SUMMARY
Translational control permits cells to respond swiftly to changing environment. Rapid attenuation of global protein synthesis under stress conditions has been largely ascribed to the inhibition of translation initiation. Here we report that intracellular proteotoxic stress reduces global protein synthesis by halting ribosomes on transcripts during elongation. Deep sequencing of ribosome-protected mRNA fragments reveals an early elongation pausing, roughly at the site where nascent polypeptide chains emerge from the ribosomal exit tunnel. Inhibiting endogenous chaperone molecules by a dominant-negative mutant or chemical inhibitors recapitulates the early elongation pausing, suggesting a dual role of molecular chaperones in facilitating polypeptide elongation and co-translational folding. Our results further support the chaperone “trapping” mechanism in promoting the passage of nascent chains. Our study reveals that translating ribosomes fine-tune the elongation rate by sensing the intracellular folding environment. The early elongation pausing represents a co-translational stress response to maintain the intracellular protein homeostasis.
Keywords: Translation, elongation, ribosome, pausing, chaperone, stress response
INTRODUCTION
Protein misfolding imposes a major risk to the health of cells and organisms. An elaborate protein quality control (PQC) system has been laid down during evolution to maintain protein homeostasis – a delicate balance between protein synthesis, folding, and degradation (Bukau et al., 2006; Frydman, 2001; Hartl et al., 2011). Molecular chaperones are “cellular lifeguards” that govern the integrity of proteome. By interacting with different co-chaperones and co-factors, Hsp70 family proteins actively participate in protein triage decisions from folding, degradation, to aggregation (McClellan et al., 2005; Zhang and Qian, 2011). Most recent studies highlighted the robust network of chaperones acting co-translationally on nascent chains in eukaryotic (del Alamo et al., 2011) as well as prokaryotic cells (Oh et al., 2011). Interestingly, prokaryotes and eukaryotes have evolved distinct ribosome-associated chaperone systems (Kramer et al., 2009). In S. cerevisiae, two ribosome-associated systems interact with newly synthesized polypeptides, the nascent chain-associated complex (NAC) and the Hsp70-based Ssb/Ssz/Zuo triad system (Kampinga and Craig, 2010). Both systems are physically located in close proximity at the peptide exit tunnel of ribosomes. The ribosome-associated chaperone system also exists in mammals, although its functionality is not fully understood (Jaiswal et al., 2011). Despite the wide appreciation of the impact that this chaperone system may have on co-translational folding, little is known about how the ribosome-associated chaperone system regulates the process of translation per se
mRNA translation can be divided into three stages - initiation, elongation and termination. Regulation of translation occurs predominantly during initiation phase (Sonenberg and Hinnebusch, 2009; Spriggs et al., 2010). The initiation is a complex multi-step process governed by a large number of protein factors and involves mRNA 5′-cap recognition, scanning and start codon recognition (Gray and Wickens, 1998; Jackson et al., 2010). Much attention has been focused on the role of translation initiation factors (eIFs) in the assembly of elongation-competent ribosome complexes. However, after the commitment of polypeptide synthesis, the regulatory steps during elongation remain poorly understood.
Given the fact that translation consumes a lion’s share of energy, cells often reduce global protein synthesis under most, if not all, types of adverse conditions. The global repression of protein synthesis not only saves the cellular energy, but also relieves the burden of the PQC system due to the less protein production (Holcik and Sonenberg, 2005). Current models for the mechanism governing this translational attenuation are largely limited to the initiation stage. For instance, eIF4F complex-mediated cap recognition and eIF2-controlled ternary complex formation are key initiation targets in controlling global mRNA translation (Ma and Blenis, 2009; Ron and Walter, 2007). In response to stresses, the shutdown of protein synthesis is, in general, mediated either by the inhibition of 43S complex loading to the 5′ end cap and/or reducing the amount of ternary complex that is available. Despite the well-documented role of these initiation regulators, it remains surprisingly obscure whether the 80S ribosome, once assembled on the mRNA, maintains the responsiveness to protein misfolding during elongation.
Here we report that proteotoxic stress triggers ribosomal pausing during elongation. Remarkably, the pausing occurs primarily near the site where nascent polypeptide chains emerge from the ribosomal exit tunnel. We demonstrate that the early elongation pausing is induced by the sequestration of chaperone molecules by misfolded proteins. Our results expand the critical role of chaperone molecules form co-translational folding to polypeptide elongation. The early elongation pausing of ribosomes thus represents a mechanism of co-translational stress response to maintain intracellular protein homeostasis.
