SUMMARY
Transcription activator-like effector nucleases (TALENs) are a new class of engineered nucleases that are easier to design to cleave at desired sites in a genome than previous types of nucleases. We report the use of TALENs to rapidly and efficiently generate mutant alleles of 15 genes in cultured somatic cells or human pluripotent stem cells, the latter of which we differentiated both the targeted lines and isogenic control lines into various metabolic cell types. We demonstrate cell-autonomous phenotypes directly linked to disease—dyslipidemia, insulin resistance, hypoglycemia, lipodystrophy, motor neuron death, and hepatitis C infection. We find little evidence of TALEN off-target effects, but each clonal line nevertheless harbors a significant number of unique mutations. Given the speed and ease with which we were able to derive and characterize these cell lines, we anticipate TALEN-mediated genome editing of human cells becoming a mainstay for the investigation of human biology and disease.
INTRODUCTION
The study of human disease has been facilitated by the ability to identify responsible gene mutations; at the same time, it has been hampered by the lack of an inexhaustible supply of easily accessible tissues from patients bearing those mutations. Another limitation is that many gene mutations that would be informative for disease biology if they could be studied in isolated cells are incompatible with human life (i.e., embryonic lethal). Classical gene targeting technology via homologous recombination has proven to be an invaluable tool of experimental biology through its use in mouse embryonic stem cells to generate germline knockout and knock-in mice; however, its use in mammalian systems has been limited primarily to studies in mice. In many cases, mice do not faithfully phenocopy human physiology and disease, e.g., cholesterol metabolism, coronary artery disease, and human hepatitis C virus (HCV) infection. The emergence of genome editing with engineered nucleases, as well as human pluripotent stem cell (hPSC) technology and differentiation protocols to obtain a variety of cell and tissue types in vitro, now make it possible to rapidly interrogate the effects of genetic modification in otherwise isogenic human model systems.
Transcription activator-like effector nucleases (TALENs) are a new class of engineered nucleases that due to their modular domain structure have proven more straightforward to design and construct to perform genome editing than other types of nucleases (Bogdanove and Voytas, 2011). TALENs are typically designed as a pair that bind to genomic sequences flanking a target site and generate a double-strand break (DSB), which is repaired by the cell using either homology-directed repair (HDR) or the error-prone process of non-homologous end-joining (NHEJ) (Christian et al., 2010; Li et al., 2011; Miller et al., 2011; Hockemeyer et al., 2011). NHEJ can be exploited to introduce small insertions or deletions (indels) resulting in frameshift mutations that effectively knock out a protein-coding gene. An exogenously introduced double-stranded DNA or single-stranded DNA oligonucleotide (ssODN) can serve as a repair template for HDR to incorporate an alteration into the genome (Soldner et al., 2011). In principle, TALEN pairs can be generated de novo with standard molecular biology techniques in a matter of days (Cermak et al., 2011; Sanjana et al., 2012). To demonstrate the utility, efficiency, and rapidity of TALEN technology in generating human cellular models with which to derive new biological insights, we created mutations in 15 genes and performed detailed phenotypic analysis of four genes for which novel roles in disease biology have emerged in recent years—APOB, SORT1, AKT2, and PLIN1.
RESULTS
Modular Assembly and Use of TALENs for Efficient and Rapid Genome Editing
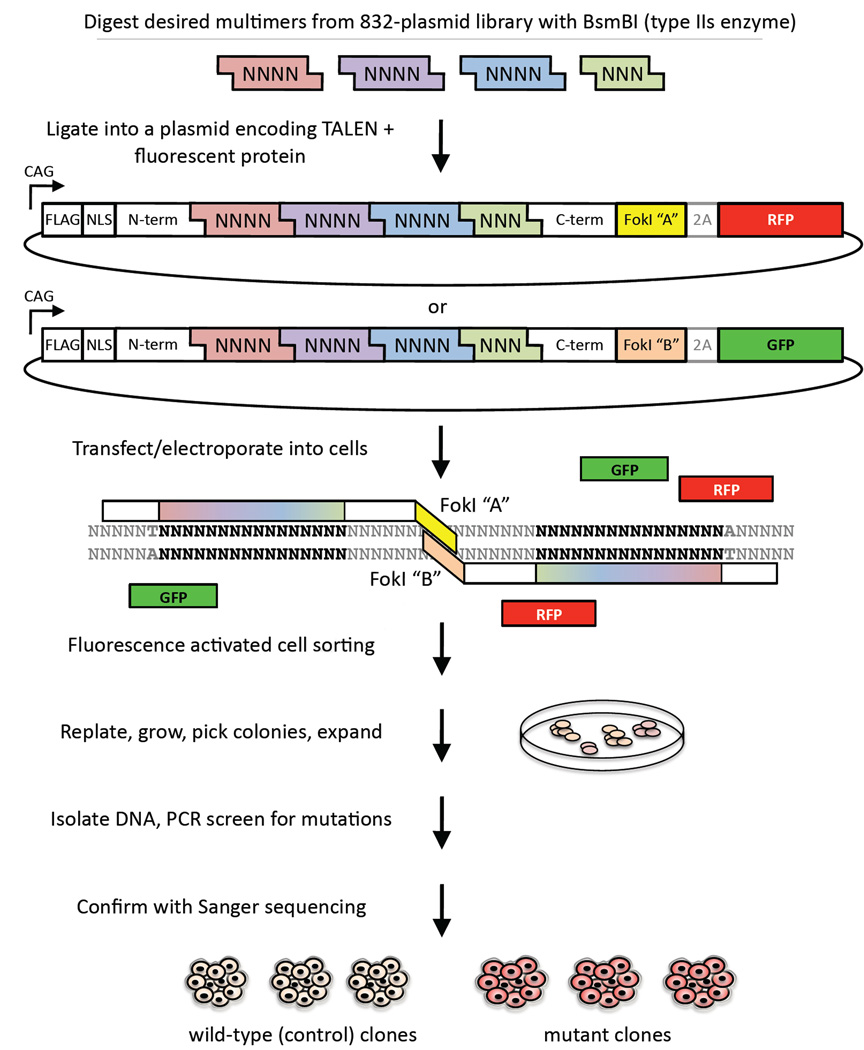
The DNA-binding domain of a TALEN comprises an array of 33- to 35-amino-acid monomers that are “coded” to recognize and bind specific DNA basepairs in a 1:1 fashion (Moscou and Bogdanove, 2009; Boch et al., 2009). We built upon previously described modular Golden Gate metholodogies to allow assembly of multiple DNA fragments in an ordered fashion (Li et al., 2011; Cermak et al., 2011) such that a single ligation of pre-assembled tetramers/trimers generates TALENs that recognize any 15-bp recognition site in the genome (Figure 1 and Figure S1). This assembly method requires only 1–2 days for completion and is not prone to errors that complicate methods that rely on polymerase chain reaction (PCR) amplification of monomers. Further, we have developed a set of optimized vectors and methods for the delivery of TALENs into mammalian cells and, in particular, hPSCs. Briefly, we transfect or electroporate TALEN pairs into cells and then subject them to fluorescence activated cell sorting (FACS) 48 hours post-transfection based on fluorescent marker expression. We replate the sorted cells at low density and allow them to recover and grow for 1 week, resulting in the formation of distinct single colonies. Colonies are expanded, genomic DNA purified, and mutations analyzed by PCR, agarose gel screening, and Sanger sequencing (Figure 1 and Figure S1). The entire process from start to finish can be completed in less than one month.
Figure 1. Schematic of System for Efficient and Rapid Genome Editing with TALENs.
GFP = green fluorescent protein; RFP = red fluorescent protein. See also Figure S1.
Utilizing these methods we generated TALEN pairs to target 16 distinct sites in 15 genes in human somatic cell lines, human embryonic stem cell (hESC) lines, or human induced pluripotent stem cell (iPSC) lines; the alterations included a variety of knockout mutations as well as a knock-in missense mutation and a functional frameshift mutation (Table 1). We observed that the efficiency of mutation varied by genomic location as well as among different cell lines, with indels from NHEJ occurring in roughly 2% to 34% of clones screened and the efficiency of knock-in by HDR occurring at a frequency of 1.6%. We then proceeded to perform detailed phenotypic analyses of cells harboring mutations in four human disease-related genes—APOB, SORT1, AKT2, and PLIN1.
Table 1.
