Abstract
Recent studies have shown that recombinant adeno-associated virus (rAAV) can persist in episomal form; however, factors affecting rAAV persistence are poorly understood. DNA-dependent PK (DNA-PK) is a DNA repair enzyme, which we previously found played an important role in determining the molecular fate of the rAAV genome in mouse skeletal muscle. In the present study, we tested the effect of DNA-PK on AAV serotype 2 integration in vitro and in vivo in mouse liver. In an in vitro integration system, addition of DNA-PK decreased AAV integration, whereas antibody against DNA-PKcs increased integration. In vivo, matched doses of a recombinant AAV serotype 2 vector were injected into the portal vein of either C57BL/6 (DNA-PKcs+/+) or severe combined immunodeficient (DNA-PKcs-/-) mice. After partial hepatectomy to stimulate hepatocyte proliferation, retention of vector genomes and of transgene expression was substantially higher in severe combined immunodeficient mice, indicating that in the absence of DNA-PKcs, a greater proportion of genomes integrated into the cellular genome. In summary, we have provided evidence that DNA-PK inhibits AAV integration both in vitro and in vivo.
Recombinant adeno-associated virus (rAAV) vectors have been increasingly used for gene therapy because they are relatively nontoxic and can mediate long-term transgene expression. Because this vector is used more frequently in clinical trials, it has become crucial to attain an improved understanding of its potential for insertional mutagenesis. Recent studies have shown that the majority of rAAV genomes persist in episomal form after in vivo delivery and that the hallmark of these episomal forms is the production of vector-to-vector junctions, resulting in either circular episomes or high molecular weight concatamers (1-3). Little is known about the cellular factors required for the maturation of rAAV DNA into these stable episomal forms. We previously demonstrated that in skeletal muscle of severe combined immunodeficient (SCID) [DNA-dependent protein kinase catalytic subunit (DNA-PKcs)-negative] mice, some rAAV serotype 2 (rAAV2) genomes persist as linear episomes and then gradually integrate into the cellular genome, whereas in C57BL/6 (DNA-PKcs-positive) mice, they form circular episomes (2). Most recently, Duan et al. (4) also have shown that SCID skeletal muscle retains both circular and linear forms of rAAV genomes, whereas C57BL/6 muscle retains only circular forms of rAAV.
The DNA-PK is composed of a DNA-binding Ku70/Ku80 heterodimer and a large catalytic subunit (DNA-PKcs) and functions as a nuclear serine/threonine protein kinase (5). The Ku protein was first identified as an autoantigen in patients with lupus. It is a heterodimer composed of two tightly associated subunits, Ku70 and Ku80, and is the most abundant DNA end-binding protein in mammalian cells. It recognizes a variety of DNA structures (blunt, overhanging, or hairpin) and binds with high affinity in a DNA sequence-independent manner. In the present studies, we show that the DNA-PKcs inhibits AAV integration both in a cell-free integration system and in vivo in murine liver. The extent of vector DNA integration is confirmed by using a partial hepatectomy/liver regeneration model. This work suggests that host factors will affect the potential risk for rAAV-mediated insertional mutagenesis in the in vivo setting and implies the potential of modulation of AAV integration by regulating host factors, such as DNA-PK.
Methods
In Vitro Integration. A previously described model for in vitro integration was modified (6). Briefly, a linear AAV substrate was generated by BglII digestion of the AAV2 proviral plasmid, pAV2. The integration target used for these studies was p220.2/AAV integration site 1 (AAVS1, 0-1.6 kb). The standard reaction contained 40 mM Hepes (pH 7.7); 7 mM MgCl2; 4 mM ATP; 200 μM each CTP, GTP, and UTP; 100 μM each dATP, dCTP, dGTP, and dTTP; 2 mM DTT; 40 mM creatine phosphoate; 0.5 μg of creatine phosphokinase; 30 μg of cell extract proteins (from adenovirus-infected HeLa, M059J, or M059K); 50 ng of AAV substrate; 100 ng of AAVS1 substrate; and 50 or 0 ng of His-Tag-purified Rep68 (molar ratio:AAV/AAVS1/Rep68 = 1:1:50 or 1:1:0). Reactions were incubated at 34°C for 16 h. After proteinase K digestion, phenol-chloroform extraction, ethanol precipitation, and heat denaturation (95°C for 10 min), integration junctions were amplified by PCR with the Advantage-GC PCR kit (Clontech) by using AAV right-end (BAAV4) and AAVS1 primers (H4d1). The PCR conditions were 94°C for 5 min, followed by 30 cycles of 94°C for 45 sec and 72°C for 3 min. The PCR products were analyzed on 1.5% agarose gel. DNA junctions were detected by Southern blot analysis by using an AAV-inverted terminal repeat (ITR)-specific probe (AAVD) or AAVS1 probe (H4d1). The probes were labeled with the digoxigenin oligonucelotide 3′-End Labeling Kit (Roche Diagnostics).
