Abstract
To understand the evolutionary events and possible selection mechanisms involved in the emergence of pathogenic Vibrio cholerae, we analyzed diverse strains of V. cholerae isolated from environmental waters in Bangladesh by direct enrichment in the intestines of adult rabbits and by conventional laboratory culture. Strains isolated by conventional culture were mostly (99.2%) negative for the major virulence gene clusters encoding toxin-coregulated pilus (TCP) and cholera toxin (CT) and were nonpathogenic in animal models. In contrast, all strains selected in rabbits were competent for colonizing infant mice, and 56.8% of these strains carried genes encoding TCP alone or both TCP and CT. Ribotypes of toxigenic O1 and O139 strains from the environment were similar to pandemic strains, whereas ribotypes of non-O1 non-O139 strains and TCP- nontoxigenic O1 strains diverged widely from the seventh pandemic O1 and the O139 strains. Results of this study suggest that (i) the environmental V. cholerae population in a cholera-endemic area is highly heterogeneous, (ii) selection in the mammalian intestine can cause enrichment of environmental strains with virulence potential, (iii) pathogenicity of V. cholerae involves more virulence genes than currently appreciated, and (iv) most environmental V. cholerae strains are unlikely to attain a pandemic potential by acquisition of TCP and CT genes alone. Because most of the recorded cholera pandemics originated in the Ganges Delta region, this ecological setting presumably favors extensive genetic exchange among V. cholerae strains and thus promotes the rare, multiple-gene transfer events needed to assemble the critical combination of genes required for pandemic spread.
The Gram-negative bacterium Vibrio cholerae belongs to a group of organisms whose natural habitat is the aquatic ecosystem (1, 2), although some strains of this species are associated with severe enteric infections in humans. Toxigenic strains of V. cholerae belonging to the O1 and O139 serogroups cause cholera, a devastating watery diarrhea that occurs frequently as epidemics in many developing countries (3, 4). Strains belonging to other serogroups, collectively referred to as non-O1 non-139, have also been implicated as etiologic agents of moderate to severe human gastroenteritis (5-8). V. cholerae O1 and O139 are commonly known to carry a set of virulence genes necessary for pathogenesis in humans. Recent studies have indicated that virulence genes or their homologues are also dispersed among environmental strains of V. cholerae belonging to diverse serogroups that appear to constitute an environmental reservoir of virulence genes (9, 10). Although the roles of virulence-associated factors in the environment and the selection pressures for environmental V. cholerae carrying virulence genes is not clear, it is possible that these strains may be precursors of pathogenic strains or may participate in gene-transfer events leading to the origination of pathogenic strains.
To track the evolutionary events in the origination of pathogenic V. cholerae from their nonpathogenic progenitors, it is important to identify intermediate strains that are likely to have a lower virulence potential than the epidemic strains. Because cholera is a water-born disease, environmental monitoring for the presence of V. cholerae strains with pathogenic potential is also important to identify the source of strains causing either epidemics of cholera or sporadic episodes of gastroenteritis. However, isolation of pathogenic V. cholerae from the environment is often limited by the lack of a suitable technique to selectively enrich pathogenic strains from the vast majority of nonpathogenic strains normally found in the environment. Several animal models, including infant mice and adult rabbits, have been used to assay for colonization and diarrheagenic ability of clinical and environmental isolates of V. cholerae (11-13). In this study, we show that environmental V. cholerae strains with virulence potential are selectively enriched in the rabbit intestine from the majority of genetically diverse nonpathogenic strains. This has implications in understanding the possible roles of the human host in the enrichment of strains with epidemic potential, leading to the initiation of seasonal cholera epidemics in an endemic area.
Materials and Methods
A total of 129 water samples collected from six different sites of two major rivers in Dhaka, Bangladesh were analyzed. Sampling was done every 2-3 weeks between July 2002 and August 2003. V. cholerae were isolated from the water samples by using two different enrichment techniques. These included conventional culture on selective media after enrichment in alkaline peptone water (14) and culture after enrichment in the ileal loops of adult rabbits. Clinical strains used as controls in different assays or for comparison with the environmental isolates were obtained from our culture collection.
Enrichment in Rabbit Ileal Loops. An aliquot (35 ml) of each water sample was centrifuged at 4,500 × g, and the pellet was resuspended in 3.5 ml of 10 mM PBS (pH 7.4). The suspension was vortexed to dislodge any bacteria adhering to solid particles and then centrifuged at low speed (1,000 × g) to precipitate particulate matter. The supernatant fluid containing suspended microorganisms was inoculated in ileal loops of adult New Zealand White rabbits. One milliliter of the suspension was inoculated into each loop as described (11). After 18 h rabbits were killed and the loops were examined for fluid accumulation. Ileal loop fluids were collected, and dilutions of the fluid in 10 mM PBS (pH 7.5) were plated on taurocholate-tellurite-gelatin agar plates (14). Suspected Vibrio colonies were picked and tested by biochemical and serological methods (15). Rabbit Ileal loops showing no fluid accumulation were also washed internally with PBS, and the washings were plated on taurocholate-tellurite-gelatin agar plates. For examining fluid accumulation by isolated strains, cell-free culture supernatants of the strains grown overnight in AKI medium at 30°C and live cultures were tested in rabbit ileal loops as described (11). Production of cholera toxin (CT) by V. cholerae isolate was also assayed by the GM1-ganglioside-dependent ELISA by using a rabbit anti-CT monoclonal antibody (Sigma) as described (16).
