapatinib and neratinib inhibited the growth of these invasive colonies. We also tested the ability of these HER2 mutations to promote the anchorage independent growth of MCF10A cells by performing soft agar colony assays and quantifying the number of colonies using image analysis software (Supplementary Fig. 5 and Fig.4C). V777L, V842I, and G309A increased the number of colonies formed in soft agar as compared to WT (Fig. 4C). While D769H did not produce a statistically significant increase in number of colonies, it did result in the formation of larger colonies, as seen in the photomicrographs of the soft agar colonies (Fig. 4D). MCF10A cells transduced with L755S, del.755-759, and R678Q formed fewer colonies than HER2 WT cells. Lapatinib treatment decreased the number of colonies formed in all mutations, except for L755S. In fact, a small but statistically significant increase in colony number was seen with lapatinib treatment of L755S as compared to vehicle (Supplementary Fig. 5), which may be due to persistent EGFR signaling in the presence of a lapatinib resistant form of HER2. In contrast, neratinib produced a strong inhibition of colony formation for all mutations, including L755S (Fig. 4C-D).
Effect on Tumor Growth in Xenograft Models
The ability of the HER2 mutations to increase tumor formation in xenografts was tested (Fig. 5). NIH3T3-HER2 WT cells served as the control cell line. NIH3T3-HER2 V777L, D769H, and G309A mutant cell lines had more rapid tumor growth than the HER2 WT control (p-values all <0.01). The L755S, V842I, and R678Q cells showed tumor growth indistinguishable from the HER2 WT control cells (Fig. 5A and B). The NIH3T3-HER2 del.755-759 cells formed tumors slower than NIH3T3-HER2 WT cells (Fig. 5C and D). NIH3T3 cells have low levels of endogenous EGFR and HER3 (36, 37) and since the del.755-759 mutation seems to be more dependent on a heterodimer pattern like EGFR, the tumor formation ability of del.755-759 may be decreased in NIH3T3 cells. Similarly, MCF7-HER2 V777L cells formed tumors more rapidly in nude mice than MCF7-HER2 WT cells (Supplementary Fig. 6), indicating that HER2 mutations can increase tumor formation in this cell line as well. The strong effect of HER2 G309A seen in the tissue culture and xenograft experiments (Fig 4. and 5) was surprising given the small effect of this mutation on HER2 phosphorylation (Fig. 3B-C). However, Greulich et al. recently showed that HER2 G309E can increase HER2 dimerization without increasing HER2 phosphorylation (30).
Figure 5.
HER2 mutants V777L, G309A, D769H formed tumors more rapidly than HER2 WT. A, 0.5×106 cells of NIH3T3 expressing either HER2 WT or mutant were injected in the flanks of nude mice. N=5 mice for each construct. Tumor size was measured every 2-3 days. A linear mixed effects model was fit to the logarithm of tumor volumes to statistically test whether HER2 mutants grew faster than HER2 WT. * = p<0.01. B, Photographs were taken on day 11. C, NIH3T3 cells expressing HER2 WT, G309A, or del.755-759 were injected into nude mice as in a. N=3 mice for G309A and WT and N=4 mice for del.755-759. * = p<0.01. D, Photographs taken on day 12.
Inhibition of Cell Growth by Neratinib and Lapatinib
Given the effectiveness of neratinib and lapatinib seen above, we measured the IC50 values of these two HER2/EGFR tyrosine kinase inhibitors on MCF10A-HER2 cells (Table 1). BT474 cells, a HER2 gene amplified cell line, served as the positive control and the IC50 value of <2nM with neratinib and 31 nM with lapatinib match previously published values (38, 39). MCF7 cells served as the negative control and were not sensitive to either inhibitor. MCF10A cells bearing mutant or WT HER2 were potently growth inhibited by neratinib (Table 1). In contrast, lapatinib doses of 400 – 1000 nM were required to inhibit growth of most of these mutations. The MCF10A-HER2 L755S cells were resistant to lapatinib (IC50 > 10 μM) but could be readily inhibited by neratinib (IC50 of 15 nM). Neratinib is an irreversible inhibitor of EGFR and HER2 and it has been used to treat gefitinib-resistant lung cancers that contain the EGFR T790M mutation (40).
TABLE 1.