RESULTS
Proteotoxic Stress Attenuates Global Protein Synthesis
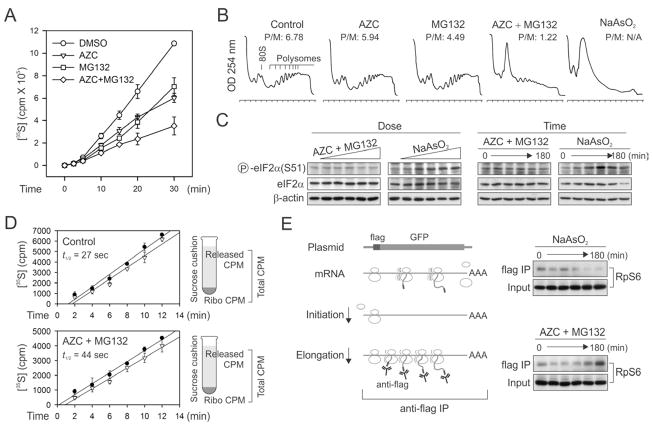
Intracellular accumulation of misfolded proteins is a common feature of a variety of stress conditions. To induce misfolding of newly synthesized polypeptides without massively perturbing cellular functions, we used an amino acid analog L-azetidine-2-carboxylic acid (AZC) that competes with proline during amino acid incorporation (Goldberg and Dice, 1974). Once incorporated into proteins in place of proline, AZC potently induces protein misfolding and degradation (Qian et al., 2010; Trotter et al., 2002). Pre-exposure of HEK293 cells to 10 mM AZC resulted in a marked reduction of [35S] incorporation (Figure 1A). In agreement with the enhanced degradation of AZC-incorporated polypeptides, pulse-chase analysis showed an increased turnover of [35S] labeled proteins in the presence of AZC (Figure S1A). We asked whether proteasome inhibition would prevent the loss of [35S] incorporation by blocking the degradation. To our surprise, adding proteasome inhibitor MG132 further decreased the total amount of [35S] incorporation (Figure 1A). This was not due to the side effects of MG132 because adding this inhibitor alone only partially reduced the level of [35S] incorporation. Since the AZC-induced misfolded polypeptides progressively accumulate under proteasome inhibition, it appears that the intracellular proteotoxic stress triggers a rapid attenuation of translation. To substantiate this finding further, we analyzed the polysome profiles by velocity sedimentation of lysates in sucrose gradients. Treatment with either AZC or MG132 alone had minor effects on the polysome formation (Figure 1B). In contrast, the presence of both AZC and MG132 markedly disassembled the polysomes with an approximately 5 fold decrease in the polysome/monosome (P/M) ratio.
Figure 1. Proteotoxic stress attenuates protein synthesis by affecting translation elongation.
(A)Global protein synthesis in HEK293 cells treated with either 10 mM AZC, or 20 μM MG132, or both. [35S] radioactivity of trichloroacetic acid (TCA)-insoluble material was measured at given times. Means ± SEM of four experiments are shown.
(B) Polysome profiles were determined using sucrose gradient sedimentation. HEK293 cells were pre-treated with either 10 mM AZC, or 20 μM MG132, or both for 60 min followed by polysome preparation. P/M ratio is calculated by comparing areas under the polysome and 80S peak.
(C) HEK293 cells were treated with increasing doses of AZC (from 0 to 25 mM with 5-fold dilution) in the presence of 20 μM MG132 for 60 min, or increasing doses of NaAsO2 (from 0 to 1 mM with 2-fold dilution) for 60 min (left two panels), followed by immunoblotting using antibodies as indicated. The right two panels show the immunoblotting results of cells treated with 10 mM AZC and 20 μM MG132 or 500 μM NaAsO2 for various times (0, 10, 30, 60, 120, and 180 min)
(D) The ribosomal half-transit time was determined in the absence or presence of 10 mM AZC and 20 μM MG132. Fitting lines of [35S] incorporation into total (filled circle) and completed (open triangle) protein synthesis are obtained by linear regression. Means ± SEM of three experiments are shown.
(E) Schematic for nascent chain immunoprecipitation assay to differentiate elongation defect from initiation deficiency (left panel). HEK293 cells expressing Flag-GFP were pre-treated with 10 mM AZC and 20 μM MG132 or 500 μM NaAsO2 for various times (0, 10, 30, 60, 120, and 180 min). Immuno-precipitation was performed using anti-Flag antibody-coated beads followed by immunoblotting with anti-RpS6 antibody. The 0 time point serves as the control condition without any drug treatment. See also Figure S1.
Proteotoxic Stress Affects Primarily Translation Elongation
To investigate the mechanisms underlying the proteotoxic stress-induced translational attenuation, we examined the phosphorylation status of eIF2α, a prominent initiation regulator in the unfolded protein response (Ron and Walter, 2007). In contrast to sodium arsenite (NaAsO2), a known inducer of eIF2α phosphorylation, treating cells with both AZC and MG132 at increasing doses and for extended times had little effect on eIF2α phosphorylation (Figure 1C). Additionally, we observed no change in the phosphorylation of S6 and its kinase S6K1, one of the downstream targets of mammalian target of rapamycin complex 1 (mTORC1) (Jackson et al., 2010; Ma and Blenis, 2009) (Figure S1B). Thus, the intracellular proteotoxic stress does not affect primarily the translation initiation regulators, at least in the early stage.
We next examined whether proteotoxic stress inhibits protein synthesis by interfering with post-initiation events, such as elongation. One way to distinguish elongation from initiation is the formation of stress granules (SG). Inhibiting translation initiation triggers SG formation, whereas blocking translation elongation prevents this process (Buchan and Parker, 2009; Kedersha et al., 2000). Unlike sodium arsenite treatment that induced an evident SG formation, adding both AZC and MG132 to cells failed to induce any discernible SG formation (Figure S1C). Thus proteotoxic stress likely affects translation elongation rather than initiation. To assess independently whether the reduced protein synthesis under proteotoxic stress was primarily due to defective elongation, we determined ribosomal transit times in these cells. The ribosomal transit time refers to the time required for a ribosome, after initiation, to traverse an average-sized mRNA and release the completed polypeptide chain (Nielsen and McConkey, 1980). The estimated half-transit time (t1/2) in the presence of both AZC and MG132 (44s) was ~1.6 fold longer than that in control cells (27s) (Figure 1D), confirming that proteotoxic stress significantly reduced the elongation rate of polypeptide synthesis. Additionally, we conducted an elongation chase experiment using a synthesized firefly luciferase (Fluc) mRNA in lysates programmed from cells with or without proteotoxic stress. Compared to the control, the stressed cell lysates showed a delayed accumulation of Fluc activity (Figure S1D), further indicating a slowdown of elongation process under proteotoxic stress.