Mouse Colonization Assay. Colonization of infant mice by V. cholerae strains was tested by a competition assay with a streptomycin-resistant reference strain Bah-2 as described (10, 13). In brief, the test strain and the reference strain were mixed at a 1:1 ratio, and ≈105 colony-forming units of the bacterial mix was inoculated intragastrically in groups of 5-day-old Swiss Albino mice. The mice were then killed, and bacteria were recovered from the small intestines by homogenization in PBS (pH 7.4). Serial dilutions of the homogenates were plated on LB agar containing streptomycin (100 μg/ml) and on plates devoid of the antibiotic to determine the ratio of the test and reference strains. Competitive indices were calculated by dividing the output ratios by the precise inoculum input ratio of the test and reference strains.
Culture of Environmental Samples. An aliquot of each PBS extract prepared from environmental samples as described above were also tested for detection of V. cholerae by conventional culture. One milliliter of the suspension was added to 10 ml of alkaline peptone water [APW; peptone 1% (wt/vol), NaCl 1% (wt/vol), pH 8.5] contained in 20-ml screw-cap glass tubes and incubated at 37°C with shaking (100 rpm) for 6-8 h. Dilutions of this APW culture were streaked on taurocholate-tellurite-gelatin agar plates (14). Suspected Vibrio colonies were picked and subjected to biochemical and serological tests (14, 15).
Probes and PCR Assays. All V. cholerae O1 and O139 strains and one representative V. cholerae non-O1 non-O139 strain (when present) derived from each water sample were analyzed for different virulence-associated genes by using specific DNA probes or PCR assays. The gene probe for cholera toxin was a 0.5-kb EcoRI fragment of pCVD27 (17), and the NAG-ST probe was a 0.27-kb EcoRI-BamHI fragment of pAO111 (18). The rRNA gene probe consisted of a 7.5-kb BamHI fragment of the Escherichia coli rRNA clone pKK3535 (19, 20). Probes were labeled by using a random primers DNA-labeling kit (Invitrogen) and deoxycytidine [α-32P]triphosphate (3,000 Ci/mmol, Amersham Pharmacia Biosciences). Colony blots or Southern blots were prepared and hybridized with the labeled probes by standard methods (21).
PCR assays used in this study have been described previously. Included are PCR assays specific for the tcpA, tcpI, and acfB genes of the toxin-coregulated pilus (TCP) pathogenicity island (22-24), environmental tcpA variant (9), genes of the RS1-element (rstR and rstC), hemolysin (hlyA), RTX toxin gene cluster (rtxA and rtxC), and mshA for mannose-sensitive hemagglutinin (25-29). PCR reagents and kits were obtained either from Perkin-Elmer or Invitrogen, and PCR was performed essentially as described previously.
Numerical Analysis of rDNA Bands. Banding profiles from HindIII and BglI digests of rRNA genes were analyzed as described (20) by using the PHP-2 software (Version 2, Biosys Inova, Luntmakargatan, Stockholm). In brief, the similarity of rDNA bands between each pair of isolates expressed as correlation coefficient was calculated to yield a similarity matrix consisting of n × (n - 1)/2 correlation coefficients, where n is the number of isolates. The similarity matrix was clustered according to the unweighed-pair-group method with arithmetic averages to produce a dendrogram. Clusters were defined as groups of isolates with >98% similarity and were designated as ribotypes.
Results
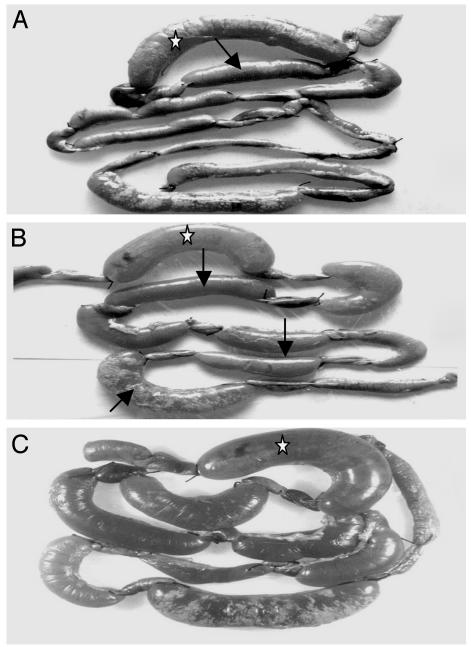
Rabbit intestine enriches V. cholerae strains with virulence potential. The rabbit ileal loop assay initially identified 33 of 129 water samples (25.5%) that caused a fluid accumulation (FA) response and 96 samples (74.4%) that were FA-negative. Fluid accumulation varied between 0.75 and 2.96 ml/cm of ileal loop for different FA-positive samples and were clearly distinguishable from the negative samples (Fig. 1). Culture of ileal loop fluids derived from the 33 positive samples led to isolation of a fluid-causing V. cholerae strain from 30 samples. This finding was further confirmed by ileal loop assays with isolated strains (Fig. 1). Analysis of the contents of ileal loops that were FA-negative also showed the presence of V. cholerae in 19 (19.7%) samples. Representative isolates from seven of these ileal loops caused an FA response in rabbits in subsequent assays, although the water samples were negative in the initial ileal loop assay. This result could be due to the presence of low numbers of fluid-causing organisms in the water samples, which were not enough to induce an FA response in the initial assay. Details of different V. cholerae strains isolated in this study are presented in Table 1.