Targeting Efficiency of TALENs at 16 Loci in 15 Genes in Various Cell Types
| Gene | Target Sequence (Flanked by Underlined TALEN Binding Sites) | Cell Linea | Number of Clones Screened |
Number of Mutants |
Efficiency of Mutation |
|---|---|---|---|---|---|
| AKT2 | TCCCTTCCTGCCTCATTTCAGGTGAATACATCAAGACCTGGAGGCCA | HUES 9 | 192 | 17 | 8.9% |
| AKT2 E17K | TCCCTTCCTGCCTCATTTCAGGTGAATACATCAAGACCTGGAGGCCA | HUES 9 | 192 | 3b | 1.6% |
| ANGPTL3 | TCAAAACTTGAAAGCCTCCTAGAAGAAAAAATTCTACTTCAACAAAA | HUES 9 | 424 | 84 | 19.8% |
| APOB | TAAGCGACTGGCTGCCTATCTTATGTTGATGAGGAGTCCTTCACA | HuH-7 | 126 | 9 | 7.1% |
| ATGL | TTGCTGGCGTTGCCCCCCGCCCGCCCCCACGGCCCAGAGGACAAGGA | HUES 9 | 192 | 28 | 14.6% |
| C6orf106 | TGGAGGGCATGGACGTAGACCTGGACCCGGAGCTGATGCAGAAGTTCA | HUES 9 | 216 | 13 | 6.0% |
| CIITA | TAACAGCGATGCTGACCCCCTGTGCCTCTACCACTTCTATGACCAGA | BJRiPS | 292 | 37 | 12.7% |
| CELSR2 | TGCTGGCTCGGCTGCCCTGAGGTTGCTCAATCAAGCACAGGTTTCAA | HUES 1 | 506 | 18 | 3.5% |
| CFTR | TGGAATCACACTGAGTGGAGGTCAACGAGCAAGAATTTCTTTAGCA | CF-RiPS | 140 | 3 | 2.1% |
| GLUT4 | TGGTCCTTGCTGTGTTCTCTGCGGTGCTTGGCTCCCTGCAGTTTGGGTA | HUES9 | 155 | 52 | 33.5% |
| LINC00116 | TCAGAGAGGACACTGCAGTTGTCCGTGCTAGTAGCCTTCGCTTCTGGA | HUES 9 | 88 | 26 | 29.5% |
| NLRC5 | TGGAGCTGTCTTGTGAGGCTGCTCACCAAAGACCCAGAATGGCTGAA | BJ-RiPS | 83 | 6 | 7.2% |
| PLIN1 | TGACAACGTGGTGGACACAGTGGTGCATTACGTGCCGGTGAGTACCA | HUES 9 | 293 | 70 | 23.8% |
| PLIN1 | TGACAACGTGGTGGACACAGTGGTGCATTACGTGCCGGTGAGTACCA | BJ-RiPS | 439 | 29 | 6.6% |
| SORT1 (exon 2) | TGATGATCTCAGAGGCTCAGTATCCTTGTCCTGGGTTGGAGATAGCA | HUES 1 | 576 | 128 | 22.2% |
| SORT1 (exon 3) | TGGTAATTATGACTTTTGGACAGTCCAAGCTATATCGAAGGTGAGATCA | HUES 9 | 192 | 21 | 10.9% |
| TRIB1 | TCTTCAAGCAGATTGTCTCCGCCGTCGCCCACTGCCACCAGTCAGCCA | HUES 9 | 169 | 41 | 24.2% |
| TTN | TTGAGGCTCCTTAATGATGACAGAAGAGAAGGCTTCTCTGGGTTCA | BJ-RiPS | 250 | 17 | 6.8% |
HUES 1 and HUES 9 are human embryonic stem cell lines (Cowan et al., 2004); BJ-RiPS and CF-RiPS are induced pluripotent stem cell lines (Warren et al., 2010); HuH-7 cells are cultured human hepatocellular carcinoma cells
Successfully inserted E17K knock-in mutation using single-stranded DNA oligonucleotide
APOB is Required for HCV Replication
APOB, which encodes apolipoprotein B, the core protein of very-low-density lipoprotein (VLDL) and low-density lipoprotein (LDL) particles that transport cholesterol and triglycerides from the liver to other tissues via the bloodstream, has been suggested to play a critical role in hepatitis C virus (HCV) infection. In HCV models using cultured human HuH-7 hepatoma cells, RNA interference resulting in partial knockdown of APOB expression has been reported to reduce HCV secretion, albeit not HCV replication (Huang et al., 2007; Nahmias et al., 2008); however, another report has suggested that apolipoprotein E, but not apolipoprotein B, is necessary for HCV production (Jiang and Luo, 2009). Thus, the importance of APOB and precise points of interaction with the HCV lifecycle remain to be determined. We sought to address this question by generating APOB knockout HuH-7 cells.
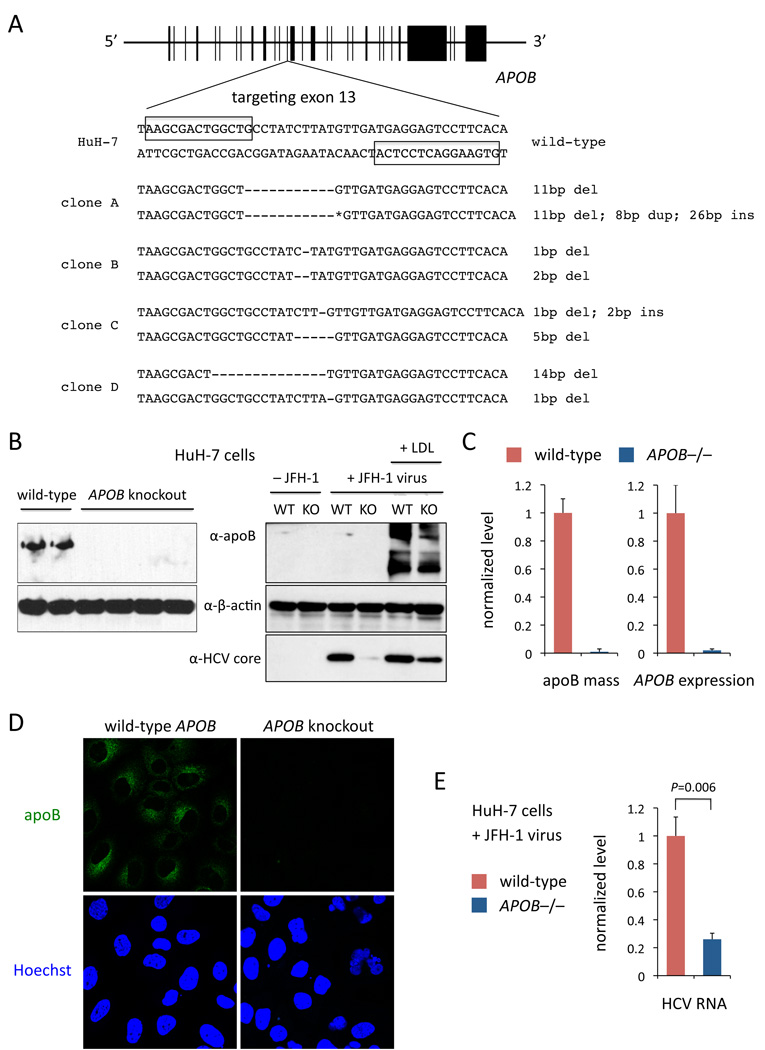
The human APOB gene encodes a 512 kDa protein termed apoB-100. We designed a TALEN pair targeting a site in exon 13 (Figure 2A); frameshift mutations at the site would generate truncated proteins about 12.5% of the size of apoB-100 (apoB-12.5). We transfected a clonal line of HuH-7 with high expression of CD81 (a co-receptor for HCV entry; HuH-7/CD81high) with the APOB TALEN pair. Following FACS with a co-translated fluorescent marker, replating of sorted cells at limiting dilution, and expansion of single clones, we found that of 126 screened clones, indels were present in nine clones (Figure S2A), of which four had exon 13 frameshift mutations in both alleles (Figure 2A). Compared to wild-type controls from the same set of screened clones, APOB knockout cells had no detectable intracellular apoB protein, no secreted apoB mass in the media, and <3% APOB mRNA expression, consistent with nonsense-mediated mRNA decay (Figures 2B, 2C, and 2D).
Figure 2. APOB is Important for HCV Replication.
(A) Generation of APOB knockout HuH-7 clones with TALENs targeting exon 13. Boxes indicate the TALEN binding sites. Deletions, insertions, and duplications in the two alleles of each clone are indicated. The 26-bp insertion and 8-bp duplication (asterisk) in clone A are 5’-GAGTCGCTTCTCCGGGAGATAAGTCA-3’ and 5’-GACTGGCT-3’, respectively.
(B) Left panel, Western blot using whole cell lysates from two wild-type and four knockout HuH-7 cell lines (clones A–D). Right panel, Western blot from a wild-type clone and a knockout clone (clone A) infected with or without JFH-1 virus and incubated with or without LDL particles. The same wild-type clone and knockout clone (clone A) were used for all subsequent experiments.
(C) Left panel, apoB ELISAs performed on conditioned media from cells; values are normalized to level from wild-type clone. Right panel, APOB mRNA expression by qRT-PCR from whole cell lysates; expression is indicated as fold change of 2−ΔΔCt with reference to 18S rRNA, normalized to level in wild-type clone.
(D) Immunocytochemistry for apoB.
(E) HCV RNA levels by qRT-PCR from clones infected with JFH-1 virus; expression is indicated as fold change of 2−ΔΔCt with reference to GAPDH, normalized to level in wild-type clone.
Error bars show s.e.m. from experiments with biological replicates, N = 3. P values calculated with unpaired t test. See also Figure S2.
