Abstract
The diphosphoinositol polyphosphates (“inositol pyrophosphates”; PP-InsPs) regulate many cellular processes in eukaryotes, including stress responses, apoptosis, vesicle trafficking, cytoskeletal dynamics, exocytosis, telomere maintenance, insulin signaling and neutrophil activation. Thus, the enzymes that control the metabolism of the PP-InsPs serve important cell signaling roles. In order to fully characterize how these enzymes are regulated, we need to determine the atomic-level architecture of their active sites. Only then can we fully appreciate reaction mechanisms and their modes of regulation. In this review, we summarize published information obtained from the structural analysis of a human diphosphoinositol polyphosphate phosphohydrolase (DIPP), and a human diphosphoinositol polyphosphate kinase (PPIP5K). This work includes the analysis of crystal complexes with substrates, products, transition state analogues, and a novel phosphonoacetate substrate analogue.
Keywords: inositol pyrophosphates, structure, analogues, diphosphoinositol polyphosphates, cell-signaling, kinase, phosphorylation
Introduction
The phosphate group is a ubiquitous signaling device that establishes specificity in ligand-protein and protein-protein interactions. The phosphate’s bulk imposes geometric constraints upon these interactions. The phosphate’s negative charge also bestows specificity through ionic and hydrogen bonds with certain amino acid residues at physiological pH. The negative charges on the phosphate group also make soluble, phosphorylated molecules lipid-impermeant, so that they can be retained inside cells.
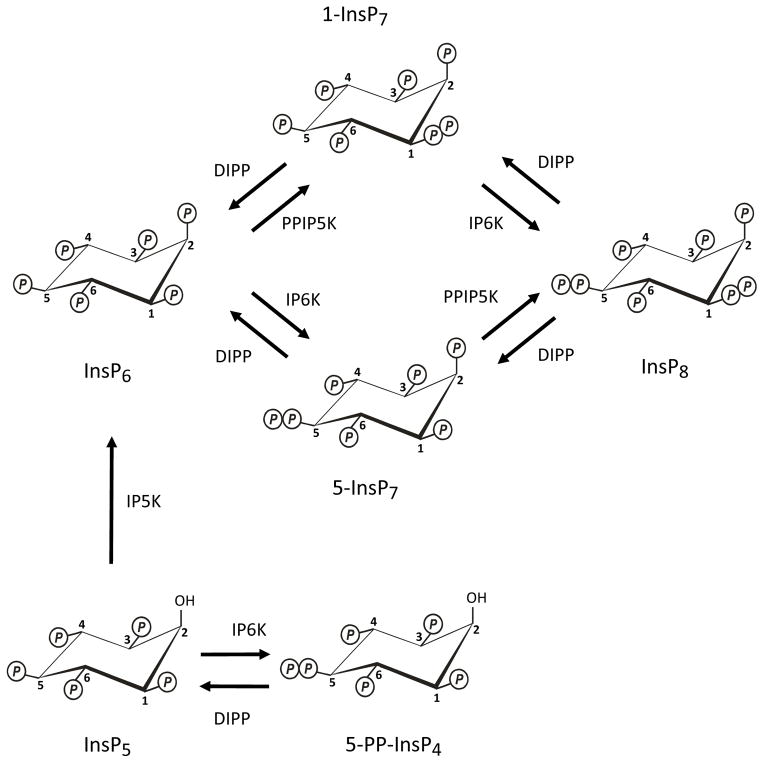
An extreme example of these applications for the phosphate group is provided by the diphosphoinositol polyphosphates (PP-InsPs), also known as “inositol pyrophosphates”. These molecules possess the most crowded three-dimensional array of phosphate groups that are found throughout Nature. The most studied of the PP-InsPs are those that are formed from InsP6, namely, 1-InsP7, 5-InsP7 and InsP8 (Figure 1). Two groups of enzymes participate in these reactions. The 5-kinase activities of IP6K1/2/3 (E.C.2.7.4.21) (Draskovic et al., 2008; Saiardi et al., 2001) convert InsP6 and 1-InsP7 to 5-InsP7 and 1,5-InsP8 respectively (Figure 1). Second, the 1-kinase activities of PPIP5K1/2 (E.C.2.7.4.24) (Choi et al., 2007; Fridy et al., 2007; Wang et al., 2012) phosphorylate InsP6 and 5-InsP7 to 1-InsP7 and InsP8 respectively (Figure 1). These PP-InsPs regulate many cellular processes in eukaryotes, including stress responses, apoptosis, vesicle trafficking, cytoskeletal dynamics, exocytosis, insulin signaling and neutrophil activation (for reviews see (Barker et al., 2009; Burton et al., 2009; Chakraborty et al., 2011; Shears 2009)). In addition, the IP6Ks can also phosphorylate Ins(1,3,4,5,6)P5 (Figure 1) to a compound that is usually annotated as PP-InsP4 (Saiardi et al., 2000). The latter appears to modulate telomere maintenance (Ponnusamy et al., 2008; Saiardi et al., 2005; Seeds et al., 2004; York et al., 2005). Thus, the IP6Ks and PPIP5Ks are extremely important enzymes in cell signaling.
Fig. 1. Synthesis and metabolism of the PP-InsPs.