Inhibition of Cell Growth by Neratinib and Lapatinib. Cells were incubated with drugs for 6 days and cell viability and number was measured using Alamar Blue.
| IC50 (nM) | ||
|---|---|---|
| Neratinib | Lapatinib | |
| MCF10A - HER2 WT | <2 | 400 ± 60 |
| G309A | <2 | 470 ± 50 |
| V777L | <2 | 1,040 ± 570 |
| D769H | <2 | 980 ± 950 |
| V842I | <2 | 650 ± 210 |
| Del755-759 | 2.1 ± 0.2 | 660 ± 90 |
| L755S | 15 ± 6 | > 10,000 |
|
| ||
| BT474 cells | <2 | 31 ± 2 |
| MCF7 cells | > 3,000 | > 10,000 |
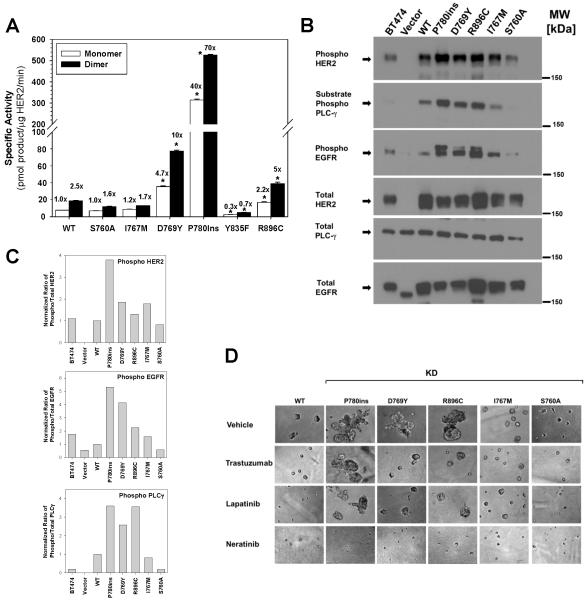
Characterization of Six Additional HER2 Somatic Mutations
In June 2012, additional breast cancer sequencing studies were published and several new HER2 somatic mutations were identified (Supplementary Table 1) (10-12). Therefore, we tested six more HER2 somatic mutations using the above assays. D769Y, P780-Y781insertionGSP (abbreviated P780ins), and R896C increased the in vitro kinase activity of HER2 (Fig. 6A). All six mutations were introduced into MCF10A cells using retroviral vectors. HER2 Y835F showed decreased in vitro kinase activity and cells bearing this mutation showed no indication for oncogenic transformation and grew slower than HER2 WT cells. Therefore, this mutation was not analyzed further. The D769Y, P780ins, and R896C mutations increased HER2, EGFR, and PLCγ phosphorylation in MCF10A cells (Fig. 6B-C) and these mutations produced colonies with irregular structures and spiculated, invading protrusions in 3D Matrigel culture (Fig. 6D). D769Y and R896C mutations were sensitive to trastuzumab and lapatinib, but the growth and invasive appearance of cells bearing the P780ins mutation was not affected by either of these drugs. In contrast, neratinib inhibited the growth of all the mutations tested here (Fig. 6D).
Figure 6.
HER2 kinase domain mutations differentially activate HER2 signaling, induce an invasive phenotype and respond to HER2-targeted drugs. A, Six additional HER2 kinase domain mutations were tested by in vitro kinase assay. Fold change relative to HER2 WT monomer is indicated above each bar. * = p<0.01. B, MCF10A cells were retrovirally transduced with HER2 WT or the respective mutants and lysates were probed with the indicated antibodies. C, Quantitation of western blots from panel B. D, HER2 WT or mutants were seeded on 3D Matrigel culture in the presence of DMSO vehicle (0.5%), trastuzumab (100 μg/ml), lapatinib (0.5 μM), or neratinib (0.5μM). Phase contrast images of acini or invasive structures were obtained at 200x magnification on day 6.