To examine whether the stalled ribosome during elongation was still associated with the newly synthesized polypeptide, we performed nascent chain immunoprecipitation followed by detection of ribosomal small subunit S6 (RpS6). We established a HEK293 cell line stably expressing a GFP reporter with an NH2-terminal Flag-tag (Figure 1E). We enriched the ribosome complexes bearing the partially synthesized GFP by anti-Flag immunoprecipitation. Arsenite treatment led to a progressive loss of the associated RpS6 in a time course-dependent manner (Figure 1E, right top panel), which is consistent with the inhibition of translation initiation. Remarkably, treating cells with both AZC and MG132 resulted in an accumulation of RpS6 in the anti-Flag precipitates, a clear evidence of paused ribosomes on the mRNA during elongation. The prolonged ribosome association with the nascent chain persists in the polysome fractions of these cells (Figures S1E and S1F). Taken together, our results strongly indicate that proteotoxic stress acts at the level of translation elongation to suppress protein synthesis.
Proteotoxic Stress Triggers Early Elongation Pausing of Ribosomes
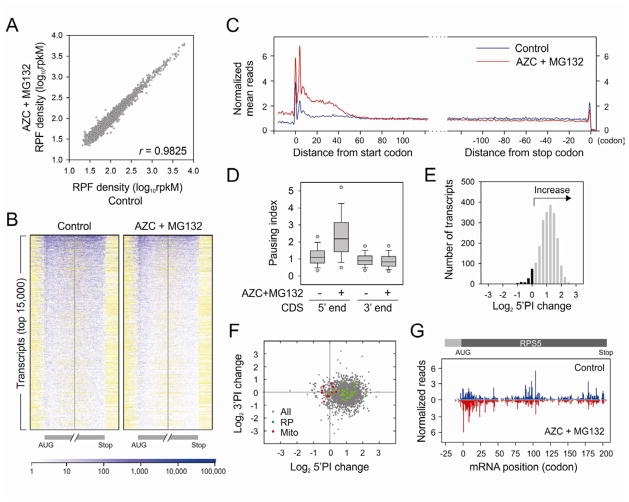
A defective translation elongation should result in slower ribosome run-off and the retention of polysomes (Saini et al., 2009). It is surprising to find that the polysomes were largely disassembled in cells treated with both AZC and MG132 (Figure 1B). We considered the possibility that proteotoxic stress primarily induced ribosomal pausing at the early stage of elongation, thereby creating a road block for following ribosomes. To provide a definitive assessment of ribosome positions on mRNAs under proteotoxic stress, we isolated the ribosome protected mRNA fragments (RPFs) and performed deep-sequencing using methods reported previously (Ingolia et al., 2009). RPF reads obtained from cells with or without proteotoxic stress were of equal quality as evidenced by the similar size distribution and strong 3-nt periodicity after alignment. Notably, AZC and MG132 treatment did not result in global variation in overall ribosome density along each transcript (r = 0.9825) (Figure 2A). To directly visualize the pattern of RPF distribution on individual transcripts, we built a ribosome density map across the entire transcriptome (Figure 2B). Compared to control cells, the presence of both AZC and MG132 led to a clear enrichment of RPF density at the 5′ end of coding sequences (CDS) on the vast majority of mRNAs. Meta-gene analysis revealed a pronounced accumulation of RPF reads within the first 50 codon region of transcripts in cells treated with both AZC and MG132 (Figure 2C). We defined the ribosome pausing index (PI) of individual transcript by calculating the normalized ribosome density within a 50-codon window from start codon (5′PI) or stop codon (3′PI) respectively. In cells under proteotoxic stress, the median 5′PI showed more than 2-fold increase as compared to control cells (Figure 2D). Intriguingly, proteotoxic stress also caused an elevation of RPF density in the 5′ untranslated region (5′UTR) (Figure 2C), an indication of wide-spread alternative initiation under stress conditions.
Figure 2. Intracellular proteotoxic stress triggers early elongation pausing of ribosomes.
(A) HEK293cells were treated with 10 mM AZC and 20 μM MG132 for 1 h before ribosome profiling. Ribosome densities of cells with or without treatment are plotted for comparison. The density in reads per kilobase of coding sequence per million mapped reads (rpkM) is a measure of overall translation along each transcript.
(B) Ribosome density heat-maps of cells with or without treatment. The entire transcriptome is sorted based on total RPF reads and the top 15,000 transcripts are aligned in row. Both the first and last 160-codon regions of CDS are shown, together with flanking 40-codon untranslated regions. Reads density is represented in blue. White color indicates regions without reads, whereas yellow for regions without sequence. A short 5′UTR has yellow region before the AUG, whereas a short 3′UTR has yellow region after the stop codon.
(C) Meta-gene analysis of early ribosome pausing of cells with or without treatment. Normalized RPF reads are averaged across the entire transcriptome, aligned at either their start (left panel) or stop (right panel) codon, and plotted as smoothed lines.