Fig. 1.
Accumulation of fluid in the ileal loops of adult rabbits by microorganisms present in environmental water samples. (A and B) Fluid accumulation shown in several ileal loops (arrow) in response to environmental samples. (C) Ileal loop contents from the primary assays cultured to isolate bacteria that were again tested for fluid accumulation in subsequent ileal loop assays. Ileal loops marked with a star were inoculated with a toxigenic V. cholerae strain used as a positive control.
Table 1. Presence of virulence-associated genes and virulence potential of V. cholerae strains isolated from environmental water samples.
| Presence of virulence-associated genes
|
Strains positive in animal assays, n
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Description | Serogroup | Isolates, n | ctxA | rstR | tcpA | tcpl | acfB | rtxA | rtxC | hlyA | mshA | stn | Mouse colonization | Rabbit ileal loop assay |
| Strains isolated by enrichment in rabbit ileal loops | O1 El Tor | 3 | + | + | + | + | + | + | + | + | + | − | 3 | 3 |
| O1 El Tor | 2 | − | − | + | + | + | + | + | + | + | − | 2 | 0 | |
| O139 | 2 | + | + | + | + | + | + | + | + | + | − | 2 | 2 | |
| Non-O1 non-O139 | 3 | + | + | + | + | + | + | + | − | + | − | 3 | 3 | |
| Non-O1 non-O139 | 19 | − | − | + | + | + | + | + | + | + | − | 19 | 14 | |
| Non-O1 non-O139 | 21 | − | − | − | − | − | + | + | + | + | − | 21 | 15 | |
| Non-O1 non-O139 | 1 | − | − | − | − | − | − | − | + | + | − | 1 | 0 | |
| Total (%) | 51 | 51 (100) | 37 (72.5) | |||||||||||
| Strains Isolated by conventional culture* | O1 El Tor | 3 | − | − | − | − | − | + | + | + | + | − | 0 | 0 |
| O1 El Tor | 1 | + | + | + | + | + | + | + | + | + | − | 1 | 1 | |
| Non-O1 non-O139 | 3 | − | − | − | − | − | + | + | + | + | − | 0 | 0 | |
| Non-O1 non-O139 | 4 | − | − | − | − | − | + | − | + | + | − | 0 | 0 | |
| Non-O1 non-O139 | 2 | − | − | − | − | − | + | + | + | + | + | 2 | 2 | |
| Non-O1 non-O139 | 116 | − | − | − | − | − | + | + | + | + | − | 0 | 0 | |
| Total (%) | 129 | 3 (2.3) | 3 (2.3) | |||||||||||
Conventional culture refers to enrichment in alkaline peptone water followed by culture on selective media (see text for details).
The rabbit ileal loop assay has been widely used to test the ability of organisms to produce enterotoxins and hence their diarrheagenic potential (11, 23, 24). Because colonization of the intestine is assumed to be an important step in the pathogenesis of V. cholerae (3, 4), possible intermediate strains with the ability to colonize but unable to produce enterotoxins are also likely to be enriched in the rabbit intestinal loops. This assumption was supported by the observation that all strains isolated from the rabbits, irrespective of their fluid-causing ability, colonized infant mice in competition with a known TCP+ CT- strain Bah-2. The competitive index of colonization varied from 0.57 to 2.45. In contrast, TCP- control strain TCP-2 included in the study was strongly out-competed by the reference TCP+ strain (competition index, 0.07).
To further verify that the rabbit intestinal environment could selectively enrich for potentially pathogenic strains in the presence of nonpathogenic strains, we also attempted to identify the whole range of Vibrio-related species present in the water samples, irrespective of their virulence potential. Conventional culture of water samples after enrichment in alkaline peptone broth allowed isolation of V. cholerae O1 or non-O1 non-O139 strains from 125 (96.8%) samples. Of these samples, 116 (89.9%) were found to contain non-O1 non-O139 strains alone, and four samples contained V. cholerae O1 in addition to non-O1 non-O139 strains. The remaining five samples contained other species, including Vibrio mimicus, Vibrio fluvialis, and Aeromonas hydrophila in addition to V. cholerae non-O1 non-O139. Subsequent analysis of representative V. cholerae isolates showed that, except for one V. cholerae O1 strain and two non-O1 non-O139 strains, none of the other strains isolated by conventional culture colonized infant mice or caused an FA response in the rabbit ileal loop assay. Thus, although culturable V. cholerae were present in at least 96.8% of the water samples, only 2.3% of strains isolated by conventional methods had any virulence potential. In contrast, all strains selected in rabbits colonized infant mice, and 72.5% caused fluid accumulation in rabbit ileal loops (Table 1). Together these findings suggested that strains with pathogenic potential were selectively enriched in the rabbit intestinal environment, and the enrichment possibly occurs primarily because of the ability of these strains to colonize rabbits.