Structural visualizations of these mutations show that P780ins, D769Y, I767M, and S760A flank or occur in the αC helix (Fig 2A-B). The S760A mutation is not an independent mutation and in fact, it occurs as part of the del.755-759 mutation, thus it is not surprising that S760A has no effect on its own. The finding of strong activation by D769Y is very revealing. When combined with the results on D769H shown above (Fig. 3-5), we can infer that either the loss of the acidic side chain or the addition of an aromatic ring (either Y or H) at aa 769 must activate the HER2 tyrosine kinase. Visualization of a HER2-HER3 kinase domain dimer (Supplementary Fig. 7) shows several hydrophobic residues in close proximity to HER2 D769. These residues include HER2 Y772, and HER3 residues I919 and M923 (HER3 numbering as per PDB: 3KEX). We postulate that the D769H/Y mutations may increase hydrophobic contacts in this area, which would thereby increase HER2 dimerization.
DISCUSSION
In this study, we functionally characterized thirteen HER2 somatic mutations and we can group them into several different phenotypes. Seven of these thirteen are activating mutations, including G309A, D769H, D769Y, V777L, P780ins, V842I, and R896C. When we combine these results with a recent functional study on the HER2 G309E and S310F mutations (30), we estimate that 13 out of the 25 patients identified here have activating HER2 mutations that are likely driver events in their cancer. One mutation, L755S, showed a lapatinib resistant phenotype, but it did not cause oncogenic transformation. Since 6 out of the 25 patients (about 25%) have L755S somatic mutations, we anticipate that these cases will be lapatinib resistant, but may respond to neratinib. One mutation, del. 755-759, had a neomorphic phenotype, with increased phosphorylation of HER2’s heterodimerization partners, EGFR and HER3. This somatic mutation was found in 3 of the 25 patients and its clinical effect on these breast cancer cases will need to be determined. HER2 del.755-759 is homologous to EGFR exon 19 deletions, which produce gefitinib sensitive NSCLC, and therefore, it is likely that this HER2 mutation is also influencing breast cancer biology. Three mutations (R678Q, I767M, and Y835F) showed no functional effect in our assays and these mutations are found in two patients (the R678Q mutation co-occurred with L755W in patient TCGA-A2-A0T6). Two mutations (L755W and G1201V) have not been functional tested yet, and we anticipate that with further HER2 gene sequencing, the number of mutations that require characterization will grow.
Can we predict a priori which of these HER2 mutations will have a functional effect? These results suggest that recurrence is a good predictor of functional effect, since all of the recurrent mutations examined here had an effect on HER2 function and cell behavior. However, some of the mutations that were just found in one patient, such as P780ins, V842I, and R896C, exhibited functional effects as well. Further, recurrence did not predict the phenotype of the mutation (activating, drug resistant, or neomorphic). Therefore, the presence of recurrence in these HER2 mutations tends to predict a functional effect, but lack of recurrence cannot be used to rule-out an effect by the mutation. Another criterion that can be considered is whether the mutation is a conservative amino acid substitution. Amongst this panel of HER2 mutations, we find that whether the amino acid change is a conservative substitution does not predict whether the mutation will have a functional effect. Val to Leu (V777L), Gly to Ala (G309A), and Val to Ile (V842I) were all activating mutations, while Tyr to Phe (Y835F), Ile to Met (I767M), and Ser to Ala (S760A) all had no functional effect. It appears that protein structure and conformation makes a large contribution to the functional effect of the mutation, with conservative substitutions occurring in critical locations for the protein (such as V777L in the proximity of the kinase domain DFG motif, and G309A in the ECD dimerization arm contact surface) having a large effect on HER2. We acknowledge that these inferences are based only on the HER2 results obtained here, and are not necessarily generalizable.
Several HER2 targeted drugs were tested on these mutations. We observed that neratinib was a very potent inhibitor for all of the HER2 mutations. Lapatinib was less potent and was not effective in reducing the growth of cells bearing L755S or P780ins mutation. Trastuzumab did reduce the formation of spiculated, invasive appearing colonies in the Matrigel assays on MCF10A cells, but there did not appear to be a cytotoxic effect from this drug. Trastuzumab has a complex mechanism of action, which includes activation of the immune system (41), and we cannot adequately model this in our tissue culture assays. The effect of pertuzumab was not tested here, but HER2 residues 309-310 are known to be part of the pertuzumab binding epitope (9, 42), suggesting that pertuzumab binding could be affected by G309A/E and S310F mutations.