The figure describes the metabolic reactions that account for the turnover of the P-InsPs in both yeasts and mammalian cells. The positions of the diphosphate groups were determined in the following publications: (Albert et al., 1997; Draskovic et al., 2008; Wang et al., 2012). DIPP (E.C. 3.6.1.52; Ddp1 in yeast), diphosphoinositol polyphosphate phosphohydrolase; IP5K (E.C. 2.7.1.158; Ipk1 in yeast), inositol pentakisphosphate kinase; IP6K (E.C.2.7.4.21; Kcs1 in yeast), inositol hexakisphosphate kinase, PPIP5K (E.C.2.7.4.24; Vip1 in yeast), diphosphoinositol pentakisphophate kinase.
The diphosphoinositol polyphosphate phosphohydrolases (DIPP; E.C. 3.6.1.52) that dephosphorylate PP-InsPs (Figure 1) are just as critical to cell function as are the kinases. The high activity of these DIPPs ensures that PP-InsPs turnover rapidly inside cells (Menniti et al., 1993). These activities also appear to be a major reason why mammalian cells contain such low steady-state concentrations of diphosphoinositol polyphosphates in most circumstances. Levels of total InsP7 usually lie in the 1 to 5 μM range (Barker et al., 2004; Bennett et al., 2006; Fisher et al., 2002; Illies et al., 2007; Ingram et al., 2003), most of which appears to be the 5-isomer in mammalian cells (Albert et al., 1997). The concentrations of PP-InsP4 and InsP8 in yeast and mammalian cells are each about 10–20% of those of InsP7 (Choi et al., 2005; Choi et al., 2008; Glennon et al., 1993). This places the levels of individual PP-InsPs in roughly the same range as those of Ins(1,4,5)P3, the Ca2+-mobilizing second messenger (Streb et al., 1983). Five mammalian DIPPs are known: type 1 (Chu et al., 2004; Safrany et al., 1998), types 2α/2β (Caffrey et al., 2000; Hua et al., 2001) and types 3α/3β (Hidaka et al., 2002; Hua et al., 2003; Leslie et al., 2002).
The PP-InsPs have been shown to non-enzymatically diphosphorylate certain proteins (see below and (Bhandari et al., 2007; Saiardi et al., 2004)). As discussed elsewhere (Shears et al., 2011), it has not yet been shown that protein diphosphorylation occurs in vivo. Furthermore, there is as yet no evidence for the existence of an “off-switch” for this putative signaling process, namely, phosphatase-directed cleavage of the diphosphorylated protein (Bhandari et al., 2008; Burton et al., 2009). So it is uncertain what the contributions of these process to PP-InsPs turnover might be in vivo.
Central to the characterization of PP-InsPs turnover is to determine the atomic-level architecture of the active sites of the participating enzymes. Only then can we fully appreciate reaction mechanisms and their modes of regulation. Recent developments in this area represent the focus of this review.
The biological importance of the diphosphate groups in the PP-InsPs
Despite the presence of multiple phosphate groups in the PP-InsPs, it is their diphosphates that are their key, functionally significant feature. For example, it is the diphosphate group which facilitate the ability of 5-InsP7 to compete with phosphatidylinositol-3,4,5-trisphosphate for binding to PH domains (Chakraborty et al., 2010; Prasad et al., 2011). PtdIns(3,4,5)P3-binding proteins that are captured at the plasma membrane following stimulus-dependent PI3K activation promote assembly of multiprotein complexes and priming of kinase cascades; there are multiple biological consequences to these signaling events. Thus, the potential that 5-InsP7 has for inhibiting these signaling pathways is far reaching. For example, this phenomenon has been proposed to explain how changes in expression or activity of IP6K can modulate signaling by insulin (Chakraborty et al., 2010), and also regulate neutrophil activity (Prasad et al., 2011).
A separate study provided an example of a more classical InsP7 “receptor”, the Pho80/Pho85/Pho81 cyclin-dependent kinase complex (Lee et al., 2008). By binding to this complex, 1-InsP7 augments the ability of Pho81 to inhibit kinase activity, and the ligand’s diphosphate group contributes to the specificity of the interaction (Lee et al., 2008). (To avoid confusion, we should note that in the latter report the structure of the active isomer was erroneously described as 4/6-InsP7, based on an earlier, tentative suggestion (Mulugu et al., 2007)).
A third, but more provocative mechanism of action of the PP-InsPs has also been put forward by Snyder and colleagues (Azevedo et al., 2009; Bhandari et al., 2007; Saiardi et al., 2004; Szijgyarto et al., 2011). In vitro, at least, the diphosphate groups may also be utilized for the non-enzymatic phosphorylation of a range of different proteins. It is the β-phosphate of the diphosphate groups on the PP-InsPs that are donated. They are added to a pre-existing Ser-phosphate that is initially provided by a casein kinase II dependent phosphorylation event. That is, a diphosphate group is formed on the target protein. Since CK2 phosphorylates so many proteins (Ruzzene et al., 2010), this particular mechanism of action of PP-InsPs offers a potential explanation for their multifunctionality (Saiardi et al., 2004). However, a number of questions remain concerning the viability of this as a physiologically-relevant process in vivo (Shears et al., 2011).