We infected APOB−/− and wild-type cells with the tissue-culture-infectious HCV strain JFH-1. The APOB−/− cells had significantly lower intracellular HCV RNA levels (74% reduction, P = 0.006), with minimal detectable HCV core protein (Figures 2B and 2E). Reintroduction of apoB-100 protein into the APOB−/− cells by adding LDL particles to the media, allowing for cellular LDL uptake, resulted in partial restoration of HCV core protein levels, arguing that the HCV replication defect was the result of loss of APOB function rather than an off-target effect of the TALENs (i.e., mutagenesis at other sites in the genome) (Figure 2B). Together, these data suggest that apoB-100 is integral to the HCV viral lifecycle and that APOB-targeting therapeutics (e.g., mipomersen) may have efficacy in treating HCV-infected patients.
Isogenic Disease Models in hPSCs
We found that the karyotype of the HuH-7 cells was severely abnormal (Figure S2B)—fortuitously, it harbored two APOB alleles, in contrast to SORT1, with at least five alleles—highlighting the disadvantages of cultured tumor cell lines for rigorous genetic studies. hPSCs offer several advantages: they can maintain stable genomes with normal karyotypes while propagated in culture (Figure S2B), preserving correct gene dosage; they can be differentiated into a variety of cell types, extending studies beyond a single cell type; and they can yield human cell types that are not available as cultured cell lines, e.g., adipocytes and motor neurons.
These advantages are mitigated by the significant variability in differentiation capacity and phenotypic characteristics among different hPSC lines, particularly among iPSC lines. This variability is attributed to differences in genetic background, in epigenetic state, and in derivation of the cell lines and adaptation to culture, among other factors. In this variability lies the potential for confounding of any phenotypic differences observed among differentiated cell lines generated to serve as disease models or controls—a significant weakness of studies in which a few iPSC lines from patients are compared with a few iPSC lines from healthy individuals, as has been the case with most published studies to date, since any observed differences cannot be reliably attributed to the effects of disease mutations. We demonstrated this cell line-to-cell line variability by differentiating two hESC lines, HUES 1 and HUES 9, into hepatocyte-like cells (HLCs) using an adapted protocol (Si-Tayeb et al., 2010) (Figure S3). We found that there were significant differences in the amounts of apoB and albumin secreted by the two cell lines and retained in the media (Figure S4A); when apoB mass was normalized to albumin mass, there was a two-fold difference between the two lines (P = 0.0001).
Using genome editing to generate isogenic cell lines that differ only with respect to a single mutation of interest provides a superior study design, since the cell lines would have the same origin and would thus be matched in genetic background, epigenetic state, differentiation capacity, derivation and adaptation to culture, etc. This would minimize confounding of the experiment and allow for more confidence in concluding that any phenotypic differences are secondary to the mutation. For these reasons, our subsequent studies were all performed in genome-edited hESCs.
SORT1 Mediates Diverse Cellular Functions in Hepatocytes, Adipocytes, and Neurons
SORT1 (encoding sortilin) was recently discovered by genome-wide association studies to regulate human blood LDL cholesterol levels and risk for coronary artery disease, via the modulation of the hepatic secretion of apoB-100-containing particles into the bloodstream; however, conflicting studies in humans and mice disagree about the direction of the effect of sortilin on apoB secretion (Musunuru et al., 2010; Kjolby et al., 2010). Human genetic studies have found that single nucleotide polymorphisms (SNPs) associated with increased hepatic SORT1 expression are also associated with decreased blood LDL cholesterol levels (Musunuru et al., 2010). Knockdown and overexpression of Sort1 in mouse liver suggested that sortilin functions to decrease hepatocyte apoB secretion (Musunuru et al., 2010). In contrast, a study of Sort1 knockout mice suggested that sortilin increases hepatocyte apoB secretion (Kjolby et al., 2010).
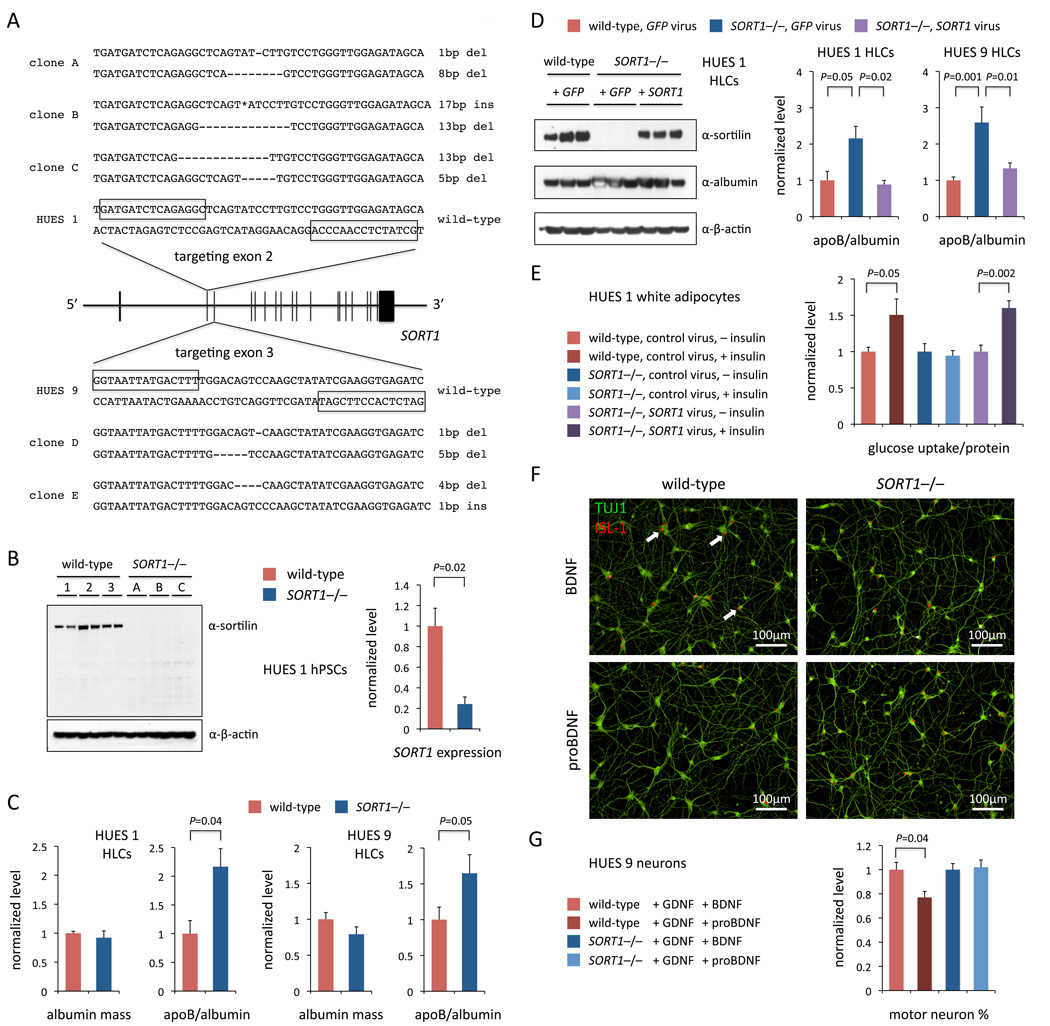
We targeted exon 2 in the hESC line HUES 1 and, in a single round of TALEN targeting, generated three clones that were compound heterozygous for frameshift mutations (out of 576 clones screened) and confirmed that they lacked sortilin protein (Figures 3A and 3B). In parallel, we targeted exon 3 in the hESC line HUES 9 and obtained two knockout clones (out of 192 clones screened). We differentiated two SORT1−/− and two wild-type HUES 1 clones or two SORT1−/− and two wild-type HUES 9 clones into hepatocyte-like cells (HLCs) using an adapted protocol (Si-Tayeb et al., 2010) (Figure S3). Measuring the levels of apoB as well as albumin and apoA-I (reference controls) secreted from the HLCs and retained in the media, we found that knockout cells had significantly increased apoB mass (HUES 1: 117% increase in apoB/albumin ratio, P = 0.04; HUES 9: 65% increase in apoB/albumin ratio, P = 0.05) (Figure 3C and Figure S4B). We infected knockout HUES 1 HLCs or HUES 9 HLCs with a lentivirus expressing the SORT1 cDNA or a control lentivirus and found that reconstitution of SORT1 to the levels observed in wild-type HUES 1 or HUES 9 HLCs resulted in normalization of the apoB mass (Figure 3D and Figure S4C), confirming that the observed differences in apoB mass are specific to SORT1 function and not the result of off-target effects. We found that secreted levels of additional hepatic proteins—ANGPTL4, ANGPTL6, HGF, and FGF-19—did not differ among the various experimental conditions (Figure S4D), nor did mRNA levels of APOB and other lipid-related genes such as HMGCR, LDLR, and SREBP1 (Figure S4E). Our data suggest that, in humans, sortilin acts in hepatocytes to reduce apoB-containing particle levels in the blood, resulting in lower cholesterol levels and reduced risk of coronary artery disease—consistent with human genetic studies (Musunuru et al., 2010) and, notably, contradicting the results reported from Sort1 knockout mice (Kjolby et al., 2010).