In order to estimate the prevalence of these HER2 mutations in breast cancer, we reviewed the breast cancer sequencing studies available to date (Fig. 1A and Supplementary table 1) (5-12). The 25 patients with HER2 mutation positive breast cancer were identified from a total of 1,499 patients. This suggests an overall HER2 mutation rate of approximately 1.6%. Given that the annual incidence of breast cancer in the U.S. was 232,620 cases in 2011 (43), the number of HER2 mutation positive patients may be approximately 4,000 patients annually. This is on par with the annual U.S. incidence of chronic myelogenous leukemia (5,150 patients annually) (43), a disease for which extensive drug development and clinical trials have been performed. Lobular breast cancer may have an increased frequency of HER2 somatic mutations, but the numbers of cases sequenced to date is small (3 lobular breast cancer patients with HER2 somatic mutation among 39 lobular breast cases in the TCGA study and 3 patients with HER2 mutations among 113 lobular cases in Shah et al.) (6, 8). Furthermore, the breast cancer genome sequencing studies cited here have focused on newly diagnosed patients. The HER2 mutation frequency in relapsed or metastatic breast cancer patients is currently unknown and potentially could be higher than 1.6%. Because of the low mutation rate, prospective clinical trials utilizing HER2 gene sequencing results will need to screen a large number of patients and the cooperation of many academic institutions and treatment centers is essential.
Potential limitations of this study include the fact that retroviral transduction of HER2 mutations does produce HER2 protein overexpression. We compared the level of HER2 expression in the virally transduced cells to BT474 cells, which have HER2 gene amplification, in order to ensure that our experiments were performed under HER2 levels that are appropriate for breast cancer studies. Further, all experiments compared the effect of the mutant allele to WT HER2 to determine if there was an effect of the mutation, above and beyond that seen with increased HER2 expression alone. A second issue is that this study does not address whether HER2 mutations occur in HER2 gene amplified patients. The majority of breast cancer patients sequenced to date are HER2 gene amplification negative (Fig. 1A and Supplementary table 1), and no conclusions can be drawn about the HER2 gene amplification positive patients (5-12).
In conclusion, these data demonstrate that HER2 somatic mutations are an alternative mechanism to activate HER2 in breast cancer. Most of these HER2 mutations are likely driver events in the breast cancer and this suggests that HER2 mutation positive patients constitute a breast cancer subpopulation that will likely benefit from HER2-targeted drugs, particularly irreversible inhibitors such as neratinib. Functional testing of new HER2 mutations should be considered in clinical trial design or when making patient care decisions. A prospective, multi-institutional clinical trial is being launched to screen stage IV breast cancer patients for HER2 somatic mutation and determine the clinical outcome of treating them with HER2-targeted drugs.
METHODS
Cell lines
MCF10A and NIH3T3 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA). MCF7 cells were a gift from Dr. Saraswati Sukumar (Johns Hopkins School of Medicine) and were received from her lab in 2003. The MCF10A and MCF7 cells lines were authenticated by short tandem repeat (STR) profiling performed by ATCC on Oct. 15, 2012. The NIH3T3 cells were passaged for fewer than 6 months after receipt and were not reauthenticated. Culture medium for these cell lines was as follows: MCF10A cells - DMEM/F12 supplemented with 5% horse serum, 20ng/ml epidermal growth factor (EGF), 10μg/ml insulin, 0.5μg/ml hydrocortisone, and 1% penicillin/streptomycin (P/S) as per (33), MCF7 cells - RPMI supplemented with 10% fetal bovine serum, 10mM Hepes, 10mM sodium pyruvate, 2mM Glutamine and 1% P/S, and NIH-3T3 cells – DMEM high glucose supplemented with 10% bovine calf serum and 1% P/S. Inhibition of cell growth by lapatinib and neratinib was measured by Alamar Blue, as per (44) and IC50 values calculated by a 4 parameter nonlinear regression performed using SigmaPlot 2001 software (Systat Software, Inc., San Jose, CA).
Antibodies and Inhibitors
Antibodies were purchased from Cell Signaling Technologies: phospho PLCγ (Tyr783), PLCγ phospho HER3 (Y1289), phospho EGFR (Tyr1173), EGFR, phospho p44/42 MAPK (Thr202/Tyr204) and p44/42 MAPK. Phospho HER2 (pY1248) and HER3 antibody was from Millipore. HER2 monoclonal antibody (clone is from Thermo Fisher. Trastuzumab was obtained from the Barnes-Jewish Hospital pharmacy. Lapatinib and canertinib were obtained from Axon Medchem (Groningen, The Netherlands). Neratinib was obtained from Selleckchem (Houston, TX).