rAAV Vectors. The rAAV2-CB-AT vector was produced as described (7). Briefly, this vector contains human alpha 1-antitrypsin (hAAT) cDNA driven by cytomegalovirus (CMV) enhancer and chicken β actin promoter. AAV serotype 2 vector was packaged and purified by the plasmid cotransfection method described previously (8, 9). The physical titers of vector preparations were assessed by quantitative competitive PCR and dot blot analysis, and the biological titers were assessed by infectious center assay. All vector preparations lacked any detectable WT AAV by either physical particle titer or infectious unit measurements.
Portal Vein Injection and Partial Hepatectomy. Eight-week-old male C57BL/6 and C57BL/6-SCID mice were purchased from The Jackson Laboratory. All animals were housed in an specific pathogen-free room and handled as approved by the University of Florida Institutional Animal Care and Use Committee. The portal vein injection procedure has been described (7). For partial hepatectomy, animals were anesthetized with isoflurane inhalation. A median-line incision was made to expose the liver. The large median lobe of the liver and the left lateral lobe were securely ligated and then excised. In this way, 65-70% of the total liver was removed.
DNA Extraction and Southern Blot Analysis. Total liver DNA from vector or saline-injected mice was isolated and subjected to Southern blot analysis, as described (7).
Real-Time PCR. The total copies of rAAV genomes in the liver were quantified by real-time PCR, as described (10).
Results
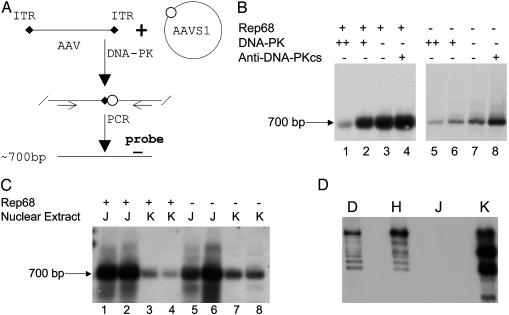
DNA-PKcs Inhibits AAV Integration in Vitro. Based on previous observations that the abundance of different episomal forms of rAAV DNA was affected by the presence of DNA-PKcs, we hypothesized that DNA-PK may impact AAV integration. To test this hypothesis, we used an in vitro assay system for AAV integration (6). This system was designed to examine the effect of cellular proteins on AAV integration (Fig. 1A). When partially purified DNA-PK from HeLa cells (Promega) was added to this in vitro integration system, AAV integration decreased in a dose-dependent manner (Fig. 1B, lanes 1 and 2), as compared with a similar preparation without additional DNA-PK (lane 3). Although not strictly quantitative, the degree of inhibition was ≈1.5- to 15-fold based on the relative intensity of the PCR signal. In contrast, when antibody against DNA-PKcs (a known inhibitor of DNA-PK activity) was added to the reaction, integration junctions increased by ≈1.5-fold (lane 4). It should be noted that HeLa cell nuclear extract was used to provide the cellular environment for the in vitro system. Because the commercial DNA-PK was also isolated from HeLa nuclear extract (as a multicomponent complex consisting of the catalytic subunit (Fig. 1D, lane H) and the heterodimeric DNA-binding Ku (Ku70 and Ku80), we sought to eliminate the possibility that the inhibitory effect was simply from the additional cellular proteins (i.e., a nonspecific effect). To confirm the inhibition was from DNA-PK, we replaced the HeLa cell nuclear extract with nuclear extracts from the M59J cell line, which is DNA-PKcs-negative (Fig. 1D, lane J), or the matched M059K cell line, which is DNA-PKcs-positive (Fig. 1D, lane D). As shown in Fig. 1C, integration junctions were substantially less abundant in the presence of DNA-PKcs (lanes 3 and 4) than in the absence of DNA-PKcs (lanes 1 and 2). Taken together, these results clearly showed that DNA-PK inhibits AAV integration in this cell-free assay system.