Figure 3. SORT1 Reduces Hepatocyte Secreted ApoB Mass, is Important for Insulin-Responsive Glucose Transport in Adipocytes, and Mediates proBDNF-Induced Motor Neuron Death.
(A) Generation of SORT1 knockout hPSC clones with TALENs targeting exon 2 or exon 3. Boxes indicate the TALEN binding sites. Deletions and insertions in the two alleles of each clone are indicated. The 17-bp insertion (asterisk) in clone B is 5’-TGCTATCTCCAACCAGG-3’.
(B) Western blot for sortilin and qRT-PCR for SORT1 mRNA in wild-type and knockout HUES 1 clones (clones A–C); mRNA expression is indicated as fold change of 2−ΔΔCt with reference to 18S rRNA, normalized to mean level in wild-type clones.
(C) Albumin and apoB mass measured by ELISA in media collected from wild-type and knockout HLCs (two clones each; A and B for HUES 1, D and E for HUES 9), normalized to mean levels in wild-type HLCs. N = 3 for HUES 1, N = 6 for HUES 9.
(D) Western blots of lysates and ELISAs in media from wild-type and knockout HLCs (one clone each, A for HUES 1; two clones each, D and E for HUES 9) infected with SORT1- or GFP-expressing lentivirus, normalized to mean levels in wild-type HLCs. N = 2 for HUES 1, N = 6 for HUES 9.
(E) Ratios of glucose uptake to total protein content in wild-type and knockout HUES 1 adipocytes (one clone each; clone A) infected with SORT1-expressing or control lentivirus and treated with or without insulin, all normalized to mean ratio in wild-type adipocytes without insulin. N = 6.
(F) Immunocytochemistry for TUJ1 and ISL-1 in wild-type and knockout HUES 9 motor neurons. Arrows indicate representative double-positive cells.
(G) Counts of wild-type and knockout HUES 9 (2 clones each, D and E) motor neurons (TUJ1/ISL-1 double-positive cells) treated with BDNF vs. proBDNF. N = 12.
Error bars show s.e.m. from experiments with biological replicates. P values calculated with unpaired t test. See also Figure S3, Figure S4, and Figure S5.
SORT1 has also been suggested to play an important role in regulating blood glucose levels by modulating insulin-dependent translocation of the fat- and muscle-specific glucose transporter, Glut4, to the plasma membrane via the formation and transport of Glut4 storage vesicles, based on studies in cultured mouse 3T3-L1 cells (Shi and Kandror, 2005). We differentiated two SORT1−/− and two wild-type HUES 1 clones into white adipocytes using a recently published protocol (Ahfeldt et al., 2012), and we observed a substantial increase in glucose uptake in wild-type adipocytes upon treatment with insulin (63% increase, P = 0.009) but not in SORT1−/− adipocytes (Figure S5A). We infected the knockout adipocytes with a SORT1 or control lentivirus and found that reconstitution of SORT1 restored insulin-responsive glucose uptake (60% increase, P = 0.002), confirming that the loss of insulin response in the knockout adipocytes was specific to SORT1 function and not the result of off-target effects (Figure 3E and Figure S5B). Thus, SORT1 appears to be critical for insulin-responsive glucose uptake in human adipocytes and may play a role in insulin sensitivity in humans.
Finally, SORT1 has also been implicated in the viability and function of neurons (Nykjaer and Willnow, 2012). In motor neurons, sortilin has been found to regulate neuronal survival during a temporally and spatially specific period of programmed cell death. Specifically, induction of motor neuron cell death by the pro-form of brain-derived neurotrophic factor (proBDNF) has been reported to be dependent on the presence of sortilin (Teng et al., 2005; Taylor et al., 2011). We differentiated two SORT1−/− and two wild-type HUES 9 clones into TUJ1+/ISL-1+ motor neurons using an adapted protocol (Di Giorgio et al., 2008; Chambers et al., 2009) and observed that while both SORT1−/− and wild-type hPSCs generated similar numbers of motor neurons (Figure S5C), wild-type motor neurons exhibited a substantial reduction after three days of proBDNF treatment (23% reduction, P = 0.004), whereas SORT1−/− motor neurons were unaffected (Figures 3F and 3G). These data agree with the reported requirement of SORT1 for proBDNF-induced programmed cell death in human motor neurons.
AKT2 Regulates Insulin Signaling and Glucose Metabolism in Hepatocytes and Adipocytes
The human AKT2 gene (encoding serine/threonine-protein kinase AKT2/PKBβ) has also been implicated in the regulation of insulin sensitivity. Loss of function of AKT2 in humans has been reported to result in severe insulin resistance as well as decreased body fat and partial lipodystrophy attributed to reduced adipocyte differentiation (George et al., 2004; Agarwal and Garg, 2006), and Akt2 knockout mice are resistant to the effects of insulin on glucose metabolism in liver and muscle and manifest lipoatrophy (Cho et al., 2001; Garofalo et al., 2003). Recently, three patients with severe hypoglycemia, hypoinsulinemia, and increased body fat were reported to bear a missense mutation in AKT2, p.Glu17Lys (E17K) (Hussain et al., 2011). Although the function of the mutant AKT2 E17K protein was assessed by heterologous overexpression studies in cultured cell lines (HeLa and 3T3-L1) and interpreted as being activated, the inability to study a physiologically relevant phenotype (e.g., glucose metabolism) in physiologically relevant tissues (e.g., human liver) precluded the conclusion that the AKT2 mutation was causal for the metabolic disorder in the patients.
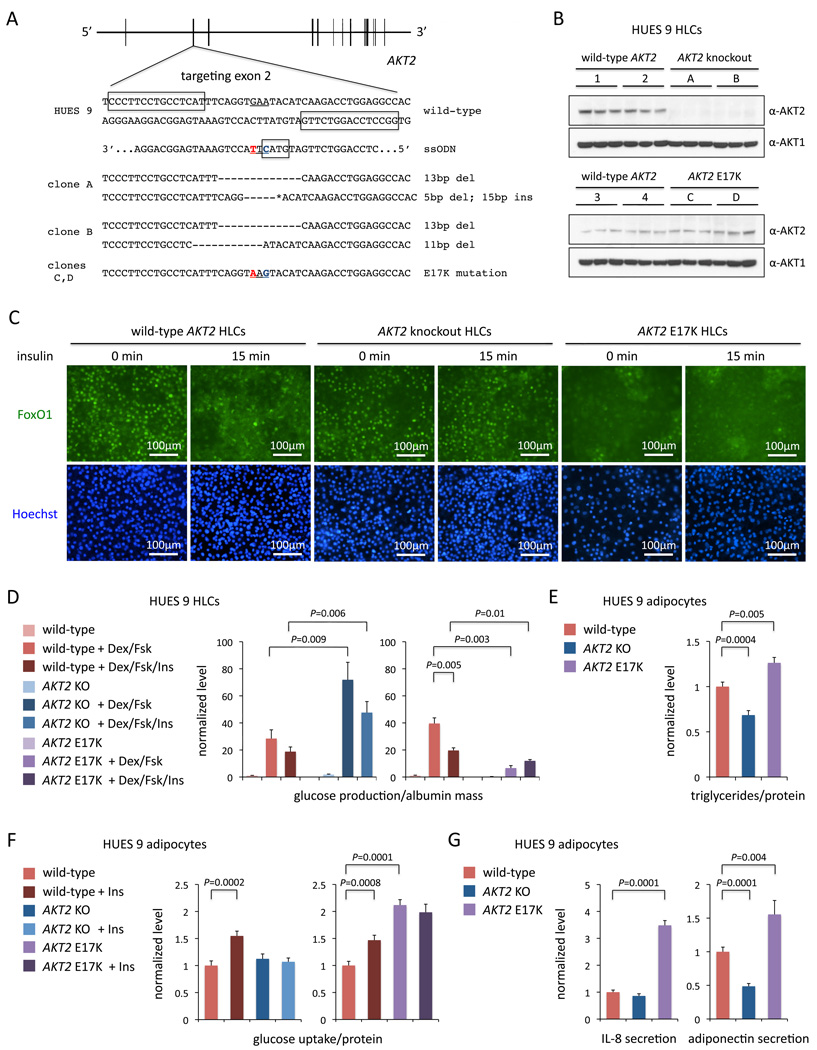
We sought to unequivocally establish a dominant, activated function of the AKT2 E17K mutant on glucose metabolism by generating an allelic series of isogenic hPSC lines with wild-type AKT2, knockout of AKT2, or a single AKT2E17K allele. We designed TALENs to target the site of the E17K mutation in the second coding exon (Figure 4A). In one round of targeting of HUES 9 cells with the TALEN pair alone, we obtained 17 clones with indels (out of 192 clones screened), none of which was compound heterozygous for frameshift mutations. A second round of TALEN targeting with a clone with one frameshift allele yielded two clones compound heterozygous for frameshift mutations (out of 96 clones screened). In parallel, we co-electroporated wild-type HUES 9 cells with the TALEN pair and a 67-nt antisense ssODN harboring the E17K missense variant, yielding three AKT2E17K heterozygous clones (out of 192 clones screened) (Figure S6A).