(D) Ribosome pausing index (PI) is determined in a 50-codon window at the beginning (5′ end) and end (3′ end) of CDS, respectively. Both the 5′ and 3′ PI of each transcript in cells with or without treatment are shown in box plots with single dots as 5th/95th percentile.
(E) Distribution of 5′PI changes in cells with proteotoxic stress. The log2 change of 5′PI after AZC and MG132 treatment is plotted, with the increase shown in grey bar and the decrease in black.
(F) Changes of 5′PI and 3′PI after AZC and MG132 treatment. The log2 change is computed across the entire transcriptome and presented as a scatter plot with green dots for genes encoding ribosome subunits (RP) and red dots for mitochondria-encoded genes (Mito).
(G) A typical example of early elongation pausing under proteotoxic stress. RPF reads density is shown on the CDS of RPS5 with or without AZC and MG132 treatment. See also Figure S2 – S4.
A large portion of mRNAs showed an increased 5′PI in response to proteotoxic stress (Figure 2E, grey bar). However, a small group of transcripts showed less change or even decreased 5′PI (Figure 2E, black bar). At the transcriptome level, neither the CDS length nor the overall translation had any strong correlation with the changes of 5′PI (Figures S2A and S2B). Gene ontology (GO) analysis revealed that genes involving ATP synthesis (e.g., mitochondria-encoded genes) were enriched in the group with decreased 5′PI in response to proteotoxic stress (Figures S2C, S2D and Figure 2F). In contrast, genes with increased 5′PI were involved in cellular processes like RNA metabolism and translation (e.g., ribosomal proteins). As a typical example, proteotoxic stress led to a clear ribosome accumulation near the beginning of RPS5 CDS region (Figure 2G).
Through an independent biological replicate, we confirmed the early elongation pausing of ribosomes in response to proteotoxic stress (Figure S3). The global RPF distribution was highly reproducible across the replicates (Figure S3E). Notably, treatment with either AZC or MG132 alone had little effects on the ribosome dynamics (FigureS S4A and S4B). In particular, we saw no unique pausing sites at individual codon in the presence of AZC (Figure S4C). These results argue that the presence of AZC-charged tRNA neither perturbs the intracellular pool of amino acids nor alters the behavior of translating ribosomes. Therefore, it is the accumulation of misfolded proteins that triggers the early elongation pausing.
A Dominant-Negative Hsc70 Mutant Induces Early Elongation Pausing of Ribosomes
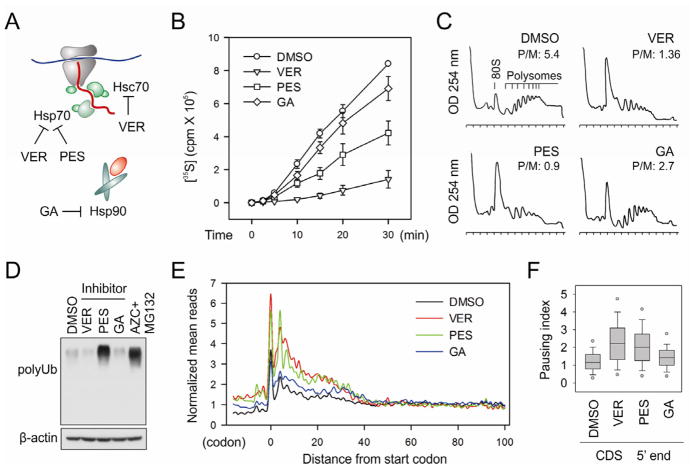
The approximate 50 codon region where the elongation pausing occurs under proteotoxic stress corresponds remarkably well to the length of polypeptide needed to fill the exit tunnel of the ribosome (approximately 30–40 amino acids in extended conformation) (Kramer et al., 2009). This raises an intriguing possibility that the changing environment of nascent polypeptides from the ribosome tunnel to the cytosol might influence the dynamics of translating ribosomes. Within the cellular environment, the emerging nascent chains interact with molecular chaperones that guide their folding process. At the forefront is Hsc/Hsp70 that transiently associates with a large fraction of nascent chains (Frydman, 2001; Hansen et al., 1999; Kampinga and Craig, 2010). This led us to hypothesize that the accumulated misfolded proteins titrate out the intracellular chaperone pool and the lack of chaperone association might prevent nascent chains from protruding out of the ribosome exit tunnel. The elongation slowdown at this position likely causes ribosomes to pile up over the first 50-codon region. Supporting the notion that proteotoxic stress sequesters intracellular chaperone molecules, we observed a progressive loss of ribosome associated Hsc70 along with AZC and MG132 treatment (Figure 3A). To test the hypothesis that reduced chaperone availability leads to an early elongation pausing, we first used a dominant-negative mutant Hsc70 (K71M), which sequesters and inactivates the endogenous Hsc70 molecules (Newmyer and Schmid, 2001). The integrated “tet-off” system allows a rapid induction of the transgene expression in HeLa-tTA cells after removal of doxycycline (Dox) (Figure S5A). After 12h of transgene induction by removal of Dox, [35S] metabolic labeling revealed ~ 40% decrease of the global protein synthesis (Figure 3B). Similar to cells treated with both AZC and MG132, Hsc70(K71M) expression caused disassembly of polysomes with a concomitant increase of 80S peak (Figure 3C).
Figure 3. Disrupting endogenous Hsc70 recapitulates the effects of proteotoxic stress on early elongation pausing.