Environmental V. cholerae Strains Carry Diverse Combinations of Virulence Genes. Distribution of different virulence-associated genes among the environmental V. cholerae isolates are presented in Table 1. The most well characterized virulence genes in V. cholerae are those carried by strains of the O1 and O139 serogroups associated with cholera epidemics. These genes include the TCP pathogenicity island, which encodes the major colonization factor TCP, and the CTX prophage, which encodes CT. The present study identified four V. cholerae O1, two V. cholerae O139, and four non-O1 non-O139 strains that were positive for both the CTX prophage and the TCP island (Table 1). More interesting, however, was the isolation of two O1 strains and 19 non-O1 non-O139 strains that were positive for the TCP island but negative for the CTX prophage. These strains appear to be intermediate strains that are likely to be competent for toxigenic conversion by CTXϕ. This study also identified three strains of V. cholerae O1 that were negative for both TCP and CT genes. TCP-island-specific genes tcpA, tcpI, and acfB and presumably the entire TCP island and the CTX prophage were absent in most of the non-O1 non-O139 strains (Table 1). Included were 15 strains that colonized mice and caused fluid accumulation in rabbits despite the absence of TCP and CT genes. Recent studies have identified genetic variants of tcpA gene encoding the major pilus subunit in environmental strains (9). These 15 strains were also negative for the known environmental variants of tcpA gene. These results demonstrate that the aquatic environment in a cholera endemic area harbors V. cholerae strains carrying various combinations of known and undefined virulence-associated genes.
Non-O1 Non-O139 V. cholerae May Produce Unknown Virulence Factors. As noted above, ileal loop enrichment allowed us to isolate V. cholerae non-O1 non-O139 strains that colonize infant mice and can cause fluid accumulation in rabbits despite the absence of genes encoding TCP and CT (Table 2). To examine whether the apparent virulence properties were due to the presence of other genes encoding colonization factors or toxins, all strains were analyzed with DNA probes or PCR assays for a variety of genes encoding previously described putative additional virulence-associated factors (Table 1). These included the RTX toxin gene cluster (30), which has been shown to encode cytotoxic activity for Hep-2 cells, the hlyA gene (26) encoding a hemolysin, the mshA gene (29) for mannose-sensitive hemagglutinin pilus, and the stn gene encoding a heat-stable enterotoxin of non-O1 vibrios (NAG-ST) (28). We found that these genes were distributed among strains irrespective of their ability to cause an FA response in rabbits (Tables 1 and 2). Therefore, in the present study, none of the previously described virulence-related genes were found to be specifically associated with TCP- CTX- non-O1 non-O139 strains that colonized mice and caused fluid accumulation in rabbits.
Table 2. Ribotypes of environmental V. cholerae isolates carrying different combinations of horizontally acquired gene clusters and possessing virulence characteristics.
| Serotype | Ribotypes | Presence of horizontally acquired gene clusters | Fluid accumulation in rabbit ileal loops* | Colonization of infant mice† |
|---|---|---|---|---|
| O1 | R-1, R-2, R-3, R-4, | TCP+ CTX+ RTX+ | + | + |
| O1 | R-8 | TCP+ CTX− RTX+ | − | + |
| O1 | R-26, R-34 | TCP− CTX− RTX+ | − | − |
| O139 | R-14, R15 | TCP+ CTX+ RTX+ | + | + |
| Non-O1 non-O139 | R-11, R-12, R-13, R-33 | TCP+ CTX+ RTX+ | + | + |
| Non-O1 non-O139 | R-5, R-10, R-25, R-27, R-28, R-30, R-32 | TCP+ CTX− RTX+ | + | + |
| Non-O1 non-O139 | R-22, R-23, R-29, R-30, R-31, R-35, R-36, R-37, R-38, R-39, R-42 | TCP− CTX− RTX+ | + | + |
| Non-O1 non-O139 | R-40, R-41, R-43 | TCP− CTX− RTX+ | − | + |
| Non-O1 non-O139 | R-24 | TCP− CTX− RTX− | + | + |
The fluid accumulation varied between 0.75 and 2.96 ml/cm of ileal loop for different strains.
Colonization of infant mice was assayed in competition with a known TCP− CT− strain Bah-2 (see text for details). The competitive index of colonization varied between 0.57 and 2.45 for different strains.