Figure 4. AKT2 E17K is a Dominant, Activating Mutation.
(A) Generation of AKT2 knockout and E17K knock-in hPSC clones with TALENs targeting exon 2. The underline indicates the codon encoding E17K change. Boxes in wild-type sequence indicate the TALEN binding sites. The box in the ssODN sequence indicates the RsaI restriction site created by the synonymous mutation in blue; the nucleotide in red indicates the missense mutation for E17K. Deletions and insertions in the two alleles of each clone are indicated. The 15-bp insertion (asterisk) in clone A is 5’-GACCTCCAGGTCCTG-3’.
(B) Western blot for AKT2 and AKT1 in wild-type, knockout, and E17K HUES 9 HLCs (A and B for knockout; C and D for E17K).
(C) Immunocytochemistry for FoxO1 in HLCs (clones A and C) at baseline and after 15 min insulin stimulation.
(D) Ratios of glucose production to secreted albumin mass (ELISA) in wild-type, knockout, and E17K HUES 9 HLCs (two clones each; A and B for knockout; C and D for E17K) treated with or without dexamethasone and forskolin with or without insulin, all normalized to mean ratios in wild-type HLCs without additives. Left panel, N = 4; right panel, N = 2.
(E) Ratios of triglyceride content to total protein content in wild-type, knockout, and E17K HUES 9 adipocytes (two clones each; A and B for knockout; C and D for E17K), normalized to mean ratio in wild-type adipocytes. N = 5.
(F) Ratios of glucose uptake to total protein content in wild-type, knockout, and E17K HUES 9 adipocytes (two clones each; A and B for knockout; C and D for E17K) treated with or without insulin, all normalized to mean ratio in wild-type adipocytes without insulin. N = 6.
(G) IL-8 and adiponectin mass measured by ELISA in media collected from wild-type, knockout, and E17K HUES 9 adipocytes (two clones each; A and B for knockout; C and D for E17K), normalized to mean levels from wild-type adipocytes. N = 4.
Error bars show s.e.m. from experiments with biological replicates. P values calculated with unpaired t test. See also Figure S3 and Figure S6.
We differentiated the allelic series of hPSC clones (two clones each) into HLCs. No AKT2 protein was apparent in the knockout cells, with comparable levels of AKT2 observed in the wild-type and E17K cells (Figure 4B). We assessed the regulation of the FoxO1 transcription factor, an AKT2 substrate that upon phosphorylation is translocated from the nucleus to the cytoplasm. In wild-type HLCs, FoxO1 was predominantly nuclear at baseline and cytoplasmic after insulin stimulation; in AKT2−/− HLCs, predominantly nuclear both at baseline and after stimulation; in AKT2E17K HLCs, predominantly cytoplasmic both at baseline and after stimulation (Figure 4C). We assessed glucose production in the allelic series of HLCs and found that with all three genotypes, addition of dexamethasone and forskolin to the media dramatically increased glucose production; the further addition of insulin decreased glucose production in the wild-type HLCs but not in the mutant HLCs (Figure 4D). In all media conditions, glucose production was significantly higher in AKT2−/− HLCs and lower in AKT2E17K HLCs compared to wild-type HLCs. Similar trends were observed in the mRNA expression levels of two genes involved in gluconeogenesis, G6PC and PCK1 (Figure S6B).
We also differentiated the AKT2 allelic series of hPSC clones into white adipocytes and found that AKT2−/− adipocytes had significantly decreased triglyceride content (32% reduction, P = 0.0004) and AKT2E17K adipocytes had significantly increased triglyceride content (26% increase, P = 0.005) (Figure 4E), consistent with the fat-related phenotypes observed in patients with AKT2 mutations. We observed a substantial increase in glucose uptake in wild-type adipocytes upon treatment with insulin (~50% increase in two different experiments) but, as with SORT1−/− adipocytes, we observed no significant change in glucose uptake in AKT2−/− adipocytes with insulin (Figure 4F). In contrast, AKT2E17K adipocytes displayed higher glucose uptake at baseline compared to wild-type adipocytes (111% increase, P = 0.0001); upon treatment with insulin, there was no further increase in glucose uptake, presumably because the cells were in a constitutively active state with respect to insulin signaling (Figure 4F). In the same vein, AKT2E17K adipocytes displayed substantially increased secretion of inflammatory adipokines such as IL-8 (Figure 4G), MCP1, and PAI-1 (Figure S6C). Finally, AKT2−/− and AKT2E17K adipocytes showed decreased and increased secretion of adiponectin, respectively (Figure 4G).
The opposing effects of the knockout and AKT2E17K alleles, in addition to indicating that the effects were specific to AKT2 function and not the result of off-target effects, establish that E17K is indeed a dominant, activating mutation in AKT2 and causal for the hypoglycemia and increased body fat observed in the three patients.
PLIN1 Frameshift Mutations Dominantly Alter Lipolysis in Adipocytes
PLIN1 encodes the protein perilipin, the most abundant protein coating lipid droplets in adipocytes, where it is required for droplet formation and maturation, optimal triglyceride storage, and the release of free fatty acids from the droplet (Brasaemle et al., 2009). Frameshift mutations in PLIN1 have recently been identified in patients with a novel autosomal dominant subtype of partial lipodystrophy (Gandotra et al., 2011). The frameshift mutations found in patients result in a C-terminal elongation of perilipin, with a significantly altered amino acid sequence. Mice lacking Plin1 exhibit elevated levels of basal lipolysis in adipocytes (Tansey et al., 2001; Zhai et al., 2010), which has been suggested as the mechanism by which patients harboring the frameshift mutations develop lipodystrophy (Gandotra et al., 2011). Mechanistic studies of these disease-causing mutations have been limited to the overexpression of mutant and wild-type human cDNAs in mouse 3T3-L1-derived adipocytes, with the conclusion being that wild-type but not mutant PLIN1 is able to inhibit basal lipolysis (Gandotra et al., 2011).
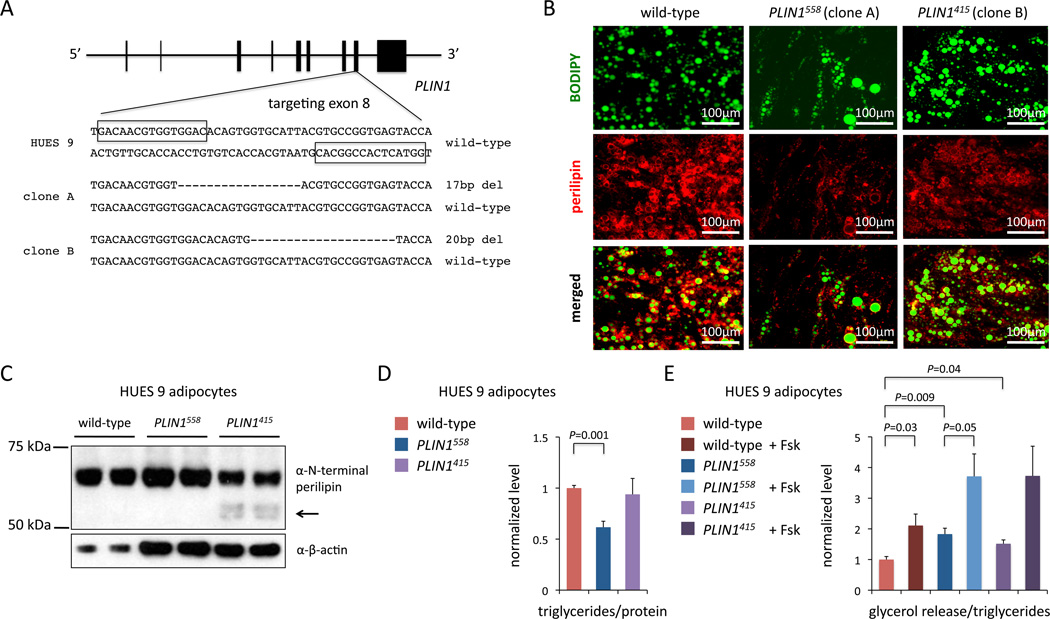
We designed TALENs to target the site of one of the naturally occurring patient-specific mutations (Val398fs) in the eighth coding exon of PLIN1 (Figure 5A). In a single round of targeting of HUES 9 cells we identified 70 mutant clones (out of 293 clones screened). We characterized two mutant clones, one of which harbors a frameshift mutation that elongates perilipin to a length of 558 amino acids (designated PLIN1558; wild-type perilipin has 522 amino acids)—very similar to the effect of the naturally occurring Val398fs mutation—and the other of which has a frameshift resulting in a C-terminal truncation of the protein (415 amino acids; designated PLIN1415) (Figure S7A).
Figure 5. PLIN1 Frameshift Mutations Produce Dominant-Acting and Truncated Proteins.