ERBB2 gene copy and expression level in TCGA Breast Cancer project data
Level 3 gene expression values from the Agilent 244K Custom Gene Expression 4502A-07-03 platform were downloaded from the TCGA data portal on February 2, 2012. Exome-based copy number values for the ERBB2 gene were determined for 515 TCGA breast carcinoma tumor-normal pairs using exome data (available at dbGAP) and the VarScan 2 algorithm (45). The plot for supplementary figure 1 was generated with the R statistical environment, version 2.13.1 (2011-07-08).
RT-PCR and Sanger sequencing of patient RNA
Total RNA was isolated and DNAse treated as described in (5). SuperScript III First-Strand Synthesis System (Invitrogen) with oligo(dT)20 was used to reverse transcribe 200 ng RNA according to the manufacturer’s recommendations. The HER2 cDNA fragment surrounding the mutation site was amplified using Phusion High-Fidelity DNA Polymerase (New England BioLabs) and the following primers: Forward: CAGAAGATCCGGAAGTACAC, Reverse: ATACACCAGTTCAGCAGGTC. Agarose gel electrophoresis confirmed PCR product of expected size and the lack of amplification in the no reverse transcription and no template controls. The amplicons were gel purified and submitted for Sanger sequencing.
In vitro kinase assays and protein structure visualization
HER2 somatic mutations were introduced into the HER2 kinase domain using the QuikChange II kit (Agilient, Santa Clara, CA) and the desired constructs were verified by Sanger sequencing. His6-tagged HER2 WT and mutant kinase domain constructs were expressed using baculovirus vectors as per the Bac-to-Bac system (Life Technologies, Carlsbad, CA). Recombinant HER2 kinase domain proteins were purified to 70-80% purity using Ni-NTA-agarose beads, as per (31). Radiometric kinase assays were performed either with kinases in solution (monomers) or attached to the surface of a liposomes to induce in vitro dimerization, as detailed in our prior publication (31). Statistical analysis of the in vitro kinase assay was performed with Student’s t-test. Protein structural alignments and visualizations were performed using PyMol (DeLano Scientific, San Carlos, CA).
Cloning and retroviral transduction of mutant and WT HER2 into cell lines
HER2 somatic mutations were introduced into the pcDNA3 vector bearing the HER2 cDNA using the QuikChange II kit (Agilient, Santa Clara, CA). These constructs were then shuttled into the pCFG5 retroviral vector, which contains a zeocin resistance marker and an IRES-GFP sequence, using the In-Fusion HD cloning system kit (Clontech, Mountain View, CA). The desired constructs were verified using Sanger sequencing. Retroviral particles were produced using φNX amphotrophic packaging cell line and cell lines were infected as per prior publications (33, 46, 47). Cell lines were subjected to 1-2 weeks of zeocin selection and transgene expression was verified by flow cytometry analysis for GFP expression (always >95%, data not shown) and western blot analysis for HER2. HER2 cell surface expression was measured by flow cytometry using a 1:20 dilution of anti-human HER2-APC (BioLegend, clone 24D2). Data were collected at medium sheath pressure on a BD FACSCalibur custom modified by Cytek (Fremont, CA) to contain a second excitation laser (630 nm).
Three-dimensional culture in Matrigel
Overlay three-dimensional culture of MCF-10A cells in Matrigel was performed as described previously (33). Briefly, 4×103 MCF10A cells expressing mutant or WT HER2 WT were seeded in Growth Factor Reduced Matrigel (BD Biosciences, San Jose, CA) in 8-well chamber slides. Lapatinib, neratinib, trastuzumab, or DMSO vehicle was added at time of seeding or on day 4. Media was changed every 4 days and photomicrographs were obtained on day 5-8.