For CT+ strains, both cell-free culture supernatants and live cells induced an FA response when inoculated in rabbit ileal loops. The culture supernatants of these strains were also positive when tested by an ELISA for CT (data not shown). In contrast, the FA response induced by the CT- non-O1 non-O139 strains was apparently not due to an extracellular toxin, because culture supernatant fluids of these strains did not induce an FA response. However, inoculation of whole live cells of these strains caused a strong FA response in rabbits. Although the epidemiology and pathogenesis of non-O1 non-O139 gastroenteritis is incompletely understood, colonization of the intestinal cells is assumed to be an important step in establishing a productive infection by enteric pathogens. Given that these strains also caused a strong FA response in rabbits, we conclude that these non-O1 non-O139 strains produce previously undiscovered colonization factors and further induce fluid accumulation by unknown mechanisms. These results also suggest that nonpathogenic environmental V. cholerae have evolved into potentially pathogenic forms by more than one evolutionary pathway and involving more virulence genes than currently appreciated.
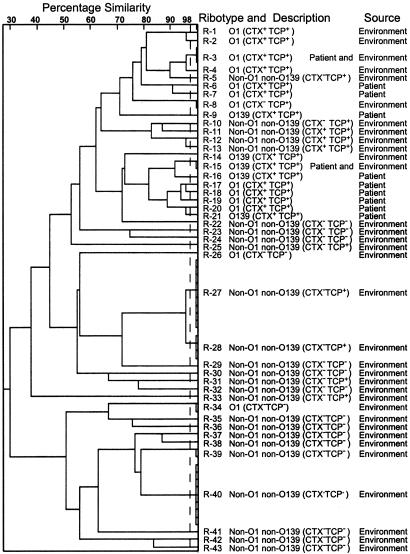
Clonal Diversity of Environmental V. cholerae. All strains were analyzed for HindIII and BglI restriction patterns of their rRNA genes to understand their clonal relationship. Previous studies have shown that ribotyping of V. cholerae by using BglI affords maximum discrimination among different clones, because BglI sites are known to vary more frequently, whereas HindIII sites are known to be highly conserved and thus allow efficient detection of ancestral clones (20, 31). In this study we used a numerical analysis of HindIII and BglI restriction patterns taken together to calculate overall similarity and divergence among ribotype patterns produced by different strains. Representative strains isolated by conventional culture from different water samples were found to be very diverse in their ribotype patterns, with 127 different ribotypes among 129 strains (data not shown). On the other hand, strains isolated by enrichment in rabbit ileal loops included four distinct clusters or ribotypes designated as R-27, R-28, R-39, and R-40 shared by 25 strains and 26 unique ribotypes produced by individual strains (Fig. 2). Ribotypes of pathogenic non-O1 non-O139 strains generally differed widely (mean similarity index, <0.63) from the ribotypes of toxigenic O1 and O139 strains. However, ribotype R-5 produced by one TCP+ non-O1 non-O139 strain was quite similar (mean similarity index, 0.82) to ribotypes R-1, R-2, R-3, and R-4 produced by toxigenic O1 strains (Fig. 2). Ribotypes of different toxigenic O1 and O139 strains isolated from either clinical or environmental sources was largely similar, but ribotypes of three nontoxigenic TCP- O1 strains isolated from the environment were very different from the toxigenic strains and the TCP+ nontoxigenic strains (Fig. 2). Strains with closely related ribotypes but carrying a different combination of virulence genes were also identified. These strains appear to represent intermediates in the evolution to pathogenic forms and are probably derived from common progenitors. Taken together, the ribotype data suggest that the environmental V. cholerae population is extensively heterogeneous, and relatively few strains have evolved into pathogenic clones. Furthermore, the ribotypes of pandemic O1 and O139 strains, in general, appear to be distinct from the ribotypes of diverse environmental strains.
Fig. 2.
Dendrogram showing cluster analysis of rRNA gene restriction fragment profiles of selected environmental and clinical strains of V. cholerae O1, O139, and non-O1 non-O139. Presence or absence of the CTX prophage and TCP island are shown in brackets. Of the 57 environmental strains shown, 51 strains were isolated by enrichment in rabbit ileal loops. Three TCP- CT- O1 strains, two NAG-ST-positive strains, and one toxigenic O1 strain were isolated from surface water by conventional culture. The clinical strains used for comparison are from our collection.
Discussion
The aquatic environment in a cholera-endemic area appears to harbor an immensely diverse population of V. cholerae strains. These include both nonpathogenic and pathogenic strains with different levels of virulence potential and belonging to different serogroups and ribotypes. Although strains isolated by conventional culture likely represented the entire population of vibrios, including O1 and O139 V. cholerae strains and the more abundant non-O1 non-O139 strains, enrichment in rabbits allowed isolation of strains with virulence potential. In a previous study, genetic profiles of toxigenic V. cholerae O1 strains isolated from the environment and from clinical cases of cholera were compared by enterobacterial repetitive intergenic consensus sequence PCR (32). This study showed similarities between environmental and clinical isolates in different areas. In the present study we analyzed strains representing the entire spectrum of V. cholerae population in the environment. This study showed that, although ribotypes of toxigenic O1 and O139 strains from the environment were similar to the pandemic strains, the ribotypes of the non-O1 non-O139 strains and three nontoxigenic O1 strains diverged widely from the seventh pandemic O1 and the O139 strains (Fig. 2). Furthermore, ribotypes of 125 non-O1 non-O139 strains isolated by conventional culture were different from the pathogenic non-O1 non-O139 strains isolated from the same water samples by enrichment in rabbits (data not shown). Thus, the aquatic environment clearly supports the viability of both nonpathogenic and pathogenic clones, but the former strains are more readily isolated from water by direct cultivation.