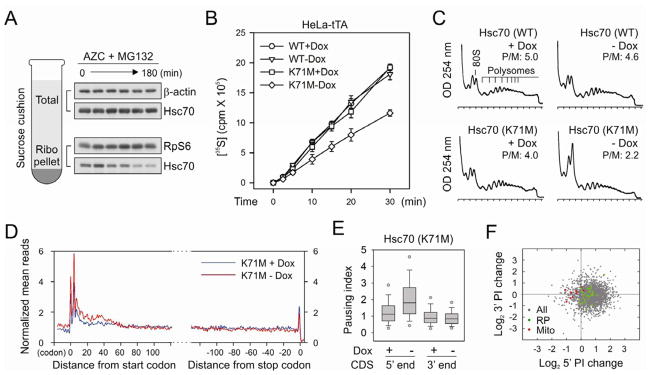
(A) Sucrose cushion analysis of ribosome-associated Hsc70 along with AZC and MG132 treatment. Both the total and ribosome pellet were immunoblotted using antibodies as indicated.
(B) Global protein synthesis was analyzed in HeLa-tTA cells infected with adenoviruses expressing Hsc70(WT) and Hsc70(K71M). Transgene expression was induced by 12 h Dox removal. [35S] radioactivity of TCA-insoluble material was measured at given times. Means ± SEM of three experiments are shown.
(C) Polysome profiles were determined from cells as in (B) using sucrose gradient sedimentation.
(D) Meta-gene analysis for early elongation pausing in cells with or without Hsc70(K71M) expression. Normalized RPF reads are averaged across the entire transcriptome, aligned at either their start (left panel) or stop (right panel) codon.
(E) Both the 5′ and 3′ PI of each transcript in cells with or without Hsc70(K71M) expression are shown in box plots.
(F) Changes of 5′PI and 3′PI after Hsc70(K71M) expression. The log2 change is computed across the entire transcriptome and presented as a scatter plot with green dots for genes encoding ribosome subunits and red dots for mitochondria- encoded genes. See also Figure S5.
To evaluate whether Hsc70(K71M) expression leads to an early elongation pausing, we performed deep sequencing of RPFs extracted from the polysomes of HeLa-tTA cells with or without transgene induction. Meta-gene analysis revealed a modest excess (~1.6-fold) in density over the initial 50 codons after Hsc70(K71M) expression (Figure 3D and 3E). This is similar in pattern, but of smaller magnitude of early elongation pausing seen in cells treated with both AZC and MG132 (Figure 2B). We repeated the experiment and obtained the similar extent of elongation pausing in the presence of Hsc70(K71M) (Figure S5). Similar to AZC and MG132 treatment, there was an evident separation between genes encoding ribosome subunits and mitochondria proteins in response to the Hsc70(K71M) expression (Figure 3F). The 5′PI changes also showed a good correlation between the two conditions (r = 0.65), although different cell lines were used (Figure S5F). Thus, interfering with endogenous Hsc70 recapitulates the effects of the proteotoxic stress in triggering early elongation pausing.
Direct Hsc/Hsp70 Inhibition Induces Early Elongation Pausing of Ribosomes
The dominant-negative Hsc70(K71M) mutant induced a rather weak elongation pausing when compared to AZC and MG132 treatment. It was likely due to an adaptive stress response under 12 h of Hsc70(K71M) expression, in which the subsequent induction of Hsp70 compromised the early elongation pausing (Figure S5A). In contrast, 1 h of AZC and MG132 treatment did not yet trigger Hsp70 expression due to the time lag. Additionally, the continuous presence of the analog prevents the production of functional chaperones, if any. To address whether chaperones play a direct role in translation elongation, we applied several specific chaperone inhibitors to HEK293 cells and monitored global protein synthesis (Figure 4A). VER-155008 is a potent inhibitor of the Hsp70 family chaperones (Massey et al., 2010), whereas 2-phenylethyenesulfo-namide (PES) acts as a direct inhibitor of stress-inducible Hsp70 (Leu et al., 2011). We also included a specific Hsp90 inhibitor geldanamycin (GA) to examine the role of different chaperones in ribosome behavior. To minimize the compensatory stress response, we only treated cells with these inhibitors for 1 h. This short treatment allows us to capture direct effects of chaperone inhibition without inducing massive accumulation of misfolded proteins.
Figure 4. Direct Hsc/Hsp70 inhibition induces early elongation pausing of ribosomes.
(A) Schematic for chaperone targets of small molecule inhibitors. VER155008 inhibits Hsc70, Hsp70 and Grp78 (not shown); PES selectively inhibits Hsp70; whereas geldanamycin (GA) is a specific inhibitor of Hsp90.
(B) Global protein synthesis was analyzed in HEK293 cells treated with 100 μM VER, 50 μM PES, or 1 μM GA for 1 h. [35S] radioactivity of TCA-insoluble material was measured at given times. Means ± SEM of three experiments are shown.
(C) Polysome profiles were determined from cells treated with chaperone inhibitors as in (B) using sucrose gradient sedimentation.
(D) Immunoblotting of whole cell lysates from cells treated with chaperone inhibitors as in (B).
(E) Meta-gene analysis for early elongation pausing in cells treated with chaperone inhibitors as in (B). Normalized RPF reads are averaged across the entire transcriptome, aligned at their start codon.