Each of these proposed mechanisms of action of the PP-InsPs relies upon the operation of a fundamental principle in signal transduction: the cellular levels of a cellular mediator – in this case PP-InsP4, InsP7 or InsP8 (Figure 1) must be altered in a predictable manner in response to a specific intracellular or extracellular stimulus. A few notable descriptions of such phenomena have been published. For example, levels of InsP7 are elevated during nutrient stress in Saccharomyces cerevisiae (Lee et al., 2007b) and slime-molds (Luo et al., 2003). Total InsP7 levels also increase substantially following the addition of growth factors to cells that have been serum-starved overnight (Chakraborty et al., 2010). As for InsP8, its levels are elevated when cells are subjected to either hyperosmotic stress or a thermal challenge (Choi et al., 2005; Pesesse et al., 2004). On the contrary, oxidative stress reduces both InsP7 and InsP8 levels (Onnebo et al., 2009). Finally, InsP8 concentration decreases during bioenergetic stress (Choi et al., 2008). It is because of all of these signaling activities that there is a need to understand how the turnover of PP-InsPs is regulated in intact cells. Central to such studies is the characterization of the atomic-level architecture of the active sites of the responsible enzymes.
The structure of a DIPP
All five isoforms of the mammalian DIPPs (see above) are relatively small proteins of just under 20 kDa (Caffrey et al., 2000; Hidaka et al., 2002; Hua et al., 2001; Hua et al., 2003; Leslie et al., 2002; Safrany et al., 1998). The active site of each is based on the so-called Nudix motif, which is typically, although not exclusively, Gx5Ex5[UA]xREx2EExGU (U represents an aliphatic, hydrophobic residue) (McLennan 2006). The DIPPs represent an unusual context in which to find this motif; it is more usually reserved for proteins whose functions are limited to the hydrolysis of nucleoside di- and triphosphates, nucleotide sugars and dinucleoside polyphosphates (McLennan 2007). A detailed mutagenic study has revealed that the specificity of human DIPP1 towards diphosphoinositol polyphosphates is entrusted to several amino acid residues that lie outside the Nudix motif (Yang et al., 1999). The structures of several Nudix proteins have been solved, but to date only one DIPP structure has been published (human DIPP1; (Thorsell et al., 2009)).
Thorsell et al (Thorsell et al., 2009) reported that human DIPP1 adopts a canonical fold: two β-sheets flanked by short helices. The Nudix motif (see above) that normally adopts a loop-helix-loop fold is configured slightly differently in DIPP1 as a strand-loop-helix. This variation in part reflects the presence of six residues instead of five between the N-terminal Gly and the first Glu (i.e. Gx6E instead of Gx5E). There are also structurally-stabilizing interactions between the first three residues of the Nudix motif with a neighboring β-strand (Thorsell et al., 2009). Although DIPP1 was not co-crystallized with any of its substrates, Thorsell et al (Thorsell et al., 2009) did obtain crystals that contained the InsP6 product. However, there was a complication that InsP6 bound in two different conformations; their study uses just one of these to derive a putative reaction mechanism and so it remains unclear how valid are their predictions.
When Thorsell et al (Thorsell et al., 2009) published their work, the characterization of InsP8 was incomplete (see below and (Lin et al., 2009)); at that time, it was unknown if the molecule had a 1,5- or a 3,5-diphosphate grouping. Thorsell et al (Thorsell et al., 2009) were able to model both alternative InsP8 structures into their structure, and this led them to speculate that 3,5-InsP8 was the more likely of the two (Thorsell et al., 2009). However, it later turned out that it was actually 1,5-InsP8 (see below and (Wang et al., 2012)).
A 2010 review from the Prestwich group (Best et al., 2010) cites unpublished crystallographic data from York’s laboratory indicating that they have co-crystallized DIPP1 with 1-InsP7. We await the publication of these data with interest, as they promise to provide more direct information concerning the molecular basis for DIPP specificity, and the reaction mechanism.
The structure of a PPIP5K
The enzymatic synthesis of inositol pyrophosphates requires the formation of high-energy phosphoanhydride bonds (Hand et al., 2007), in a class of molecules that are already unrivaled throughout Nature in their degree of phosphate congestion. Thus, the active site of an inositol pyrophosphate kinase must accommodate considerable steric bulk and intense electronegativity, and yet retain selective substrate specificity (Choi et al., 2007; Gokhale et al., 2011). Moreover, this group of kinases must also overcome a substantial energy barrier to the transition state. We (Wang et al., 2012) recently determined how these catalytic challenges have been overcome by PPIP5K2.
Being less than 20 kDa in size, the expression and purification of recombinant DIPPs for structural work (see above) has been a relatively straightforward exercise (Safrany et al., 1998). In contrast, the mammalian type 1 and type 2 PPIP5Ks are 120 and 160 kDa, respectively (Choi et al., 2007; Fridy et al., 2007). It is extremely difficult to express and purify such a large enzyme with the sufficient degree of both the yield and the degree of purity needed to determine its crystal structure. Fortunately, however, the PPIP5Ks have been found to be modular in nature (Figure 2A). For example, the kinase domain is self contained within the N-terminal one third of these proteins (Mulugu et al., 2007). A series of different crystal structures of the kinase domain of human PPIP5K2 (PPIP5K2KD) were recently obtained (Wang et al., 2012). These included co-crystal complexes with nucleotide cofactor plus either substrates, product, or a MgF3− transition-state mimic (Graham et al., 2002). The latter was especially significant as no previous structural work with any inositol phosphate kinase has captured the transition state.