In V. cholerae, major virulence genes are clustered in several chromosomal regions and appear to have been recently acquired from phages or through undefined horizontal gene transfer events (33). Consistent with the assumption that aquatic strains have acquired different virulence gene clusters in distinct steps, this study identified groups of environmental strains carrying various combinations of virulence genes (Tables 1 and 2). However, most of these strains belonged to ribotypes that are widely different from those of pandemic strains. This suggests that, although environmental strains may acquire virulence-associated genes and become human pathogens, most of these strains are unlikely to attain pandemic potential by acquisition of TCP and CT genes alone. It should also be emphasized that ribotypes are simply a tool to probe the overall genetic relatedness among strains; comparative genomic sequencing provides the most definitive measure of similarity between strains. Nevertheless, ribotyping has been used extensively to understand genetic relatedness or clonality among V. cholerae strains (20, 23). It thus appears that the non-O1 non-O139 pathogenic strains and the pandemic strains may have distinct lineages. However, this study also identified one TCP+, non-O1 non-O139 strain with a ribotype quite similar to those of a few toxigenic O1 strains (Fig. 2). Hence, although the possibility of serogroup transformation from non-O1 to O1 involving one or more horizontal gene transfer events cannot be ruled out, our data do not support that this happens frequently in the environment. The emergence of V. cholerae O139 from an O1 El Tor strain is the only widely accepted serogroup transformation event of an already existing epidemic clone of V. cholerae.
Enrichment in rabbit ileal loops appears to be a useful technique to isolate pathogenic V. cholerae strains from the environment and intermediate strains that probably have a lower virulence potential than strains associated with pandemic disease. Because colonization is a prerequisite to establishing a productive infection by V. cholerae, and given the role of TCP in colonization, acquisition of the TCP island would provide a strong, selective advantage to any V. cholerae recipient. This assumption is further supported by the observation that the rabbit ileal loop environment enriched strains carrying the TCP island even if these strains were negative for CT. Previous studies established that TCPs are absolutely required for O1 and O139 strains to colonize humans and infant mice (29, 34). However, this study identified TCP- V. cholerae non-O1 non-O139 strains that were competent for colonization in the infant mice. These strains were also negative for the CTX prophage, which was expected because CTXϕ uses TCP as its receptor for infecting new recipient strains (3). Because these strains lack TCP but are evidently still able to colonize mice, they likely produce unknown colonization factors. Identification and characterization of such factors will provide insight into the virulence mechanisms of vibrios that cause sporadic disease and possibly also into the evolution of pathogenic vibrios. This study showed that TCP- CT- strains with pathogenic potential were equally prevalent in the environment as the usual TCP+ pathogenic strains. This finding suggests that the environmental vibrios have evolved into pathogenic forms by more than one pathway. Although the more well characterized pathway to virulence is by acquisition of TCP, apparently another pathway involving genes for at least one hypothetical colonization factor exists. Non-O1 non-O139 strains that are negative for both TCP and CT have also been associated with outbreaks of diarrhea in India (35).
The functions of virulence genes or their homologues in the environment have not been adequately explored. Several studies have suggested a role of virulence factors or their homologues in the symbiotic association of V. cholerae with a variety of aquatic organisms. Besides gene clusters associated with pandemic strains, recent studies have recognized the existence of different alleles of virulence genes in environmental V. cholerae strains, including different alleles of tcpA, tcpF, and toxT genes and different alleles of the CTXϕ prophage repressor rstR in vibrios of various nonepidemic serogroups (9, 10). It has been suggested that the different environmental alleles of virulence genes may have evolved in response to selective pressures that vary between the environment and the host (36). Because V. cholerae is a human pathogen whose natural habitat is the aquatic ecosystem, the existence of clinical and environmental alleles of different genes seems reasonable. Furthermore, the virulence genes carried by clinical strains of V. cholerae may have been derived from an environmental pool of virulence genes or their alleles. In addition to microevolution of individual genes to adapt to a mammalian host, a crucial combination of different horizontally acquired gene clusters seems to be necessary for the emergence of a strain with epidemic potential. However, all the factors controlling the accumulation of critical virulence genes in the pandemic O1 serogroup have not yet been defined.