(F) The 5′ PI of each transcript in cells treated with chaperone inhibitors as in (B) are shown in box plots. See also Figure S6.
Metabolic radiolabeling analysis revealed that both VER and PES potently inhibited [35S] incorporation, whereas the Hsp90 inhibitor GA slightly reduced the level of global protein synthesis (Figure 4B). The extent of translation repression was also reflected in the pattern of polysome profile, in which the inhibitors of Hsp70 family protein, but not Hsp90, disassembled the polysome (Figure 4C). Despite the most severe inhibition of protein synthesis, 1 h treatment of VER resulted in little accumulation of ubiquitin conjugated species in cells (Figure 4D). In addition, the steady state chaperone levels remained unchanged in the presence of these inhibitors (Figure S6), suggesting that the stress response after 1 h of chaperone inhibition was minimal. We next performed deep sequencing of RPFs derived from the cells treated with these chaperone inhibitors. Meta-gene analysis revealed a prominent excess of ribosome density over the first 50-codon region in cells treated with either VER or PES (Figure 4E, 4F). Only minor effect was observed after GA-mediated Hsp90 inhibition. Collectively, these results indicate that direct inhibition of Hsp70 family proteins triggers early elongation pausing of ribosomes.
Co-translational Interaction of Nascent Chains Influences Elongation Rate
Hsp70 family proteins, including BiP of the endoplasmic reticulum and mtHsp70 of the mitochondrion, are essential for protein translocation across the membrane via unidirectional pulling (Jensen and Johnson, 1999). It is likely that the cytosol Hsc/Hsp70 uses the similar mechanism to pull the emerging polypeptide out of the ribosome exit tunnel. Two models have been proposed to describe how Hsp70 can generate such driving force: “trapping” and “power stroking” (Goloubinoff and De Los Rios, 2007). While both models rely on direct interactions, the latter requires ATP hydrolysis. We hypothesize that the Hsc/Hsp70 “trapping” might be sufficient to exert an entropy pulling force because most nascent chains emerging the exit tunnel are unfolded. To investigate whether co-translational protein interaction would generate the “pulling” force for the ribosome-bound nascent chain, we utilized the hetero-dimerization property of FRB (FKBP12-rapamycin binding domain) and FKBP (FK506 binding protein), whose high affinity binding can be induced by the small molecule rapamycin (Choi et al., 1996; Qian et al., 2009). We constructed a fusion protein FRB-GFP in order to evaluate whether the association of the NH2-terminal FRB domain with the added FKBP protein during translation would affect the elongation rate of the carboxyl terminal GFP. In an in vitro translation system based on rabbit reticulocyte lysate (RRL), we compared the translation efficiency of FRB-GFP after supplementation with the recombinant FKBP protein. Remarkably, upon addition of 1 μM rapamycin to the RRL, the kinetics of FRB-GFP completion showed a significant acceleration (Figure 5A, left panel). We observed the similar effects after swapping the FRB and FKBP domains (Figure 5A, right panel). Thus, co-translational interaction between nascent chains and specific binding partners promotes the elongation of emerging polypeptides.
Figure 5. Co-translational interaction of nascent chains facilitates the elongation of polypeptides.
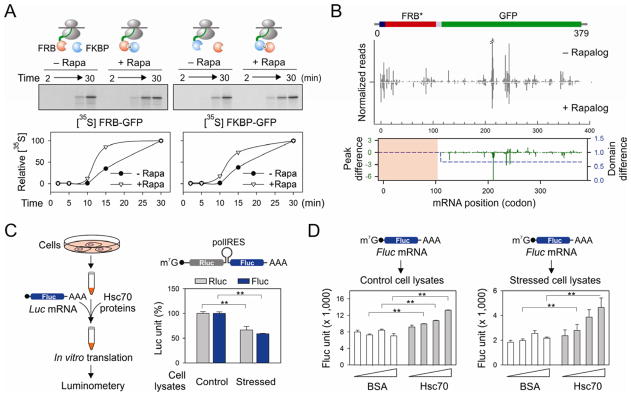
(A) Effects of FKBP (blue ball) on the in vitro translation of FRB-GFP (red ball) in the absence or presence of 1 μM rapamycin (left panel). The right panel shows the effects of FRB (red ball) on the in vitro translation of FKBP-GFP (blue ball) in the absence or presence of 1 μM rapamycin. Autoradiography of full length GFP fusion protein is quantitated and plotted as a function of time.
(B) HEK293 expressing FRB*-GFP was transfected with the plasmid encoding FKBP. Cells were pre-treated with 1 μM rapalog for 60 min before polysome profiling. The RPF density profiles are shown for the transgene FRB*-GFP with and without rapalog treatment. The RPF reads density is normalized based on the FRB* domain. The average change of RPF density over the entire GFP region (blue dot line) and single codon change (green line) are plotted together (Wilcoxon signed-rank test, p-value = 3 × 10−4). See also Figure S7
(C) Schematic of experimental design using recombinant Hsc70 protein to restore translation efficiency using an in vitro translation system programmed from cells with or without proteotoxic stress. The right panel shows the relative translation efficiency of a synthesized bicistronic mRNA containing a polio IRES element between Rluc and Fluc. Error bar: SEM. **, p < 0.001.
(D) The in vitro translation system as (C) was used to translate a synthesized Fluc mRNA in the absence or presence of recombinant Hsc70. Error bar: SEM. **, p < 0.01.