(A) Generation of PLIN1 frameshift mutant hPSC clones with TALENs targeting exon 8. Boxes indicate the TALEN binding sites. Deletions in the one allele of each clone are indicated.
(B) Immunocytochemistry for perilipin and BODIPY in wild-type, PLIN1558 (clone A), and PLIN1415 (clone B) adipocytes.
(C) Western blot for N-terminus of perilipin in wild-type, PLIN1558, and PLIN1415 adipocytes; arrow indicates truncated protein.
(D) Ratios of triglyceride content to total protein content in wild-type, PLIN1558, and PLIN1415 adipocytes. N = 3.
(E) Ratios of lipolysis activity (as measured by glycerol release) to total triglyceride content in wild-type, PLIN1558, and PLIN1415 adipocytes treated with or without 10 µM forskolin. N = 4. Error bars show s.e.m. from experiments with biological replicates. P values calculated with unpaired t test. See also Figure S7.
We differentiated the allelic series of hPSCs—wild-type, PLIN1558, and PLIN1415—into white adipocytes and observed a substantial reduction in the number of lipid droplet-containing cells as well as smaller lipid droplets in PLIN1558 adipocytes compared to either wild-type and PLIN1415 cell lines (Figure 5B and Figure S7B). We confirmed the presence of perilipin protein in the adipocytes by Western blot analysis (Figure 5C and Figure S7C). We found that the PLIN1558 adipocytes had significantly reduced triglyceride content (38% reduction compared to wild-type adipocytes, P = 0.0009), whereas the PLIN1415 adipocytes had similar triglyceride content to wild-type adipocytes (Figure 5D). We also measured basal and forskolin-stimulated lipolysis and found that both PLIN1558 and PLIN1415 adipocytes had increased basal lipolysis compared to wild-type adipocytes (83% increase, P = 0.008, and 52% increase, P = 0.04, respectively) (Figure 5E). Interestingly, both the PLIN1558 and PLIN1415 mutations resulted in increased lipolysis, though the effect was more marked with the PLIN1558 mutation; furthermore, there were significant differences in the expression levels of adipocyte-specific genes between the PLIN1558 and PLIN1415 cells, underscoring that the two different frameshifts (one leading to elongation of perilipin, the other to truncation) have distinct functional consequences (Figure S7D). Together, these data point to the C-terminal elongated form of perilipin, via a frameshift similar to naturally occurring mutations in lipodystrophy patients, acting in a dominant fashion to alter lipolysis and reduce triglyceride storage and lipid droplet formation in human adipocytes.
TALENs Exhibit Minimal Off-Target Effects, but Sequence Variants Abound
As the extent of off-target effects of TALENs (i.e., mutagenesis at other sites in the genome) in hPSCs remains to be defined, we performed exome sequencing of six cell lines: the parental HUES 1 cell line (clone X); the three SORT1 knockout HUES 1 clones (A–C in Figure 3A); a control HUES 1 clone that had been grown in parallel with the SORT1 knockout clones (i.e., had been exposed to the SORT1 exon 2 TALEN pair but retained two wild-type alleles; clone W); and a clone that had been targeted in the CELSR2 gene with a different TALEN pair (clone Y). It should be noted that TALENs virtually always induce indels by NHEJ, rather than single nucleotide variants (SNVs). Restricting our analysis to novel DNA sequence variants not found in the parental HUES 1 cell line, we identified the known on-target indels in SORT1 in clones A–C; otherwise, we identified just two indels in the exome, a 4-bp deletion in clone C in the coding sequence of LARP6, resulting in a predicted frameshift mutation, and a 1-bp deletion in clone Y in an intron near an exon-intron boundary of LUC7L3 (Table 2). Neither of these sites is flanked by sequences resembling predicted TALEN binding sites, arguing against (but not ruling out) the indels being TALEN-mediated off-target effects. More noteworthy were the 35 Sanger sequencing-confirmed SNVs we discovered across the five experimental and control clones (Table 2). None of these SNVs lay near predicted off-target TALEN binding sites (see below). It is more likely that these SNVs represent intrinsic and perhaps unavoidable heterogeneity among single cell clones of the original pool of HUES 1 cells. Arguing in favor of this interpretation, several of the SNVs were shared by both experimental and control clones, implying a common clonal origin within the original pool of HUES 1 cells. The functional significance of these SNVs is unclear, but the majority resulted in missense mutations, and at least one lay in a well-established disease gene (DMD, responsible for Duchenne muscular dystrophy).
Table 2.
Clonal Sequence Variants Detected by Exome Sequencing and Their Predicted Effects on Protein Products
| Chr | Pos | Ref | Alta | Gene | clone Ab | clone B | clone C | clone W | clone Y |
| 1 | 21014224 | G | T | KIF17 | S432Y | ||||
| 1 | 90178297 | G | A | LRRC8C | P56P | ||||
| 1 | 149921554 | C | T | OTUD7B | Q367Q | ||||
| 2 | 109347284 | T | C | RANBP2 | F39F | ||||
| 2 | 113820048 | C | A | IL36RN | L88I | ||||
| 2 | 173330424 | C | T | ITGA6 | intronic | ||||
| 3 | 132319348 | T | C | CCRL1 | V36A | ||||
| 3 | 193002674 | G | T | ATP13A5 | intronic | intronic | intronic | ||
| 4 | 113568377 | GAAGA | G | LARP7 | frameshiftc | ||||
| 6 | 152652970 | C | A | SYNE1 | A4284S | ||||
| 7 | 2260574 | C | T | MAD1L1 | G47D | ||||
| 7 | 22179660 | G | C | RAPGEF5 | L601V | ||||
| 8 | 15508246 | C | T | TUSC3 | R117C | ||||
| 9 | 33354142 | G | A | NFX1 | E130K | ||||
| 10 | 75541848 | C | T | CHCHD1 | S5S | ||||
| 11 | 124095534 | G | A | OR8G2 | G46E | ||||
| 12 | 80752469 | G | T | OTOGL | C2026F | ||||
| 12 | 101750754 | G | T | UTP20 | R1862I | R1862I | R1862I | ||
| 12 | 105445861 | G | T | ALDH1L2 | intronic | ||||
| 14 | 21967435 | C | A | METTL3 | intronic | ||||
| 15 | 43827249 | G | T | PPIP5K1 | P1071T | ||||
| 15 | 48805891 | C | A | FBN1 | intronic | ||||
| 15 | 90631649 | C | A | IDH2 | W155L | ||||
| 16 | 20043135 | C | A | GPR139 | S328S | ||||
| 17 | 11998899 | G | A | MAP24K4 | R145Q | R145Q | R145Q | ||
| 17 | 48817653 | AT | A | LUC7L3 | intronicd | ||||
| 17 | 73567174 | G | T | LLGL2 | L712L | ||||
| 17 | 79226178 | C | A | SLC38A10 | A588S | ||||
| 18 | 30992086 | G | T | C18orf34 | intronic | ||||
| 19 | 9028296 | G | T | MUC16 | L12166M | ||||
| 19 | 56552245 | G | T | NLRP5 | intronic | intronic | intronic | ||
| 20 | 60498685 | G | T | CDH44 | E425D | ||||
| 22 | 37325592 | C | A | CSF2RB | D100E | D100E | D100E | ||
| 22 | 39078278 | A | T | TOMM22 | intronic | ||||
| X | 32328257 | G | T | DMD | A1897D | ||||
| X | 48558634 | C | T | SUV39H1 | T106T | ||||
| X | 135956509 | C | A | RBMX | S323I |
Alternate allele compared to reference allele in parental HUES 1 cell line (clone X), confirmed by Sanger sequencing
Clones A, B, and C are SORT1−/− clones in HUES 1; clone W is a wild-type (SORT1+/+) clone in HUES 1 that was exposed to SORT1 TALENs, from the same pool as clones A–C; clone Y is a wild-type (SORT1+/+) clone that was exposed to a different set of TALENs (targeting CELSR2)
The surrounding sequence is: CAGAA GAGAA GAAAA AGAAA AAGAA GAAGA AAGGC CGAAT GAAAA AGGAA GACAA
The surrounding sequence is: ATAAT CACAG ATAAT TTATA CAATT ATATT TTTTC CCCCA GGTCC GTGTG AAAAA
We also performed whole-genome sequencing of the same six cell lines. Because the sequencing was performed at low coverage (6–12× coverage on average), it was not possible to perform de novo genome assembly and ascertain all sequence variation among the genomes. Instead, we used the sequencing data to interrogate the sites in the genome at which one or the other TALEN of the SORT1 exon 2 TALEN pair would be most likely to bind based on a weighted TAL monomer–nucleotide association probability matrix developed by Doyle et al. (2012) and, thus, be most likely to induce an off-target sequence change. About 100,000 potential off-target genomic sites were identified; we screened all of these sites for evidence of nearby indels. Besides the known SORT1 indels in clones A–C, we identified no indels passing our criteria.