Soft agar colony forming assay
Six-well plates were first layered with 0.6% bacto agar in MCF10A growth medium. After the solidification of bottom layer, a top layer containing 5-10 × 103 MCF10A cells per well of HER2 WT or mutants were suspended in MCF10A growth media containing 0.4% bacto agar was added as per (48). Assays were carried out in duplicates. Cells were allowed to form colonies for 7 days and photographed. Subsequently, lapatinib (0.5μM), neratinib (0.5μM), or DMSO vehicle (0.5%) was added to the cultures and they were incubated for another week. Plates were stained with crystal violet (Sigma-Aldrich) for 1 hour and photographed with a BioRad ChemiDoc XRS system. Colony number and size were determined using ImageJ version 1.46r. Every plate was processed simultaneously by adding each plate image into a single stack and converted to an 8-bit image. Background subtraction with a rolling ball with a 10 pixel radius was found to remove most background while preserving colony number. A circular ROI was formed over the first lapatinib-treated HER2 WT plate and used as input to enhance contrast assuming 0.001% saturated pixels to normalize the image stack. The image stack was thresholded to 180 and converted to a binary image. Colonies were split using watershed processing with default parameters. Each well was then pasted into slices of a new image stack and major artifacts manually removed by comparison with the original images. The “analyze particles” command was used to collect the number and areas of the processed colonies using default parameters of unrestricted particle size and circularity.
Statistical analysis of the colony count data were modeled using Poisson regression using the MCMCglmm package (version 2.16) (49), of the R statistical environment (version 2.15.1) using “Genotype” and a “Genotype:Treatment” interaction as fixed predictors. No intercept was included to force explicit measurements for each “Genotype”. MCMCglmm uses fully Bayesian modeling and parameter estimates generated using Gibbs sampling Markov Chain Monte Carlo with 100,000 iterations, a burn-in of 3000, and a thin of 10. Diagnostics revealed the lack of autocorrelation and excellent chain mixing. The default prior was used for fixed effects, which is a multivariate normal distribution with a 0 mean vector and diagonal variance matrix with variances of 10^10 and covariances of 0, as this ensures fixed effects are independent and estimated almost entirely from the data. Overdispersion and replicates were accounted for in the residual variance structure with an improper inverse-Wishart prior with nu=0 and V=1, which implicitly assumes each well is a random effect. Parameters and parameter contrasts were considered to be statistically significant when the 95% highest posterior density interval did not contain 0. The conclusions were robust to changes in the minimal colony size from 1 to 10 (the global median colony size).
Xenograft studies
Exponentially growing, retroviral transduced NIH3T3 cells were trypsinized, resuspended in serum-free medium, and 0.5×106 cells mixed with 1:2 ratio (v/v) with growth factor reduced Matrigel. For MCF7 cells, the 5-10×106 cells were similarly prepared and mixed with 1:2 ratio (v/v) with growth factor reduced Matrigel. Cell suspensions in 0.1 ml were injected subcutaneously into 8-week-old female nude mice (Jackson Laboratory, Bar Harbor, ME). Tumor volume was calculated as π/6 × (width)2 × length using digital calipers. These experiments were performed under institutional guidelines and with IACUC approval in accordance with the Washington University Animal Studies Committee.
Statistical analysis of xenograft studies was performed using a linear mixed effects model to model the tumor growth curves. The logarithm of tumor volumes increased approximately linearly with time over the measurement period and the intercept but not slope of this relationship varied within injection replicates for each HER2 genotype. Therefore, the linear mixed effects model used random intercepts for injection replicates and fixed effects for time and a time-by-genotype interaction. This was specified as lmer(log(Volume) ~ Days + Days:Genotype + (1|Injection)) using the lme4 package (50) in the R statistical environment version 2.13.2. Two-tailed p-values of the time-by-genotype interaction for HER2 mutants compared to HER2 WT were generated using 100,000 Markov Chain Monte Carlo simulations as implemented in lme4 (50). The conclusions were robust to several alternative model specifications, and all modeling assumptions were confirmed to be reasonable on diagnostic residual plots. P-values < 0.05 were considered to be statistically significant.
Supplementary Material
SIGNIFICANCE.
We demonstrate that the majority of HER2 somatic mutations in breast cancer patients are activating mutations that likely drive tumorigenesis. Several patients had mutations that are resistant to the reversible HER2 inhibitor lapatinib, but are sensitive to the irreversible HER2 inhibitor, neratinib. Our results suggest that patients with HER2 mutation positive breast cancers could benefit from existing HER2 targeted drugs.
ACKNOWLEDGEMENTS
We thank The Cancer Genome Atlas Network for data access, Martin Leverkus for providing the pCFG5 retroviral expression vector, Linda Pike and Timothy Collier for several helpful discussions, and Tom Kitchens and Maureen Highkin for technical assistance. We thank Charles Perou, Paul Goodfellow, Martin Leverkus, Linda Pike, and Timothy Ley for critical reading of this manuscript.