In order to extend these findings from RRL to mammalian cells, we utilized a well-characterized rapamycin analog AP21967 (rapalog) and a mutant FRB domain (FRB*) to avoid interfering with the endogenous mTOR function (Klemm et al., 1998). A HEK293 cell line stably expressing FRB*-GFP was transfected with plasmid-borne FKBP. After 60 min of pre-incubation with rapalog, polysome fractions were isolated followed by deep sequencing of RPFs. Notably, the presence of rapalog had little effect on the pattern of RPFs across the entire transcriptome (Figure S7). However, the FRB*-GFP transcript exhibited an altered distribution of RPF reads after rapalog treatment (Figure 5B). When the total reads mapped to the FRB* domain were normalized to be equal, rapalog treatment resulted in a 34% decrease of the average RPF density in the coding region of GFP (Figure 5B, bottom panel). Single codon comparison revealed that the reduction of RPF reads mainly occurred at the ribosome pausing sites of GFP. Since the RPF density on a given codon is proportional to the average ribosome dwell time there, the reduced ribosome density after co-translational interaction between FRB* and FKBP suggests an accelerated elongation for the remaining polypeptide.
Increasing Chaperone Availability Restores Translation Efficiency
The functional connection between chaperone availability and translation elongation underscores the central role of chaperones in protein homeostasis. Based on our results, we expected that chaperone overexpression might prevent the translation inhibition under proteotoxic stress. However, it is inherently difficult to alter the chaperone levels in cells because the chaperone concentration is controlled closely by the heat shock transcription factor 1 (HSF1) (Morimoto, 2008). Overexpression of exogenous chaperone genes inevitably suppresses the endogenous chaperone expression. To circumvent this limitation, we established an in vitro translation system programmed from cells with or without proteotoxic stress (Figure 5C, left panel). We first examined the translation efficiency using a synthesized bicistronic mRNA containing the polio internal ribosome entry site (IRES) between Renilla luciferase (Rluc) and firefly luciferase (Fluc). While the synthesis of Fluc is cap-dependent, translation of Rluc is driven by IRES via a cap-independent mechanism (Sun et al., 2011). In lysates derived from stressed cells, the synthesis of both Rluc and Fluc were equally reduced in comparison with the control cell lysates (Figure 5C, right panel). This result further supports the notion that proteotoxic stress does not primarily inhibit cap-dependent initiation.
Next we monitored the translation efficiency of a synthesized Fluc mRNA in cell lysates supplemented with recombinant chaperone molecules. Adding recombinant Hsc70, but not bovine serum albumin (BSA), increased the Fluc activities in a dose-dependent manner (Figure 5D). Co-translational folding of Fluc has been shown to be quite efficient (Kolb et al., 2000), so the translation rate is likely to be the major determinant of luciferase activity in the in vitro translation system. Notably, the chaperone-mediated rescue effect was more dramatic in the system derived from the stressed cells than the control. Therefore, increasing chaperone availability restores the translation efficiency.
DISCUSSION
The journey of a nascent polypeptide starts from the pepetidyl transferase center (PTC) of the ribosome followed by traversing the peptide exit tunnel. Once the nascent chain begins to emerge from the exit tunnel, it faces a drastic environmental change. Surprisingly, most recent ribosome profiling data did not show any specific pausing sites corresponding to this turning point (Guo et al., 2010; Ingolia et al., 2011). The smooth transition from the inside of the tunnel to the outside ribosome surface is likely due to the presence of ribosome-associated chaperone systems (Kramer et al., 2009). Our study provides strong evidence that the Hsc/Hsp70 family protein plays a crucial role in the passage of nascent chains upon emerging from the ribosome exit tunnel. Reducing chaperone availability by proteotoxic stress or chemical inhibitors unequivocally caused a pileup of ribosomes on the first 50-codon region of transcripts. Notably, we did not observe any specific RPF spikes at specific codon positions, suggesting that the lack of chaperone association slows down rather than stops the elongation. The feature of ribosome stacking at the 5′ end of the CDS further indicates that the stress-induced elongation pausing precedes the suppression of translation initiation.
mRNA translation proceeds not at a constant rate but rather in a stop-and-go traffic manner (Fredrick and Ibba, 2010). Variations of elongation speed may result from local stable mRNA structure (Gray and Hentze, 1994), or the presence of rare codons (Elf et al., 2003; Lavner and Kotlar, 2005). Interestingly, nascent chains could also induce translational pausing in a sequence-specific manner (Kramer et al., 2009). Our results uncover an additional layer of elongation regulation mediated by the ribosome-associated chaperone system. The Hsc/Hsp70 family protein, like the ER and mitochondrion counterparts, not only assists co-translational folding, but also accelerates the elongation of nascent polypeptides primarily at the site where the nascent polypeptide emerges from the ribosome exit tunnel. Early studies in S. cerevisiae reported a similar function for Ssb, although identifying the elongation pausing sites was beyond the technical ability at that time (Nelson et al., 1992). Since multiple factors constitute the chaperone network linked to protein synthesis (Albanese et al., 2006), it will be intriguing to determine whether interfering specific chaperone or co-chaperone molecules causes selective elongation pausing on a subset of transcripts.