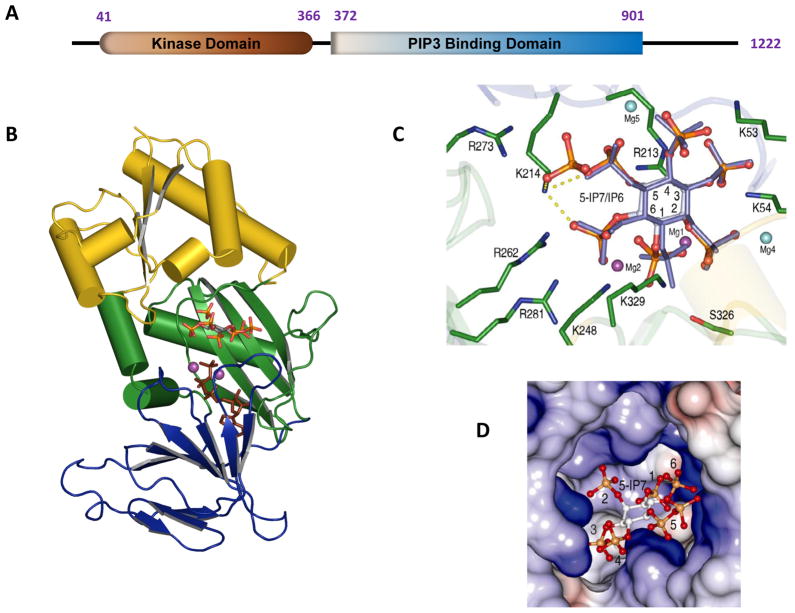
Fig. 2. Structure of the kinase domain of human PPIP5K2.
A is a graphical depiction of the modular format of PPIP5K2; the numbering of the amino acid residues depicts where the domains are considered to begin and finish. B, Ribbon diagram of residues 37-366 of PPIP5K2KD. The αβα domain (residues 42–124 and 330–366) is shown in yellow. The two antiparallel β-sheet clusters of the ATP-grasp domain are colored green (residues 125–148 and 244–329) and blue (residues 149–243). Shown in stick models are 5-InsP7 and AMP-PNP. C, Active site of PPIP5K2 KD. Shown in green are residues that make polar contacts with substrates. 5-InsP7 is shown with grey carbon and orange and red phosphate groups, while InsP6 is slate-colored. Carbon atoms on the inositol ring are numbered. The interactions of Lys214 with 5-InsP7 are highlighted by dashed yellow lines. The sky blue and purple spheres represent the magnesium atoms; The purple magnesium atomss (Mg1 and Mg2) were catalytically relevant. Mg5 was not present in the PPIP5K2KD–InsP6 complex. See (Wang et al., 2012) for full details. D, Electrostatic surface representation of the inositol phosphate substrate binding pocket of PPIP5K2KD.
Our structures (Figure 2B) reveal that nucleotide nestles between two sets of anti-parallel β-sheets, confirming the predictions of homology modeling servers (Choi et al., 2007; Mulugu et al., 2007) that hPPIP5K2KD utilizes an ATP-grasp fold. The ATP is deeply buried. Only 9% of the nucleotide was predicted to be solvent accessible. The remainder of the hPPIP5K2KD structure, mainly at its N-terminus, forms an αβα domain (Figure 2B). We propose that the structure of hPPIP5K1KD is likely very similar to hPPIP5K2KD, due to their 84% sequence identity (Choi et al., 2007). It is very possible that nucleotide binding stabilizes protein conformation, as we could not crystallize the apo-enzyme (Wang et al., 2012).
At the time of writing, it is another ATP-grasp inositol phosphate kinase, human ITPK1 (E.C. 2.7.1.134) (Chamberlain et al., 2007) that is structurally the most similar to the PPIP5Ks (Wang et al., 2012). Nevertheless, the ATP-grasp superfamily is noted for its considerable functional and structural divergence at the active site (Lee et al., 2007a). Even the amino-acid composition and architecture of the active site of ITPK1 (Chamberlain et al., 2007; Miller et al., 2005) differs considerably from that of hPPIP5K2KD. For example, hPPIP5K2KD utilizes only Arg and Lys residues to bind substrate, with the exception of S326 (Figure 2C). The human ITPK1, which phosphorylates a wider range of substrates than do the PPIP5Ks (Chamberlain et al., 2007), possesses a binding pocket that is somewhat larger, and more loosely-defined. Moreover, ITPK1 and another inositol phosphate kinase, IPMK (E.C. 2.7.1.151), phosphorylate multiple positions around the inositol ring (Shears 2004). That promiscuity in part reflects the relatively non-specific active sites (Holmes et al., 2006; Miller et al., 2005; Riley et al., 2006; Shears 2004). But there is another factor: there is a functionally significant plane of symmetry across the 2/5-axis of the inositol ring. That symmetry permits one inositol phosphate to imitate another’s three-dimensional phosphate recognition pattern, when the orientation of the inositol ring changes in relation to the protein’s ligand-binding site (Wilcox et al., 1994). This flexibility in substrate binding is not permitted by PPIP5K; it shows negligible activities towards 1-InsP7, Ins(1,3,4,5,6)P5 or PP-InsP4 (Wang et al., 2012) which, among the naturally-occurring inositol phosphates, are those that are the most similar to 5-InsP7 and InsP6. Our structural data provide an explanation for this rigid specificity of the PPIP5Ks. Its binding pocket, two near-parallel grooves that form a staggered “H”-shape (Figure 2D), makes a perfectly-tailored aperture for accommodating just one orientation of multiple phosphates placed around an inositol ring.