Financial support for this work was provided by NIH grants R01CA095614 and U01HG00651701 (to M.J.E), and U54HG003079 (to E.R.M. and Richard K. Wilson at the Washington University Genome Institute), the Edward Mallinckrodt, Jr. Foundation and the ‘Ohana Breast Cancer Research Fund (to R.B.), and the Foundation for Barnes-Jewish Hospital/ Siteman Cancer Center Cancer Frontier Fund (to C.X.M, R.B., and M.J.E).
List of Abbreviations
- ACOSOG
American College of Surgeons Oncology Group
- ECD
extracellular domain
- EGFR
epidermal growth factor receptor
- ER
estrogen receptor
- JM
juxtamembrane
- KD
kinase domain
- MAP kinase
mitogen-activated protein kinase
- NSCLC
non-small cell lung cancer
- PLCγ
phospholipase Cγ
- PR
progesterone receptor
- TCGA
The Cancer Genome Atlas
Footnotes
The authors disclose no potential conflicts of interest.
REFERENCES
- 1.Baselga J. Targeting tyrosine kinases in cancer: the second wave. Science. 2006;312:1175–8. doi: 10.1126/science.1125951. [DOI] [PubMed] [Google Scholar]
- 2.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 3.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 4.Awada A, Bozovic-Spasojevic I, Chow L. New therapies in HER2-positive breast cancer: a major step towards a cure of the disease? Cancer Treat Rev. 2012;38:494–504. doi: 10.1016/j.ctrv.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, et al. Whole Genome Sequencing to Characterise Breast Cancer Response to Aromatase Inhibition. Nature. 2012;486:353–60. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Network T. Comprehensive molecular portraits of human breast tumors. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JW, Soung YH, Seo SH, Kim SY, Park CH, Wang YP, et al. Somatic mutations of ERBB2 kinase domain in gastric, colorectal, and breast carcinomas. Clin Cancer Res. 2006;12:57–61. doi: 10.1158/1078-0432.CCR-05-0976. [DOI] [PubMed] [Google Scholar]
- 8.Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–13. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 9.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–73. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 10.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–9. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens P, Hunter C, Bignell G, Edkins S, Davies H, Teague J, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–6. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 14.Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–6. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 15.Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 16.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 17.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 18.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–4. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 20.George RE, Sanda T, Hanna M, Frohling S, Luther W, 2nd, Zhang J, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–8. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–70. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 22.Asahina H, Yamazaki K, Kinoshita I, Yokouchi H, Dosaka-Akita H, Nishimura M. Non-responsiveness to gefitinib in a patient with lung adenocarcinoma having rare EGFR mutations S768I and V769L. Lung Cancer. 2006;54:419–22. doi: 10.1016/j.lungcan.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Aertgeerts K, Skene R, Yano J, Sang BC, Zou H, Snell G, et al. Structural analysis of the mechanism of inhibition and allosteric activation of the kinase domain of HER2 protein. J Biol Chem. 2011;286:18756–65. doi: 10.1074/jbc.M110.206193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa T, Seto M, Banno H, Kawakita Y, Oorui M, Taniguchi T, et al. Design and synthesis of novel human epidermal growth factor receptor 2 (HER2)/epidermal growth factor receptor (EGFR) dual inhibitors bearing a pyrrolo[3,2-d]pyrimidine scaffold. J Med Chem. 2011;54:8030–50. doi: 10.1021/jm2008634. [DOI] [PubMed] [Google Scholar]
- 25.Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CC, Jia Y, Li N, Sun X, Ng K, Ambing E, et al. Crystal structure of the ALK (anaplastic lymphoma kinase) catalytic domain. Biochem J. 2010;430:425–37. doi: 10.1042/BJ20100609. [DOI] [PubMed] [Google Scholar]
- 27.Trowe T, Boukouvala S, Calkins K, Cutler RE, Jr., Fong R, Funke R, et al. EXEL-7647 inhibits mutant forms of ErbB2 associated with lapatinib resistance and neoplastic transformation. Clin Cancer Res. 2008;14:2465–75. doi: 10.1158/1078-0432.CCR-07-4367. [DOI] [PubMed] [Google Scholar]