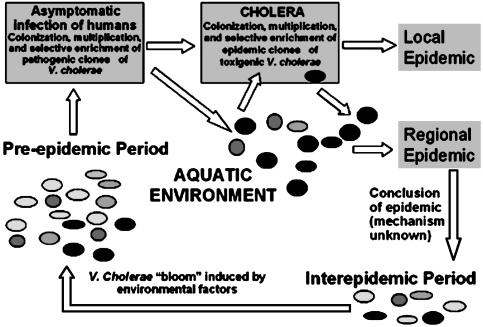
Epidemics of cholera occur with seasonal regularity in the Ganges Delta region of Bangladesh and India. Although water is clearly a vehicle for transmission of V. cholerae, the physical, chemical, and biological parameters that support this seasonal pattern of epidemics are not clear. It has been suggested that during interepidemic periods toxigenic V. cholerae strains exist in an unexplained ecological association with aquatic organisms until the next epidemic season, when environmental factors trigger the dormant bacteria to multiply and lead to cholera outbreaks (1, 2). This assumption, however, falls short of explaining how V. cholerae strains with epidemic potential are selectively enriched before an epidemic from the vast majority of environmental strains that do not appear to have epidemic potential. The present study showed that environmental V. cholerae represents an extensively heterogeneous population of which only a few strains have pathogenic characteristics. This population includes diverse strains carrying a different combination of virulence genes and relatively few virulent strains of the epidemic serogroups O1 and O139. Furthermore, environmental strains with a moderate to high level of virulence potential are enriched in the intestinal environment of a mammalian host. We propose that, in addition to possible seasonal factors causing a bloom of diverse V. cholerae in the environment, epidemics may be preceded by a gradual enrichment of pathogenic strains either in the intestine of an aquatic mammal or more likely through passage in human beings who consume surface water (Fig. 3). A corollary to this hypothesis is that, in an area of endemic disease, before an epidemic season, V. cholerae might be isolated from apparently healthy individuals with asymptomatic infection. This corollary may also constitute a means of surveillance that could predict an imminent epidemic of cholera.
Fig. 3.
Schematic diagram showing enrichment of pathogenic V. cholerae strains in humans with asymptomatic infection before a seasonal epidemic in an area of endemic cholera. Black circles represent V. cholerae strains with epidemic potential, whereas other circles represent the diverse environmental V. cholerae population.
This adaptation of V. cholerae, which is normally a marine or brackish water species, to the human intestine possibly contributed to finding a niche whereby the organism could rapidly amplify and thus ensure its continued existence. It has recently been shown that human colonization creates a hyperinfectious bacterial state that is maintained after dissemination and that may contribute to the epidemic spread of cholera (37). A comparative analysis is needed of diverse V. cholerae strains from clinical and environmental sources, including strains isolated in the present study by using genomic microarrays. These studies are likely to provide a better understanding of the events that led to the evolution of pathogenic V. cholerae clones from free-living, nonpathogenic progenitor strains.
Acknowledgments
This research was funded in part by National Institutes of Health Research Grant GM068851 under a subagreement between the Harvard Medical School and the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B), and by the Swedish International Development Agency under an agreement with ICDDR,B. The ICDDR,B is supported by the aid agencies of the governments of Australia, Bangladesh, Belgium, Canada, Japan, Kingdom of Saudi Arabia, The Netherlands, Sweden, Sri Lanka, Switzerland, and the United States.
Abbreviations: CT, cholera toxin; TCP, toxin-coregulated pilus; FA, fluid accumulation.
References
- 1.Colwell, R. R. & Spira, W. M. (1992) in Cholera, eds. Barua, D. & Greenough, W. B. (Plenum, New York), pp. 107-127.
- 2.Colwell, R. R. & Huq, A. (1994) in Vibrio cholerae and Cholera: Molecular to Global Perspectives, eds. Wachsmuth, I. K., Blake, P. A. & Olsvik O. (ASM Press, Washington, DC), pp. 117-133.
- 3.Faruque, S. M., Albert, M. J. & Mekalanos, J. J. (1998) Microbiol. Mol. Biol. Rev. 62, 1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaper, J. B., Morris, J. G. & Levine, M. M. (1995) Clin. Microbiol. Rev. 8, 48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janda, J. M., Powers, C. R., Bryant, G. & Abbott, S. L. (1988) Clin. Microbiol. Rev. 1, 245-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris, J. G. (1994) in Vibrio cholerae and Cholera: Molecular to Global Perspectives, eds. Wachsmuth, I. K., Blake, P. A. & Olsvik O. (ASM Press, Washington DC), pp. 103-116.
- 7.Mukhopadhyay, A. K., Saha, P. K., Garg, S., Bhattacharya, S. K., Shimada, T., Takeda, T., Takeda, Y. & Nair, G. B. (1995) Epidemiol. Infect. 114, 65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramamurthy, T., Bag, P. K., Pal, A., Bhattacharya, S. K., Shimada, T., Takeda, T., Karasawa, T., Kurasono, H., Takeda, Y. & Nair, G. B. (1993) J. Med. Microbiol. 39, 310-317. [DOI] [PubMed] [Google Scholar]
- 9.Mukhopadhyay, A. K., Chakraborty, S., Takeda, Y., Nair, G. B. & Berg, D. E. (2001) J. Bacteriol. 183, 4737-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque, S. M., Kamruzzaman, M., Meraz, I. M., Chowdhury, N., Nair, G. B., Sack, R. B., Colwell, R. R. & Sack, D. A. (2003) Infect. Immun. 71, 1020-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De, S. N. & Chatterje, D. N. (1953) J. Pathol. Bacteriol. 46, 559-562. [DOI] [PubMed] [Google Scholar]
- 12.Angelichio, M. J., Spector, J., Waldor, M. K. & Camilli, A. (1999) Infect. Immun. 67, 3733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spira, W. M., Sack, R. B. & Froehlich, J. L. (1981) Infect. Immun. 32, 739-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monsur, K. A. (1961) Trans. R. Soc. Trop. Med. Hyg. 55, 440-442. [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization (1974) World Health Organization Guidelines for the Laboratory Diagnosis of Cholera (Bacterial Disease Unit, World Health Organization, Geneva).