Fig. 1.
DNA-PK inhibits AAV integration in vitro. (A) Schematic diagram of the in vitro integration assay for testing the roles of the DNA-PK. (B) In vitro integration assays were performed with or without DNA-PK (200 units for lanes 1 and 5; 20 units for lanes 2 and 6) or antibody against DNA-PKcs (0.4 μg for lanes 4 and 8). HeLa nuclear extract was used in all reactions. The integration reactions were stopped and heated at 94°C for 10 min before PCR. When the integration reactions were performed with Rep68, half the amount of the reaction products was used as PCR template (lanes 1-4) to avoid saturation of the PCR and to evaluate the effects of DNA-PK and the anti-DNA-PKcs. When the integration reactions were performed without Rep68, the total reaction product was used as PCR template for enhancing amplification of the junction. An ≈700-bp PCR amplified junction (as indicated) of AAV and the AAVS1 site was detected by Southern blot with AAVS1 probe. (C) In vitro integration assay using nuclear extracts from DNA-PKcs-negative cells, M059J (J), and NDA-PKcs-positive cells, M059K (K). No HeLa nuclear extract was added in these reactions. (D) Western blot for DNA-PKcs. Antibody (Ab1, NeoMarkers, Fremont, CA) was diluted (1:200) and used to detect DNA-PKcs in the nuclear extracts from HeLa (H), M059J (J), and M059K (K) cells. DNA-PK (D) from Promega was used for positive control.
We also performed the above assays in the absence of Rep protein. Surprisingly, the integration junction was detectable but much less abundant (Fig. 1 B and C, lanes 5-8), indicating that integration can occur but with very low efficiency without rep protein. Importantly, the same inhibitory effect was observed in all conditions, indicating that the effect of DNA-PK on AAV integration is Rep-independent.
Evaluation of the Effect of DNA-PKcs on rAAV Integration in Liver. To confirm the relevance of the observed effects of DNA-PK on AAV integration, we exposed the livers of C57BL/6 (B6, DNA-PKcs positive) and C57BL/6-SCID (SCID, DNA-PKcs negative) mice to rAAV vector DNA in vivo. These two strains have an identical genetic background, with the one exception of a mutation in DNA-PKcs in this SCID mouse strain resulting in a C-terminal-truncated nonfunctional protein (11). When matched doses [1 × 1010 infectious units (i.u.)] of a rAAV2CB-AT vector were injected through the portal vein, transgene expression in the B6 mice was similar to that in SCID mice (data not shown). A separate experiment with a lower dose (3 × 109 i.u.) of the vector confirmed this result and showed dose-dependent transgene expression (data not shown).
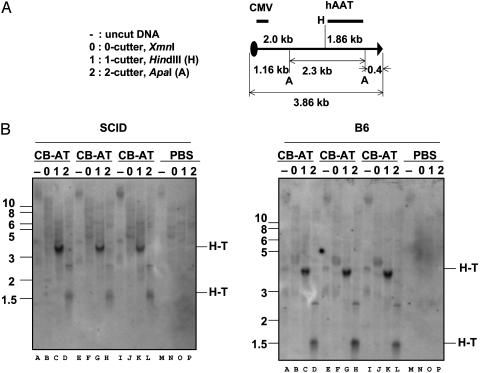
Previously, we observed that significant numbers of rAAV genomes persisted in linear episomal form in SCID mouse muscle for a prolonged period of time (at least 18 months) after injection, whereas in B6 mice, this linear rAAV DNA was undetectable by 18 weeks after injection (2). In the present study, we sought to determine whether linear rAAV genomes also persist in the liver of SCID mice. If they existed, the free-end fragments should be 2.0 kb (when digested with HindIII and hybridized by CMV enhancer probe), 1.16 kb (when digested with ApaI and probed by CMV enhancer probe), or 1.86 kb (when digested with HindIII and hybridized by hAAT probe). Surprisingly, little or no linear free-end fragments of the vector genome were detected in either SCID or B6 mice at 26 weeks after injection, as by Southern blot analysis with a CMV enhancer probe (Fig. 2), or with an hAAT probe (data not shown). In Fig. 2, the blot was overexposed for 3 weeks, causing nonspecific high Mr signal to appear even in the PBS control animals, yet there were still no free ends detectable. However, low Mr signals were detected with uncut DNA in both B6 and SCID mice (Fig. 2B, lanes A, E, and I). Consistent with previous observations, head-to-tail junction of the vector DNA was detected as the most abundant junction among all vector-vector junctions when digested with HindIII (3.86 kb, Fig. 2B, lanes C, G, and K) or ApaI (1.56 kb, Fig. 2B, lanes D, H, and L). Together, these data indicate that formation of circular episomes and perhaps integration are completed in B6 and SCID mice within 26 weeks. This suggests that the progression toward either stable vector episomal forms or integrated forms proceeds more rapidly in liver than in skeletal muscle. Importantly, these data provide clear evidence that the formation of vector-vector junctions (mainly T-H) and circular episomes can occur without DNA-PK activity.