The three-dimensional geometry of the active site of an enzyme is not the only factor that defines substrate specificity. The induced fit motion of key amino acid residues also contributes to specificity (Herschlag 1988). A comparison of the AMP-PNP/hPPIP5K2KD crystal complex with and without 5-InsP7 indicated substrate binding is accompanied by induced fit motion of side-chains of three active-site residues - R262, R281 and K329 (Wang et al., 2012). These intricate conformational dynamics within the active-site of PPIP5K help limit the degree of free-energy of activation more than would be the case if backbone re-arrangement was involved (Herschlag 1988).
Recently, we obtained an additional crystal complex of hPPIP5K2KD which contained a chemically synthesized analogue of 5-InsP7 in which the diphosphate group was replaced by an -phosphonoacetic acid (phosphonoacetate; PA) ester (Riley et al., 2012). This has significance to research into PP-InsPs; the C-P bond of the phosphonoacetate is chemically stable, and also resistant to the phosphatases that occur in higher eukaryotes (Quinn et al., 2007). Moreover, because of the much higher activation energy for cleavage of a C-P bond versus an O-P bond (Quinn et al., 2007), it is not plausible that PA-InsPs (in contrast to PP-InsPs) could act as phosphoryl donors in non-enzymatic diphosphorylation of proteins. Such a stabilized mimic should be useful in excluding protein diphosphorylation as a potential mechanism of action. For example, recent electrophysiological experiments found that PP-InsPs enhanced insulin secretion from pancreatic β-cells (Berggren et al., 2008). It would now be informative to investigate if these observations could be reproduced by the non-hydrolyzable phosphonoacetate analogue of 5-InsP7.
The synthesis of this analogue was also driven by the anticipation (Riley et al., 2012) that its terminal phosphoryl group would adequately mimic the β-phosphoryl in PP-InsPs. Such interactions could be important at the binding site of a putative receptor capable of distinguishing InsP7 from InsP6. Proof of principle for this prediction was obtained from our structural data which showed the analogue to occupy a very similar orientation to that of 5-InsP7 in the active site of PPIP5K2KD (Riley et al., 2012). Moreover, the phosphonoacetate analogue was even phosphorylated by the kinase (Riley et al., 2012), although limitations in assay sensitivity did not permit us to determine the reaction rate relative to the enzyme’s natural substrates. Nevertheless, this finding offers an opportunity to enzymatically synthesize a phosphonoacetate analogue of InsP8.
Our structural data revealed that it was the 1-phosphate of both 5-InsP7 and InsP6 that is closest to the γ-phosphate of ATP. Moreover, when human PPIP5K2KD crystals were soaked with ATP and 5-InsP7 substrate, we obtained complexes that contained ADP and 1,5-InsP8 as the products (Wang et al., 2012). These were the first published data to define the 1-kinase specificity of the PPIP5Ks. Previous work had successfully narrowed down the specificity of this reaction to either the 1- or 3-positions (Lin et al., 2009), but those two alternatives would yield products that are enantiomers, which had previously been difficult to distinguish between.
Interest in phosphoryl transfer crosses many disciplines because it is a universal mechanism for energy storage and utilization, as well as cellular communication and signaling. Yet this process operates in a variety of molecular scenarios, and there are substantial gaps in our understanding of the structural and mechanistic adaptations of key families of phosphoryl-transferases to these different situations. We gained direct insight into the reaction mechanism for PPIP5K2KD, by obtaining crystal complexes that contained both ADP and MgF3− (Wang et al., 2012). The latter matches the charge and geometry of the transition state of a phosphoryl transfer reaction (Graham et al., 2002). Indeed, in the active site, we detected MgF3− with the near planar geometry that mimics a trigonal bipyramidal phosphoryl transition state (Graham et al., 2002). The Mg atom separated the donor oxygen of ADP and the acceptor oxygen of the 1-phosphate of 5-InsP7 by a total distance of 4.2 Å (Wang et al., 2012). With a total co-ordination distance of less than 4.9 Å, a partly associative reaction mechanism is possible. That is, the new P-O bond can be formed by nucleophilic attack of the acceptor oxygen before the original P-O bond with the donor oxygen is broken (Mildvan 1997). Moreover, our structural data (Wang et al., 2012) revealed that the three negative charges of the pentacoordinate (phosphorane) transition state appear to be balanced by electrostatic interactions with two Mg2+ ions, a water molecule, and the positively charged side chains of K248 and R213. This snapshot of charge neutralization in the transition state helps explain how hPPIP5K2KD overcomes the energy barrier to efficient catalysis.