Thus, although we are not able to completely rule out TALEN off-target effects, we conclude that off-target indels rarely occur based on the results of the exome and whole-genome sequence analyses. However, extrapolating to the entire genome, we expect that each clonal cell line harbors hundreds of SNVs that distinguish it from other cell lines derived from the same pool of parental cells. Thus, it may be virtually impossible to derive truly isogenic cell lines, even with the minimized manipulation of cells entailed by our genome editing system.
DISCUSSION
With our studies, we have used human model systems to generate strong evidence that apoB-100 is critical for HCV replication in human hepatocytes; that sortilin reduces apoB secretion by human hepatocytes, facilitates insulin-mediated glucose uptake by human adipocytes, and is necessary for proBDNF-mediated motor neuron apoptosis; that AKT2 E17K is a gain-of-function mutation that leads to reduced glucose production in human hepatocytes and increased triglyceride content in human adipocytes; and that PLIN1 frameshift mutations increase basal lipolysis in human adipocytes. More generally, these findings highlight the various types of studies to which genome editing in human cells may be applied to obtain novel biological insights.
We note that genome-editing technology is rapidly advancing, and we anticipate that improvements in the engineering of TALENs will continue to make genome editing more rapid and efficient. Indeed, since we established our system, high-throughput automated assembly methods have been reported (Reyon et al., 2012; Briggs et al., 2012), as well as the characterization of TAL monomers with improved nucleotide-binding specificity (Streubel et al., 2012; Cong et al., 2012). While our specific TALEN assembly platform does not incorporate these latest advancements, in principle any up-to-date assembly platform that is paired to a delivery methodology similar to ours should be able to achieve efficient genome editing on a timescale of less than a month.
Whatever the assembly platform, our studies suggest that TALENs incur a low burden of off-target effects but that there is nevertheless significant clone-to-clone genetic variation in the form of SNVs; even if not secondary to TALEN use, they cannot be ignored. The ease and rapidity of TALEN-mediated genome editing allows for rigorous study designs that can alleviate any concerns about off-target effects or other potential confounding by clonal sequence variation. As we have demonstrated with SORT1, it is straightforward to (1) generate multiple distinct mutant cell lines with each TALEN pair, (2) use distinct TALEN pairs to target different sites in a gene, (3) generate mutant clones in different cell lines with different genetic backgrounds, and (4) perform reconstitution experiments in knockout clones. Having used all of these approaches, we are able to conclude with great confidence that the observed cellular phenotypes are indeed related to SORT1 function. We suggest that using at least one of these approaches should become de rigeur for future genetic studies in order to minimize confounding by clonal sequence variation.
With the ability to use TALENs to readily insert specific gene variants into cells, the current enthusiasm for the generation and comparison of “disease” iPSC lines from patients with genetic disorders and “control” iPSC lines from unmatched healthy individuals should shift to the use of genome editing to engineer isogenic cell lines with and without disease mutations. The time it takes to recruit a patient for the donation of tissue from which to make iPSCs (assuming such a patient is readily accessible, which may not be the case for rare disorders), to perform reprogramming to derive iPSC clones, to perform quality control to identify clones that are pluripotent and that will readily differentiate into the desired cell type, and then to undertake differentiation and phenotypic studies—in the absence of isogenic control cell lines—is a minimum of six months and usually longer. Within a shorter timeframe, we have found it to be quite feasible to use TALENs to edit a well-characterized and pre-validated (with respect to differentiation capacity) hPSC line and yield both mutant cell lines and isogenic control cell lines—allowing for a more rigorous study design—and to undertake differentiation and phenotypic studies, without any need for patient contact.
The potential advantages offered by pre-validated wild-type cell lines notwithstanding, there are many disorders for which genetic background (i.e., modifier genes) plays a significant role in determining whether disease mutations result in clinical phenotypes. In these cases, it will be important to use iPSC lines from patients with clinically apparent disease in order to have cell lines with the correct genetic backgrounds for complete disease penetrance (whereas wild-type cell lines may have non-permissive genetic backgrounds). Genome editing with TALENs could be readily applied to patient-specific iPSC lines to “cure” disease mutations and generate appropriate isogenic control lines. Indeed, the most robust possible study design may be to assess both the effect of inserting a disease mutation into a wild-type cell line—thereby testing for sufficiency of the mutation for disease—and the effect of removing a disease mutation from a patient-specific iPSC line—thereby testing for necessity of the mutation for disease. Certainly the rapidity and efficiency of genome editing with TALENs should make it feasible to test the effects of a disease mutation in a variety of genetic backgrounds.
Finally, genome editing potentially allows for the interrogation of a large number of DNA sequence variants, such as those now emerging from next-generation sequencing studies of human populations, on a single genetic background. Creating a robust allelic series of isogenic cell lines represents an approach that hitherto has only been possible in non-mammalian organisms. Such studies will represent a significant advance in our ability to dissect genotype-phenotype relationships and thereby better elucidate human biology and disease.
EXPERIMENTAL PROCEDURES
TALEN construction
TALEN genomic binding sites were chosen to be 15 bp in length or, in a few cases, 13 bp in length such that the target sequence between the two binding sites was between 14 and 18 bp in length; each binding site was anchored by a preceding T base in position “0” as has been shown to be optimal for naturally occurring TAL proteins (Moscou and Bogdanove, 2009; Boch et al., 2009). A library of 832 tetramer or trimer TAL repeats were constructed using methods based on the PCR-based protocol of Zhang et al. (2011); these multimers were designed to have complementary sticky ends when digested out of library plasmids with the type IIs restriction enzyme BsmBI. As outlined in Figure 1, multimers were assembled into an array and subcloned into a full-length TALEN harboring, in order: a N-terminal FLAG tag, a nuclear localization signal, the N-terminal portion of the TALE PthXo1 from the rice pathogen X. oryzae pv. oryzae (a kind gift of Dr. Daniel Voytas, University of Minnesota) lacking the first 176 amino acids (after Miller et al., 2011), the engineered TAL repeat array, the following 63 amino acids from the corresponding C-terminal portion of PthXo1 (after Miller et al., 2011), and one of two enhanced FokI domains. The FokI domains used were obligate heterodimers with both the Sharkey (Guo et al., 2010) and ELD:KKR (Doyon et al., 2011) mutations to enhance cleavage activity, engineered by PCR. Each TALEN was in a plasmid with the CAG promoter for optimal expression in hPSCs, with the TALEN being coexpressed with a fluorescent marker [enhanced green fluorescent protein (EGFP), mCherry (Clontech), or turbo red fluorescent protein (tRFP; Evrogen)] via an intervening viral 2A sequence. The generic TALEN protein sequences are shown in Figure S1A. All reagents, protocols, and plasmid sequences needed to generate TALENs and perform genome editing by the methods described in this manuscript will be available to academic researchers through Addgene (http://www.addgene.org/TALEN_genome_editing_collection).
Cell culture, transfection/electroporation, and sorting
HuH-7/CD81high cells were grown in adherent culture in DMEM High Glucose containing glutamine and pyruvate (Invitrogen) and supplemented with 10% FBS and penicillin/streptomycin. Transfection of the plasmids expressing the APOB TALEN pair into HuH-7 cells was performed using Fugene 6 (Roche) in 10-cm tissue culture plates according to manufacturer instructions. HUES 1 and HUES 9 cells (Cowan et al., 2004) were grown in feeder-free adherent culture in chemically defined mTeSR1 (STEMCELL Technologies) supplemented with penicillin/streptomycin on plates pre-coated with Geltrex matrix (Invitrogen). The cells were disassociated into single cells with Accutase (Invitrogen), and 10 million cells were electroporated with 50 µg of the TALEN pair (25 µg of each plasmid), or with a mix of 30 µg of the TALEN pair (15 µg of each plasmid) and 30 µg of the ssODN (5’-CAGGA AGTAC CGTGG CCTCC AGGTC TTGAT GTACT TACCT GAAAT GAGGC AGGAA GGGAG GGAGA GA-3’), in a single cuvette and replated as previously described (Schinzel et al., 2011). The cells were collected from the culture plates 48 hours post-transfection or post-electroporation by trypsin or Accutase treatment, respectively, and resuspended in PBS. Cells expressing green and/or red fluorescent markers were collected by FACS (FACSAria II; BD Biosciences) and replated on 10-cm tissue culture plates at 15,000 cells/plate to allow for recovery in growth media.
Isolation of targeted clonal cell populations
Post-FACS, the cells were allowed to recover for 7–10 days, after which single colonies were manually picked and dispersed and replated individually to wells of 96-well plates. Colonies were allowed to grow to near confluence over the next 7 days, at which point they were split using trypsin (for HuH-7 cells) or Accutase (for hESCs) and replica-plated to create a working stock and a frozen stock. The working stock was grown to confluence. Genomic DNA was extracted in 96-well format from working stocks in lysis buffer (10 mM Tris pH 7.5, 10 mM EDTA, 10 mM NaCl, 0.5% Sarcosyl) containing proteinase K at 56°C overnight in a humidified chamber. Genomic DNA was precipitated by addition of 95% ethanol containing 75 mM NaCl for 1 hr at room temperature. The DNA was then washed 2 times in 70% ethanol, allowed to dry at room temperature, and then resuspended in nuclease-free water.