Fig. 2.
Southern blot analysis for molecular fate of rAAV genome in liver. Total cellular DNA was purified from livers of rAAV2-CB-AT- (1 × 1010 i.u.) or PBS-injected mice. Samples of liver DNA were incubated either in the absence of any restriction enzyme (-) or with XmnI (no cutting site within the vector), HindIII (one site within the vector), or ApaI (two sites within the vector). (A) Schematic diagram for restriction digestion and the positions of the probes (CMV and hAAT) for hybridization. (B) DNAs from SCID (Left) and B6 (Right) mice were hybridized with 32P-labeled CMV probe. H-T indicates head-to-tail junctions.
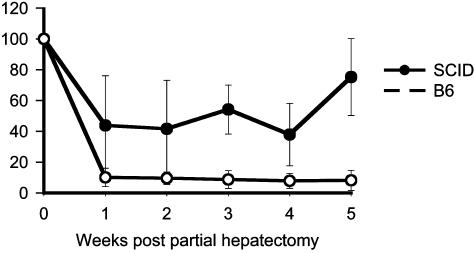
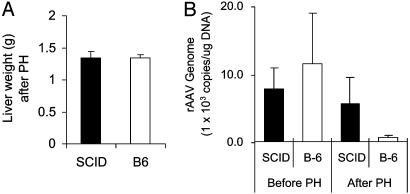
Evaluation of the Effect of DNA-PKcs on rAAV Integration in Liver. To test confirm our in vitro observation that DNA-PK inhibits AAV integration, we used partial hepatectomy, which has been previously used to stimulate hepatocyte regeneration and to evaluate rAAV integration (12). After hepatocyte regeneration, episomal forms are lost, whereas integrated forms are retained. Thus transgene expression reflects rAAV integration. Consistent with previous studies (12), <10% of transgene expression remained in C57BL/6 mice after partial hepatectomy (Fig. 3). This observation suggests that a small portion of viral genomes integrated into cellular genome and that the majority of vector genomes persisted in episomal form. However, in SCID mice, >40% of transgene expression remained after partial hepatectomy, indicating that a substantially greater proportion of vector genome had integrated into host cellular genome in the absence of DNA-PKcs (Fig. 3). Eight weeks after partial hepatectomy, animals were killed. The residual liver tissue (right lobe) from each mouse was examined and weighed. These results confirmed that livers of both SCID and B6 mice had regenerated back to normal size, and that no difference in liver weight was observed between the two strains (Fig. 4A, P < 0.01), indicating that hepatocytes proliferated equally in both strains. To test whether the levels of transgene expression truly reflect the change of vector genome in the liver, we performed real-time PCR analysis to detect the total copies of the vector genome. As shown in Fig. 4B, similar copy numbers of AAV vector were detected in both SCID and B6 mice liver before partial hepatectomy. However, <10% of the vector DNA remained in B6 liver, although >50% of the vector DNA remained in SCID liver 8 weeks after partial hepatectomy (Fig. 4B). These data closely correlated with transgene expression (Fig. 3) and strongly indicated that DNA-PK inhibited rAAV integration in mouse liver.
Fig. 3.