Finally, in the active site of the kinase the inositol ring of InsP6 and 5-InsP7 is presented perpendicular to the plane of the nucleotide’s β-phosphates, which avoids steric and electrostatic clashing between the nucleotide and the non-reacting oxygens on the 1-phosphate of the substrate (Wang et al., 2012). This innovative topology distinguishes PPIP5K2KD from all of the other inositol phosphate kinases that phosphorylate hydroxyl groups (Wang et al., 2012).
The IP6Ks face the same catalytic challenges that are overcome by the PPIP5Ks. To date, no structures of the IP6Ks have been published. Their amino acid sequences differ so substantially from the PPIP5Ks that alternative strategies may have evolved to overcome the energy barrier to PP-InsP synthesis. It will be interesting to determine what these differences are.
Acknowledgments
Work in the authors’ laboratory was supported by the Intramural Research Program of the NIH/National Institute of Environmental Health Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert C, Safrany ST, Bembenek ME, Reddy KM, Reddy KK, Falck JR, et al. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem J. 1997;327:553–560. doi: 10.1042/bj3270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Burton A, Ruiz-Mateos E, Marsh M, Saiardi A. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc Natl Acad Sci U S A. 2009;106:21161–21166. doi: 10.1073/pnas.0909176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CJ, Illies C, Gaboardi GC, Berggren PO. Inositol pyrophosphates: structure, enzymology and function. Cell Mol Life Sci. 2009;66:3851–3871. doi: 10.1007/s00018-009-0115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CJ, Wright J, Hughes PJ, Kirk CJ, Michell RH. Complex changes in cellular inositol phosphate complement accompany transit through the cell cycle. Biochem J. 2004;380:465–473. doi: 10.1042/BJ20031872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Onnebo SM, Azevedo C, Saiardi A. Inositol pyrophosphates: metabolism and signaling. Cell Mol Life Sci. 2006;63:552–564. doi: 10.1007/s00018-005-5446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggren PO, Barker CJ. A key role for phosphorylated inositol compounds in pancreatic beta-cell stimulus-secretion coupling. Adv Enzyme Regul. 2008;48:276–294. doi: 10.1016/j.advenzreg.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Best MD, Zhang H, Prestwich GD. Inositol polyphosphates, diphosphoinositol polyphosphates and phosphatidylinositol polyphosphate lipids: Structure, synthesis, and development of probes for studying biological activity. Nat Prod Rep. 2010 doi: 10.1039/b923844c. [DOI] [PubMed] [Google Scholar]
- Bhandari R, Juluri KR, Resnick AC, Snyder SH. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0712227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari R, Saiardi A, Ahmadibeni Y, Snowman AM, Resnick AC, Kristiansen TZ, et al. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc Natl Acad Sci U S A. 2007;104:15305–15310. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Hu X, Saiardi A. Are Inositol Pyrophosphates Signalling Molecules? J Cell Physiol. 2009;220:8–15. doi: 10.1002/jcp.21763. [DOI] [PubMed] [Google Scholar]
- Caffrey JJ, Safrany ST, Yang X, Shears SB. Discovery of Molecular and Catalytic Diversity Among Human Diphosphoinositol Polyphosphate Phosphohydrolases: An Expanding NUDT Family. J Biol Chem. 2000;275:12730–12736. doi: 10.1074/jbc.275.17.12730. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Kim S, Snyder SH. Inositol pyrophosphates as Mammalian cell signals. Sci Signal. 2011;4:re1. doi: 10.1126/scisignal.2001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, Juluri KR, et al. Inositol pyrophosphates inhibit akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain PP, Qian X, Stiles AR, Cho J, Jones DH, Lesley SA, et al. Integration of inositol phosphate signaling pathways via human ITPK1. J Biol Chem. 2007;282:28117–28125. doi: 10.1074/jbc.M703121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Williams J, Cho J, Falck JR, Shears SB. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem. 2007;282:30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Mollapour E, Choi JH, Shears SB. Cellular Energetic Status Supervises the Synthesis of Bis-Diphosphoinositol Tetrakisphosphate Independentlyof AMP-Activated Protein Kinase. Mol Pharmacol. 2008;74:527–536. doi: 10.1124/mol.107.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Mollapour E, Shears SB. Signal transduction during environmental stress: InsP8 operates within highly restricted contexts. Cell Signal. 2005;17:1533–1541. doi: 10.1016/j.cellsig.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Chu C, Alapat D, Wen X, Timo K, Burstein D, Lisanti M, et al. Ectopic expression of murine diphosphoinositol polyphosphate phosphohydrolase 1 attenuates signaling through the ERK1/2 pathway. Cell Signal. 2004;16:1045–1059. doi: 10.1016/j.cellsig.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Draskovic P, Saiardi A, Bhandari R, Burton A, Ilc G, Kovacevic M, et al. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem Biol. 2008;15:274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Fisher DI, Safrany ST, McLennan AG, Cartwright JL. Nudix hydrolases that degrade dinucleoside and diphosphoinositol polyphosphates also have 5-phosphoribosyl 1-pyrophosphate (PRPP) pyrophosphatase activity that generates the glycolytic activator ribose 1,5-bisphosphate. J Biol Chem. 2002;277:47313–47317. doi: 10.1074/jbc.M209795200. [DOI] [PubMed] [Google Scholar]