Genotyping at the TALEN target site was then performed for each sample by PCR amplication (94°C 30 sec; 56°C 30 sec; 68°C 30 sec) using FastStart Taq (Roche) and a primer pair designed to yield small amplicons (~150–200 bp) around the target site. Amplicons were subjected to electrophoresis on 2.5% agarose gels to discriminate clones with indels, with positive clones having a band or bands visibly shifted in size from the baseline (see Figure S1B and Figure S6A for examples); for AKT2 E17K candidate clones, the amplicons were digested with RsaI for 1 hr and subjected to electrophoresis, with positive clones displaying cleavage products (Figure S6A). For a subset of the potentially positive clones, PCR amplicons were subcloned using the TOPO TA Cloning Kit (Invitrogen) and subjected to numerous sequence reads to confirm the presence of mutant alleles; in a similar fashion, a subset of the potentially negative clones were confirmed to be wild-type. Clones with confirmed compound heterozygous mutant alleles (or the AKT2 E17K mutation) or confirmed to be wild-type were retrieved from the frozen stocks and expanded for further experiments. When no compound heterozygous clones were identified, a heterozygous clone with one mutant allele was expanded and subjected to a second round of TALEN targeting.
Differentiation of hPSCs into HLCs, white adipoctyes, and motor neurons
Differentiation was performed following the protocols of Si-Tayeb et al. (2010), Ahfeldt et al. (2012), and Di Giorgio et al. (2008) and Chambers et al., (2009). Details are given in Supplemental Experimental Procedures.
Enzyme-linked immunosorbent assays (ELISAs), immunocytochemistry, and Western blot analysis
These procedures were performed using standard methods. Details are given in Supplemental Experimental Procedures.
Glucose production, glucose uptake, triglyceride content, and lipolysis assays
Glucose production and glucose uptake were measured using protocols adapted from Hagiwara et al. (2012) and Ahfeldt et al. (2012), respectively. Details of the various procedures are given in Supplemental Experimental Procedures.
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
qRT-PCR was performed using standard methods. Details and oligonucleotide sequences are given in Supplemental Experimental Procedures.
HCV infection of HuH-7 cells
Details are given in Supplemental Experimental Procedures.
Exome and whole-genome sequence analyses
Exome sequencing and whole-genome sequencing were performed as previously described (Gnirke et al., 2009; Stransky et al., 2011). Procedural and analytical details are given in Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
A system for efficient and rapid genome editing with TALENs
Generation of isogenic human cellular models of disease
Identification of disease-associated phenotypes in multiple human cell types
Minimal TALEN off-target effects, but significant clone-to-clone sequence variation
ACKNOWLEDGMENTS
This work was supported in part by a Roche Postdoc Fellowship (Q.D.); the Sternlicht Director’s Fund Award for Graduate Students from the Harvard Stem Cell Institute (D.T.P.); the Harvard Presidential Scholars Fund of the Harvard Medical School MD/PhD Program (A.V.); grants T32-DK007191 (E.A.K.S., D.L.M.), T32-HL007604 (R.M.G.), K08-DK088951 (L.F.P.), K24-DK078772 (R.T.C.), P01-NS066888 (L.L.R.), R00-HL098364 (K.M.), U01-HL107440 (C.A.C.), and R01-DK097768 (F.Z., K.M., C.A.C.) from the United States National Institutes of Health (NIH); the New York Stem Cell Foundation (L.L.R.); the Broad Institute’s Lawrence H. Summers Fellowship and the Carlos Slim Foundation (K.M.); the Harvard Stem Cell Institute (T.B.M., L.L.R., K.M., C.A.C.), and Harvard University (L.L.R., K.M., C.A.C.). We thank David Altshuler, Noel Burtt, Guillermo del Angel, Mark DePristo, Stacey Gabriel, Namrata Gupta, J. Keith Joung, Adam Kaplan, Ami Levy-Moonshine, Heng Li, Elyse Macksoud, Khalid Shakir, Alanna Strong, Kristin Thompson, Jayaraj Rajagopal, Stephanie Regan, Jennifer Shay, and the staffs of the HSCRB-HSCI Flow Cytometry Core and the Broad Institute’s Genomics Platform for assistance and suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
A.M. is a fulltime employee of Roche Pharmaceuticals; the other authors report no relevant conflicts of interest.
REFERENCES
- Agarwal AK, Garg A. Genetic disorders of adipose tissue development, differentiation, and death. Annu. Rev. Genomics Hum. Genet. 2006;7:175–199. doi: 10.1146/annurev.genom.7.080505.115715. [DOI] [PubMed] [Google Scholar]
- Ahfeldt T, Schinzel RT, Lee YK, Hendrickson D, Kaplan A, Lum DH, Camahort R, Xia F, Shay J, Rhee EP, et al. Programming human pluripotent stem cells into white and brown adipocytes. Nat. Cell Biol. 2012;14:209–219. doi: 10.1038/ncb2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Subramanian V, Garcia A, Marcinkiewicz A, Rothenberg A. Perilipin A and the control of triacylglycerol metabolism. Mol. Cell. Biochem. 2009;326:15–21. doi: 10.1007/s11010-008-9998-8. [DOI] [PubMed] [Google Scholar]
- Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB, 3rd, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, Melton DA. Derivation of embryonic stem-cell lines from human blastocysts. N. EnglJ. Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40(Web Server issue):W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- Gandotra S, Le Dour C, Bottomley W, Cervera P, Giral P, Reznik Y, Charpentier G, Auclair M, Delépine M, Barroso I, et al. Perilipin deficiency and autosomal dominant partial lipodystrophy. N. EnglJ. Med. 2011;364:740–748. doi: 10.1056/NEJMoa1007487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo RS, Orena SJ, Rafidi K, Torchia AJ, Stock JL, Hildebrandt AL, Coskran T, Black SC, Brees DJ, Wicks JR, et al. Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKB beta. J. Clin. Invest. 2003;112:197–208. doi: 10.1172/JCI16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S, Rochford JJ, Wolfrum C, Gray SL, Schinner S, Wilson JC, Soos MA, Murgatroyd PR, Williams RM, Acerini CL, et al. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science. 2004;304:1325–1328. doi: 10.1126/science.1096706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Gaj T, Barbas CF., 3rd Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J. Mol. Biol. 2010;400:96–107. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Rüegg MA, Hall MN. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Jr, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain K, Challis B, Rocha N, Payne F, Minic M, Thompson A, Daly A, Scott C, Harris J, Smillie BJ, et al. An activating mutation of AKT2 and human hypoglycemia. Science. 2011;334:474. doi: 10.1126/science.1210878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Luo G. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 2009;83:12680–12691. doi: 10.1128/JVI.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjolby M, Andersen OM, Breiderhoff T, Fjorback AW, Pedersen KM, Madsen P, Jansen P, Heeren J, Willnow TE, Nykjaer A. Sort1, encoded by the cardiovascular risk locus 1p13.3, is a regulator of hepatic lipoprotein export. Cell Metab. 2010;12:213–223. doi: 10.1016/j.cmet.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahmias Y, Goldwasser J, Casali M, van Poll D, Wakita T, Chung RT, Yarmush ML. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology. 2008;47:1437–1445. doi: 10.1002/hep.22197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykjaer A, Willnow TE. Sortilin: a receptor to regulate neuronal viability and function. Trends Neurosci. 2012;35:261–270. doi: 10.1016/j.tins.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel RT, Ahfeldt T, Lau FH, Lee YK, Cowley A, Shen T, Peters D, Lum DH, Cowan CA. Efficient culturing and genetic manipulation of human pluripotent stem cells. PLoS One. 2011;6:e27495. doi: 10.1371/journal.pone.0027495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Kandror KV. Sortilin is essential and sufficient for the formation of Glut4 storage vesicles in 3T3-L1 adipocytes. Dev. Cell. 2005;9:99–108. doi: 10.1016/j.devcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Laganière J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel J, Blücher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- Tansey JT, Sztalryd C, Gruia-Gray J, Roush DL, Zee JV, Gavrilova O, Reitman ML, Deng CX, Li C, Kimmel AR, Londos C. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6494–6499. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AR, Gifondorwa DJ, Robinson MB, Strupe JL, Prevette D, Johnson JE, Hempstead B, Oppenheim RW, Milligan CE. Motoneuron programmed cell death in response to proBDNF. Dev. Neurobiol. 2012;72:699–712. doi: 10.1002/dneu.20964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng HK, Teng KK, Lee R, Wright S, Tevar S, Almeida RD, Kermani P, Torkin R, Chen ZY, Lee FS, et al. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J. Neurosci. 2005;25:5455–5463. doi: 10.1523/JNEUROSCI.5123-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Xu C, Ling Y, Liu S, Deng J, Qi Y, Londos C, Xu G. Increased lipolysis in adipose tissues is associated with elevation of systemic free fatty acids and insulin resistance in perilipin null mice. Horm. Metab. Res. 2010;42:247–253. doi: 10.1055/s-0029-1243599. [DOI] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.