Relative levels of serum hAAT in SCID (filled circle) and B6 (open circle) after partial hepatectomy. Sixteen weeks after portal vein injection of rAAV2-CB-AT vector (3 × 109 i.u. per mouse), mice were subjected to partial hepatectomy. Each point represents the mean of data from each group (SCID, n = 6; B6, n = 6, P < 0.01). The y axis shows the percentage of hAAT levels relative to the levels before partial hepatectomy (week 0). Serum hAAT was measured by ELISA.
Fig. 4.
Effect of DNA-PK on rAAV persistence in liver. (A) Equal generation of SCID and B6 liver after partial hepatectomy (PH). Each bar represents the mean of fresh liver weight at 8 weeks after PH (SCID, n = 6; B6, n = 5; P < 0.01). (B) Average of total copies of the vector genomes before and after partial hepatectomy (SCID, n = 6; B6, n = 5). Animals receiving 3 × 109 i.u. of rAAV2-CB-AT vector were subjected to partial hepatectomy (PH) at 16 weeks after injection. These animals were killed at 8 weeks after PH. Total rAAV DNA in all liver samples were detected by real-time PCR.
Discussion
The data presented here show clear-cut effects of DNA-PK on AAV2 DNA integration both in vivo and in vitro. Our previous report comparing the abundance of vector-vector junctions in skeletal muscle of DNA-PK-deficient and -sufficient mice suggested this effect. However, in the current study, we have taken advantage of the unique ability of liver to regenerate after partial resection to differentiate between integrated and episomal copies of vector DNA and have thereby directly demonstrated an increase in vector DNA integration in the DNA-PK-deficient animal. Furthermore, the data from the DNA-PK-deficient animals clearly indicated that integrated copies of rAAV expressed the transgene of interest.
There was another key difference between the results reported here and those seen in muscle. Free ends of linear AAV genomes were detected in DNA-PK-deficient mice in the muscle studies but not in the current liver studies. The muscle data could be interpreted as indicating a predominant effect of DNA-PK on promoting rapid end-to-end joining of vector genomes to create vector-vector junctions, thus inhibiting integration only by facilitating a competing reaction. In the liver studies, however, vector-vector junctions appeared rapidly and at high abundance even in the absence of DNA-PK, and yet a marked difference in genomic integration was still observed. This implies an alternative means of inhibition of integration. The presence of end-to-end joining in the absence of DNA-PK is also consistent with previous observations (2, 4, 13, 14). DNA-PK may have some effect on these processes, but clearly it is not absolutely required, even in muscle.
As mentioned above, DNA-PK is a multisubunit complex, which binds a range of different forms of double-stranded DNA ends (15). Although both Ku70 and Ku80 interact with DNA, Ku70 makes the more intimate interactions with its C-terminal 73-aa residues. Ku70 and Ku80 functionally depend on each other, although either subunit alone can bind DNA effectively (16-18). Interestingly, Ku has the ability to translocate along DNA molecules in an ATP-independent manner (19, 20). Consistent with this, Ku can generate footprints at internal sites as well as the termini of linear DNA molecules (21). In addition, atomic force microscopy studies have revealed the existence of internal and DNA-end-bound DNA-PK complexes and have shown that Ku can juxtapose two DNA ends via a DNA looping mechanism (22). It has been shown that Ku can actually transit directly from one linear DNA molecule to another if the termini of the two DNA ends are capable of base-pairing (23). The functional significance of these DNA end-alignment activities suggests Ku can stimulate DNA end-joining by eukaryotic DNA ligases in vitro (24). Other functions of Ku are helicase and ATPase activities. The Ku70 subunit contains the ATPase activity and is able to perform its helicase function independently of Ku80 (18). The weak ATPase activity is stimulated by DNA-PK-mediated phosphorylation. Our studies do not address which, if any, of these DNA-PK-mediated activities might interact with the AAV genome. However, that the effects are seen in the presence or absence of AAV Rep protein suggests the effects are mediated directly through the AAV2 ITRs, which are present in every system tested here.