- Fridy PC, Otto JC, Dollins DE, York JD. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J Biol Chem. 2007;282:30754–30762. doi: 10.1074/jbc.M704656200. [DOI] [PubMed] [Google Scholar]
- Glennon MC, Shears SB. Turnover of inositol pentakisphosphates, inositol hexakisphosphate and diphosphoinositol polyphosphates in primary cultured hepatocytes. Biochem J. 1993;293:583–590. doi: 10.1042/bj2930583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NA, Zaremba A, Shears SB. Receptor-dependent compartmentalization of PPIP5K1, a kinase with a cryptic polyphosphoinositide binding domain. Biochem J. 2011;434:415–426. doi: 10.1042/BJ20101437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DL, Lowe PN, Grime GW, Marsh M, Rittinger K, Smerdon SJ, et al. MgF(3)(−) as a transition state analog of phosphoryl transfer. Chem Biol. 2002;9:375–381. doi: 10.1016/s1074-5521(02)00112-6. [DOI] [PubMed] [Google Scholar]
- Hand CE, Honek JF. Phosphate transfer from inositol pyrophosphates InsP5PP and InsP4(PP)2: a semi-empirical investigation. Bioorg Med Chem Lett. 2007;17:183–188. doi: 10.1016/j.bmcl.2006.09.066. [DOI] [PubMed] [Google Scholar]
- Herschlag D. The role of induced fit and conformational changes in specificity and catalysis. Bioorganic Chemistry. 1988;16:62–96. [Google Scholar]
- Hidaka K, Caffrey JJ, Hua L, Zhang T, Falck JR, Nickel GC, et al. An Adjacent Pair of Human NUDT Genes on Chromosome X are Preferentially Expressed in Testis and Encode Two New Isoforms of Diphosphoinositol Polyphosphate Phosphohydrolase. J Biol Chem. 2002;277:32730–32738. doi: 10.1074/jbc.M205476200. [DOI] [PubMed] [Google Scholar]
- Holmes W, Jogl G. Crystal structure of inositol phosphate multikinase 2 and implications for substrate specificity. J Biol Chem. 2006;281:38109–38116. doi: 10.1074/jbc.M606883200. [DOI] [PubMed] [Google Scholar]
- Hua LV, Green M, Warsh JJ, Li PP. Molecular cloning of a novel isoform of diphosphoinositol polyphosphate phosphohydrolase: A potential target for lithium therapy. Neuropsychopharmacology. 2001;24:640–651. doi: 10.1016/S0893-133X(00)00233-5. [DOI] [PubMed] [Google Scholar]
- Hua LV, Hidaka K, Pesesse X, Barnes LD, Shears SB. Paralogous murine Nudt10 and Nudt11 genes have differential expression patterns but encode identical proteins that are physiologically competent diphosphoinositol polyphosphate phosphohydrolases. Biochem J. 2003;373:81–89. doi: 10.1042/BJ20030142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illies C, Gromada J, Fiume R, Leibiger B, Yu J, Juhl K, et al. Inositol pyrophosphates determine exocytic capacity. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- Ingram SW, Safrany ST, Barnes LD. Disruption and overexpression of the Schizosaccharomyces pombe aps1 gene and the effects on growth rate, morphology, and intracellular diadenosine 5′, 5‴-P1, P5-pentaphosphate and diphosphoinositol polyphosphate concentrations. Biochem J. 2003;369:519–528. doi: 10.1042/BJ20020733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat Rev Mol Cell Biol. 2007a;8:995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]
- Lee YS, Huang K, Quiocho FA, O’Shea EK. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat Chem Biol. 2008;4:25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007b;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie NR, McLennan AG, Safrany ST. Cloning and characterization of hAps1 and hAps2, human diadenosine polyphosphate-metabolizing Nudix hydrolases. BMC Biochemistry. 2002;3:20. doi: 10.1186/1471-2091-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Fridy PC, Ribeiro AA, Choi JH, Barma DK, Vogel G, et al. Structural analysis and detection of biological inositol pyrophosphates reveals that the VIP/PPIP5K family are 1/3-kinases. J Biol Chem. 2009;284:1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, et al. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan AG. Decapitation: poxvirus makes RNA lose its head. Trends Biochem Sci. 2007;32:297–299. doi: 10.1016/j.tibs.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Menniti FS, Miller RN, Putney JW, Jr, Shears SB. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem. 1993;268:3850–3856. [PubMed] [Google Scholar]
- Mildvan AS. Mechanisms of Signaling and Related Enzymes. Proteins. 1997;29:401–416. [PubMed] [Google Scholar]
- Miller GJ, Wilson MP, Majerus PW, Hurley JH. Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase. Mol Cell. 2005;18:201–212. doi: 10.1016/j.molcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE, et al. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316:106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- Onnebo SM, Saiardi A. Inositol pyrophosphates modulate hydrogen peroxide signalling. Biochem J. 2009;423:109–118. doi: 10.1042/BJ20090241. [DOI] [PubMed] [Google Scholar]