- 28.Kancha RK, von Bubnoff N, Bartosch N, Peschel C, Engh RA, Duyster J. Differential sensitivity of ERBB2 kinase domain mutations towards lapatinib. PLoS ONE. 2011;6:e26760. doi: 10.1371/journal.pone.0026760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes Dev. 2012;26:1131–55. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greulich H, Kaplan B, Mertins P, Chen TH, Tanaka KE, Yun CH, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012;109:14476–81. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monsey J, Shen W, Schlesinger P, Bose R. Her4 and Her2/neu Tyrosine Kinase Domains Dimerize and Activate in a Reconstituted in vitro System. Journal of Biological Chemistry. 2010;285:7035–44. doi: 10.1074/jbc.M109.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–49. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 34.Moasser MM, Basso A, Averbuch SD, Rosen N. The tyrosine kinase inhibitor ZD1839 (“Iressa”) inhibits HER2-driven signaling and suppresses the growth of HER2-overexpressing tumor cells. Cancer Res. 2001;61:7184–8. [PubMed] [Google Scholar]
- 35.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 36.Di Fiore PP, Pierce JH, Fleming TP, Hazan R, Ullrich A, King CR, et al. Overexpression of the human EGF receptor confers an EGF-dependent transformed phenotype to NIH 3T3 cells. Cell. 1987;51:1063–70. doi: 10.1016/0092-8674(87)90592-7. [DOI] [PubMed] [Google Scholar]
- 37.Bose R, Molina H, Patterson AS, Bitok JK, Periaswamy B, Bader JS, et al. Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc Natl Acad Sci U S A. 2006;103:9773–8. doi: 10.1073/pnas.0603948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabindran SK, Discafani CM, Rosfjord EC, Baxter M, Floyd MB, Golas J, et al. Antitumor activity of HKI-272, an orally active, irreversible inhibitor of the HER-2 tyrosine kinase. Cancer Res. 2004;64:3958–65. doi: 10.1158/0008-5472.CAN-03-2868. [DOI] [PubMed] [Google Scholar]
- 39.Konecny GE, Pegram MD, Venkatesan N, Finn R, Yang G, Rahmeh M, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–9. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 40.Kwak EL, Sordella R, Bell DW, Godin-Heymann N, Okimoto RA, Brannigan BW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–70. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sliwkowski MX, Lofgren JA, Lewis GD, Hotaling TE, Fendly BM, Fox JA. Nonclinical studies addressing the mechanism of action of trastuzumab (Herceptin) Semin Oncol. 1999;26:60–70. [PubMed] [Google Scholar]
- 42.Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–28. doi: 10.1016/s1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- 43.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 44.Ahmed SA, Gogal RM, Jr., Walsh JE. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: an alternative to [3H]thymidine incorporation assay. J Immunol Methods. 1994;170:211–24. doi: 10.1016/0022-1759(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 45.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diessenbacher P, Hupe M, Sprick MR, Kerstan A, Geserick P, Haas TL, et al. NF-kappaB inhibition reveals differential mechanisms of TNF versus TRAIL-induced apoptosis upstream or at the level of caspase-8 activation independent of cIAP2. J Invest Dermatol. 2008;128:1134–47. doi: 10.1038/sj.jid.5701141. [DOI] [PubMed] [Google Scholar]
- 47.Kavuri SM, Geserick P, Berg D, Dimitrova DP, Feoktistova M, Siegmund D, et al. Cellular FLICE-inhibitory Protein (cFLIP) Isoforms Block CD95- and TRAIL Death Receptor-induced Gene Induction Irrespective of Processing of Caspase-8 or cFLIP in the Death-inducing Signaling Complex. J Biol Chem. 2011;286:16631–46. doi: 10.1074/jbc.M110.148585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson SN, Towne DL, Burns DJ, Warrior U. A high-throughput soft agar assay for identification of anticancer compound. J Biomol Screen. 2007;12:938–45. doi: 10.1177/1087057107306130. [DOI] [PubMed] [Google Scholar]
- 49.Hadfield JD. MCMC Methods for Multi-Response Generalized Linear Mixed Models: The MCMCglmm R Package. Journal of Statistical Software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 50.Bates D, Maechler M, Bolker B. lme4: Linear mixed-effects models using S4 classes. R package version 0.999375-42. 2011 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.