Although the original purpose of the in vitro assay used here was to detect site-specific integration, it is capable of detecting random site integration as well, i.e., when Rep is not present. Therefore, the results suggest that at the in vitro level, DNA-PK inhibits both site-specific and random integration. This does not imply that WT AAV and rAAV are integrating through the same mechanism, merely that the AAV-ITRs are the common cis elements in both of these processes. Therefore, any cellular protein or factor that interacts with AAV-ITRs may affect ITR functions, including integration. This effect of DNA-PK on the ITR could be direct or indirect and could vary among cell types and species. For example, primate cells generally have much higher levels of DNA-PK activity than rodent cells (25). Therefore, integration behavior of rAAV observed in a rodent model may not reflect that in human cells. Finally, it should be noted that the AAV-ITR-flanked genomes, with or without active Rep, can activate a P53-dependent pathway, which is likely to activate DNA-PK (26); thus, it may be the ability of DNA-PK to be induced, rather than its basal constitutive level, that will determine the frequency of integration.
A report has indicated that rAAV vectors preferentially integrate into active genes in mice (27). This finding has brought greater attention to the potential for insertional mutagenesis from rAAV gene therapy (28, 29). This probably reflects the more open chromatin structure surrounding transcriptionally active genes and is consistent with the finding that Rep-deleted rAAV integration, to the extent to which it occurs, is relatively random (30). It must be noted, however, that our studies provided no direct evidence for integration in normal mice. Because ≈70% of the liver was removed, one would have anticipated that episomes capable of efficient segregation to daughter cell nuclei after mitosis would have resulted in a maximum residual level of DNA presence and transgene expression of ≈30%. The level of retention of genomes and of transgene expression was, in fact, much less than that, at ≈7%, suggesting that most episomal copies were lost during mitosis. Furthermore, the residual activity that was seen could be from cells that did not divide or from episomes that did happen to segregate into daughter cells rather than from integration. Thus, whereas it is clear that frequent integration occurred in the DNA-PK-deficient animals, our studies do not demonstrate that any integration occurred in the normal animals.
Indeed, this study as a whole suggests that a DNA-PK-dependent mechanism is operative to work against rAAV integration. Furthermore, in contrast with the known propensity of oncoretroviruses to mediate oncogenesis, both as WT viruses and most recently as human vectors (31), there has never been a recorded instance of AAV-mediated tumorigenesis with either WT AAV or rAAV in animals or in humans. However, if any concern about oncogenesis should arise, there are two potential mechanisms now available to decrease that risk. The first would be to increase DNA-PK activity and inhibit integration, whereas the alternative would be to harness Rep-mediated site-specific integration into the AAVS1 site, which is likewise relatively safe.
Acknowledgments
This work was supported in part by grants from the Alpha-1 Foundation, the National Institutes of Health (DK62652, DK58327, HL69877, and HL59412), and the Juvenile Diabetes Research Foundation.
Abbreviations: AAV, adeno-associated virus; rAAV, recombinant AAV; DNA-PK, DNA-dependent PK; DNA-PKcs, DNA-PK catalytic subunit; SCID, severe combined immunodeficient; CMV, cytomegalovirus; ITR, inverted terminal repeat; AAVS1, AAV integration site 1; hAAT, human alpha 1-antitrypsin; AAV2, AAV serotype 2; i.u., infectious units.
References