- Pesesse X, Choi K, Zhang T, Shears SB. Signalling by higher inositolpolyphosphates: Synthesis of bis-diphosphoinositol tetrakisphosphate (“InsP8”) is selectively activated by hyperosmotic stress. J Biol Chem. 2004;279:43378–43381. doi: 10.1074/jbc.C400286200. [DOI] [PubMed] [Google Scholar]
- Ponnusamy S, Alderson NL, Hama H, Bielawski J, Jiang JC, Bhandari R, et al. Regulation of telomere length by fatty acid elongase 3 in yeast. Involvement of inositol phosphate metabolism and Ku70/80 function. J Biol Chem. 2008;283:27514–27524. doi: 10.1074/jbc.M802980200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Jia Y, Chakraborty A, Li Y, Jain SK, Zhong J, et al. Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat Immunol. 2011;12:752–760. doi: 10.1038/ni.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JP, Kulakova AN, Cooley NA, McGrath JW. New ways to break an old bond: the bacterial carbon-phosphorus hydrolases and their role in biogeochemical phosphorus cycling. Environ Microbiol. 2007;9:2392–2400. doi: 10.1111/j.1462-2920.2007.01397.x. [DOI] [PubMed] [Google Scholar]
- Riley AM, Deleu S, Qian X, Mitchell J, Chung SK, Adelt S, et al. On the contribution of stereochemistry to human ITPK1 specificity: Ins(1,4,5,6)P(4) is not a physiologic substrate. FEBS Lett. 2006;580:324–330. doi: 10.1016/j.febslet.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Riley AM, Wang H, Weaver JD, Shears SB, Potter BVL. First synthetic analogues of diphosphoinositol polyphosphates: interaction with PPIP5 kinase. Chem Commun. 2012 doi: 10.1039/c2cc36044f. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzene M, Pinna LA. Addiction to protein kinase CK2: a common denominator of diverse cancer cells? Biochim Biophys Acta. 2010;1804:499–504. doi: 10.1016/j.bbapap.2009.07.018. [DOI] [PubMed] [Google Scholar]
- Safrany ST, Caffrey JJ, Yang X, Bembenek ME, Moyer MB, Burkhart WA, et al. A novel context for the “MutT” module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 1998;17:6599–6607. doi: 10.1093/emboj/17.22.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Bhandari A, Resnick R, Cain A, Snowman AM, Snyder SH. Inositol Pyrophosphate: Physiologic Phosphorylation of Proteins. Science. 2004;306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The Inositol Hexakisphosphate Kinase Family: Catalytic Flexibility, and Function in Yeast Vacuole Biogenesis. J Biol Chem. 2000;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Nagata E, Luo HR, Snowman AM, Snyder SH. Identification and characterization of a novel inositol hexakisphosphate kinase. J Biol Chem. 2001;276:39179–39185. doi: 10.1074/jbc.M106842200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length via PI3K-related protein kinases. Proc Nat Acad Sci USA. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeds AM, Sandquist JC, Spana EP, York JD. A molecular basis for inositol polyphosphate synthesis in Drosophila melanogaster. J Biol Chem. 2004;279:47222–47232. doi: 10.1074/jbc.M408295200. [DOI] [PubMed] [Google Scholar]
- Shears SB. How versatile are inositol phosphate kinases? Biochem J. 2004;377:265–280. doi: 10.1042/BJ20031428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB. Diphosphoinositol polyphosphates: metabolic messengers? Mol Pharmacol. 2009;76:236–252. doi: 10.1124/mol.109.055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB, Gokhale NA, Wang H, Zaremba A. Diphosphoinositol polyphosphates: what are the mechanisms? Adv Enzyme Regul. 2011;51:13–25. doi: 10.1016/j.advenzreg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb H, Irvine RF, Berridge MJ, Schulz I. Release of Ca2+ from a nonmitochondrial store in pancreatic cells by inositol-1,4,5-trisphosphate. Nature. 1983;306:67–68. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- Szijgyarto Z, Garedew A, Azevedo C, Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334:802–805. doi: 10.1126/science.1211908. [DOI] [PubMed] [Google Scholar]
- Thorsell AG, Persson C, Graslund S, Hammarstrom M, Busam RD, Hallberg BM. Crystal structure of human diphosphoinositol phosphatase 1. Proteins. 2009;77:242–246. doi: 10.1002/prot.22489. [DOI] [PubMed] [Google Scholar]
- Wang H, Falck JR, Hall TM, Shears SB. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat Chem Biol. 2012;8:111–116. doi: 10.1038/nchembio.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox RA, Safrany ST, Lampe D, Mills SJ, Nahorski SR, Potter BVL. Modification at C2 of myo-inositol 1,4,5-trisphosphate produces inositol trisphosphates and tetrakisphosphates with potent biological activities. Eur J Biochem. 1994;223:115–124. doi: 10.1111/j.1432-1033.1994.tb18972.x. [DOI] [PubMed] [Google Scholar]
- Yang X, Safrany ST, Shears SB. Site-directed mutagenesis of DIPP, a dual specificity MutT/Nudix-type hydrolase that attacks diadenosine polyphosphates and diphosphoinositol polyphosphates. J Biol Chem. 1999;274:35434–35440. doi: 10.1074/jbc.274.50.35434. [DOI] [PubMed] [Google Scholar]
- York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]