- 16.Sack, D. A., Huda, S., Neogi, P. K. B., Daniel, R. R. & Spira, W. M. (1980) J. Clin. Microbiol. 1, 35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaper, J. B., Morris, J. G., Jr., & Nishibuchi, M. (1988) in DNA Probes for Infectious Disease, ed. Tenover, F. C. (CRC, Boca Raton, FL), pp. 65-77.
- 18.Pal, A., Ramamurthy, T., Bhadra, R. K., Takeda, T., Shimada, T., Takeda, Y., Nair, G. B., Pal, S. C. & Chakrabarti, S. (1992) Appl. Environ. Microbiol. 58, 2485-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brosius, J., Ullrich, A., Raker, M. A., Gray, A., Dull, T. J., Gutell, R. R. & Noller, H. F. (1981) Plasmid 6, 112-118. [DOI] [PubMed] [Google Scholar]
- 20.Faruque, S. M., Roy, S. K., Alim, A. R. M. A., Siddique, A. K. & Albert, M. J. (1995) J. Clin. Microbiol. 33, 2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis, T., Fritsch, E. F. & Sambrook. J. (1982) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 22.Keasler, S. P. & Hall, R. H. (1993) Lancet 341, 1661 (lett.). [DOI] [PubMed] [Google Scholar]
- 23.Faruque, S. M., Siddique, A. K., Saha, M. N., Asadulghani, Rahman, M. M., Zaman, K., Albert, M. J., Sack, D. A. & Sack, R. B. (1999) J. Clin. Microbiol. 37, 1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faruque S. M., Asadulghani, Saha, M. N., Alim, A. R. M. A., Albert, M. J., Islam, K. M. N. & Mekalanos, J. J. (1998) Infect. Immun. 66, 5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faruque, S. M., Asadulghani, Kamruzzaman, M., Nandi, R. K., Ghosh, A. N., Nair, G. B., Mekalanos, J. J. & Sack, D. A. (2002) Infect. Immun. 70, 163-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivera, I. N. G., Chun, J., Huq, A., Sack, R. B. & Colwell, R. R. (2001) Appl. Environ. Microbiol. 67, 2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow, K. H., Ng, T. K., Yuen, K. Y. & Yam, W. C. (2001) J. Clin. Microbiol. 39, 2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faruque, S. M., Chowdhury, N., Kamruzzaman, M., Ahmad, Q. S., Faruque, A. S. G., Salam, M. A., Ramamurthy, T., Nair, G. B., Weintraub, A. & Sack, D. A. (2003) Emerg. Infect. Dis. 9, 1116-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thelin, K. H. & Taylor, R. K. (1996) Infect. Immun. 64, 2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, W., Fullner, K. J., Clayton, R., Sexton, J. A., Rogers, M. B., Calia, K. E., Calderwood, S. B., Fraser, C. & Mekalanos, J. J. (1999) Proc. Natl. Acad. Sci. USA 96, 1071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faruque, S. M., Saha, M. N., Asadulghani, Sack, D. A., Sack, R. B., Takeda, Y. & Nair, G. B. (2000) J. Infect. Dis. 182, 1161-1168. [DOI] [PubMed] [Google Scholar]
- 32.Zo, Y. G., Rivera, I. N. G., Russek-Cohen, E., Islam, M. S., Siddique, A. K., Yunus, M., Sack, R. B., Huq, A. & Colwell, R. R. (2002) Proc. Natl. Acad. Sci. USA 99, 12409-12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faruque, S. M. & Mekalanos, J. J. (2003) Trends Microbiol. 11, 505-510. [DOI] [PubMed] [Google Scholar]
- 34.Herrington, D. A., Hall, R. H., Losonsky, G., Mekalanos, J. J., Taylor, R. K. & Levine, M. M. (1988) J. Exp. Med. 168, 1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma, C., Thungapathra, M., Ghosh, A., Mukhopadhyay, A. K., Basu, A., Mitra, R., Basu, I., Bhattacharya, S. K., Shimada, T., Ramamurthy, T., et al. (1998) J. Clin. Microbiol. 36, 756-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyd, E. F. & Waldor, M. K. (2002) Microbiology 148, 1655-1666. [DOI] [PubMed] [Google Scholar]
- 37.Merrell, D. S., Butler, S. M., Qadri, F., Dolganov, N. A., Alam, A., Cohen, M. B., Calderwood, S. B., Schoolnik, G. K. & Camilli A. (2002) Nature 417, 642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]