- 1.Afione, S. A., Conrad, C. K., Kearns, W. G., Chunduru, S., Adams, R., Reynolds, T. C., Guggino, W. B., Cutting, G. R., Carter, B. J. & Flotte, T. R. (1996) J. Virol. 70, 3235-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song, S., Laipis, P. J., Berns, K. I. & Flotte, T. R. (2001) Proc. Natl. Acad. Sci. USA 98, 4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnepp, B. C., Clark, K. R., Klemanski, D. L., Pacak, C. A. & Johnson, P. R. (2003) J. Virol. 77, 3495-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan, D., Yue, Y. & Engelhardt, J. F. (2003) J. Virol. 77, 4751-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith, G. C. & Jackson, S. P. (1999) Genes Dev. 13, 916-934. [DOI] [PubMed] [Google Scholar]
- 6.Dyall, J., Szabo, P. & Berns, K. I. (1999) Proc. Natl. Acad. Sci. USA 96, 12849-12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song, S., Embury, J., Laipis, P. J., Berns, K. I., Crawford, J. M. & Flotte, T. R. (2001) Gene Ther. 8, 1299-1306. [DOI] [PubMed] [Google Scholar]
- 8.Song, S., Morgan, M., Ellis, T., Poirier, A., Chesnut, K., Wang, J., Brantly, M., Muzyczka, N., Byrne, B. J., Atkinson, M., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 14384-14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zolotukhin, S., Byrne, B. J., Mason, E., Zolotukhin, I., Potter, M., Chesnut, K., Summerford, C., Samulski, R. J. & Muzyczka, N. (1999) Gene Ther. 6, 973-985. [DOI] [PubMed] [Google Scholar]
- 10.Song, S., Scott-Jorgensen, M., Wang, J., Poirier, A., Crawford, J., Campbell-Thompson, M. & Flotte, T. R. (2002) Mol. Ther. 6, 329-335. [DOI] [PubMed] [Google Scholar]
- 11.Blunt, T., Gell, D., Fox, M., Taccioli, G. E., Lehmann, A. R., Jackson, S. P. & Jeggo, P. A. (1996) Proc. Natl. Acad. Sci. USA 93, 10285-10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakai, H., Yant, S. R., Storm, T. A., Fuess, S., Meuse, L. & Kay, M. A. (2001) J. Virol. 75, 6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakai, H., Storm, T. A., Fuess, S. & Kay, M. A. (2003) Hum. Gene Ther. 14, 871-881. [DOI] [PubMed] [Google Scholar]
- 14.Yue, Y. & Duan, D. (2003) Virology 313, 1-7. [DOI] [PubMed] [Google Scholar]
- 15.Dynan, W. S. & Yoo, S. (1998) Nucleic Acids Res. 26, 1551-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffith, A. J., Blier, P. R., Mimori, T. & Hardin, J. A. (1992) J. Biol. Chem. 267, 331-338. [PubMed] [Google Scholar]
- 17.Wu, X. & Lieber, M. R. (1996) Mol. Cell. Biol. 16, 5186-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochem, A. E., Skopac, D., Costa, M., Rabilloud, T., Vuillard, L., Simoncsits, A., Giacca, M. & Falaschi, A. (1997) J. Biol. Chem. 272, 29919-29926. [DOI] [PubMed] [Google Scholar]
- 19.de Vries, E., van Driel, W., Bergsma, W. G., Arnberg, A. C. & van der Vliet, P. C. (1989) J. Mol. Biol. 208, 65-78. [DOI] [PubMed] [Google Scholar]
- 20.Paillard, S. & Strauss, F. (1991) Nucleic Acids Res. 19, 5619-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mimori, T. & Hardin, J. A. (1986) J. Biol. Chem. 261, 10375-10379. [PubMed] [Google Scholar]
- 22.Cary, R. B., Peterson, S. R., Wang, J., Bear, D. G., Bradbury, E. M. & Chen, D. J. (1997) Proc. Natl. Acad. Sci. USA 94, 4267-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bliss, T. M. & Lane, D. P. (1997) J. Biol. Chem. 272, 5765-5773. [DOI] [PubMed] [Google Scholar]
- 24.Ramsden, D. A. & Gellert, M. (1998) EMBO J. 17, 609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson, C. W. & Lees-Miller, S. P. (1992) Crit. Rev. Eukaryotic Gene Expr. 2, 283-314. [PubMed] [Google Scholar]
- 26.Raj, K., Ogston, P. & Beard, P. (2001) Nature 412, 914-917. [DOI] [PubMed] [Google Scholar]
- 27.Nakai, H., Montini, E., Fuess, S., Storm, T. A., Grompe, M. & Kay, M. A. (2003) Nat. Genet. 34, 297-302. [DOI] [PubMed] [Google Scholar]
- 28.Russell, D. W. (2003) Nat. Genet. 34, 241-242. [DOI] [PubMed] [Google Scholar]
- 29.Kay, M. A. & Nakai, H. (2003) Nature 424, 251. [DOI] [PubMed] [Google Scholar]
- 30.Kearns, W. G., Afione, S. A., Fulmer, S. B., Pang, M. C., Erikson, D., Egan, M., Landrum, M. J., Flotte, T. R. & Cutting, G. R. (1996) Gene Ther. 3, 748-755. [PubMed] [Google Scholar]
- 31.Hacein-Bey-Abina, S., Von Kalle, C., Schmidt, M., McCormack, M. P., Wulffraat, N., Leboulch, P., Lim, A., Osborne, C. S., Pawliuk, R., Morillon, E., et al. (2003) Science 302, 415-419. [DOI] [PubMed] [Google Scholar]