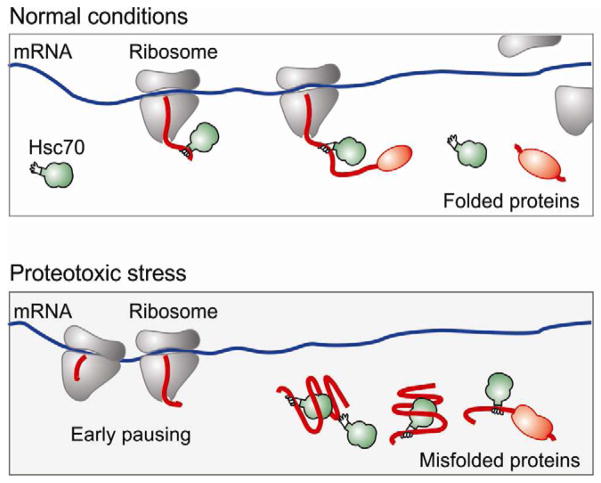
Despite the apparent abundance of chaperone molecules in cells, their concentration is titrated closely to the folding requirements within a specific cell type (Morimoto, 2008). Cells exploit chaperone availability as a sensing mechanism to induce stress response. At the level of transcription, reduced chaperone availability triggers the activation of heat shock transcription factor 1 (HSF1) (Morimoto, 1998). As a result, more chaperone molecules will be produced to restore the protein homeostasis. The functional connection between chaperone availability and translation elongation offers a novel mode of regulation in response to stress conditions (Figure 6). Intracellular accumulation of misfolded proteins, a common feature of a variety of stress conditions, sequesters molecular chaperones and the lack of chaperone association with the ribosome delays nascent chains from emerging. Our data suggest that the ribosome fine-tunes the elongation rate based on the chaperone availability to match protein production with the intracellular folding capacity. This level of control allows a rapid change in the complement of proteins prior to transcriptional regulation. The early elongation pausing under proteotoxic stress thus represents the very first line of protective response for cells to maintain intracellular protein homeostasis.
Figure 6. A model for co-translational stress response via early ribosome pausing.
The cytosolic chaperone molecules, such as Hsc70 (green), not only assist the co-translational folding, but also facilitate the elongation of emerging polypeptides (red). Under the condition of proteotoxic stress, the accumulation of misfolded proteins titrates out molecular chaperones. The lack of co-translational interaction of chaperone molecules leads to early elongation pausing and rapid suppression of global protein synthesis.
EXPERIMENTAL PROCEDURES
Ribosome Profiling and Data Analysis
Ribosome profiling was performed based on the reported protocol (Ingolia et al., 2009) with minor modifications. Cells were lysed in polysome lysis buffer (pH 7.4, 10 mM HEPES, 100 mM KCl, 5 mM MgCl2, 100 μg/ml cycloheximide and 2% Triton X-100) and cleared lysates were separated by sedimentation through sucrose gradients. Separated samples were fractionated and continually monitored OD254 values. Collected polysome fractions were digested with RNase I and the RPF fragments were size selected and purified by gel extraction. After the library construction, deep sequencing was performed using Illumina HiSEQ2000. The trimmed RPF reads were aligned to Ensembl human transcriptome reference by SOAP 2.0 allowing two mismatches. The meta-gene analysis was carried out by calculating the normalized mean reads (NMR) density at each codon position. The ribosome pausing index (5′PI) was defined as the ratio between the read density in the first 50 codon window and the immediate following 100 codon region. Additional details are available in Supplemental Experimental Procedures.
[35S] Pulse Assay and Ribosomal Half-Transit Time
After the treatment as indicated, cells were metabolically labeled in pulsing medium containing 10μCi [35S] mix (Perkin Elmer). An aliquot of cells was withdrawn at each time point and mixed with stop medium. Cells were lysed with polysome lysis buffer and lysates were cleared by centrifugation. For the measurement of ribosomal half-transit time, 100 μl lysates were mixed with 350 μl polysome buffer and 450 μl 0.14M sucrose in polysome buffer. 400 μl mixture was saved for measurement of total [35S] incorporation. Ribosomes were pelleted from the remaining 500 μl mixture by centrifugation at 60,000 rpm for 15 min at 4°C using a Beckman TLA-100.4 rotor. 400 μl of supernatant was taken to measure the [35S] incorporation into the completed polypeptide. Proteins were precipitated with 10% Trichloroacetic acid (Sigma). The precipitates were collected on GF/C filter membrane (Watman) and the [35S] incorporation was measured by scintillation counting (Beckman).
Immunoprecipitation
Cells were pre-treated with 100 μg/ml cycloheximide at 37°C for 3mins to stabilize ribosome-nascent chain complex and then scraped extensively in polysome lysis buffer supplemented with EDTA-free cocktail protease inhibitor (Roche). After clearance by centrifugation, the supernatant was collected and incubated with anti-Flag M2 affinity gel (Sigma) at 4°C for 1h. The beads were extensively washed for three times with polysome lysis buffer and the associated proteins were eluted by heating for 10 min in the sample buffer.
In vitro Translation
Fluc mRNA was synthesized through in vitro transcription using mMESSAGE mMACHINE T7 ULTRA Kit (Ambion). Programmed in vitro translation was performed following published protocol (Rakotondrafara and Hentze, 2011). Cell extracts were prepared from HEK293 cells with or without 1h treatment of 10 mM AZC and 20 μM MG132. Control and stressed cell lysates were adjusted to be equal based on protein concentration. In vitro translation was incubated at 37°C for 2 hour and luciferase substrate (Promega) was added to measure the Fluc activity by luminometery. For in vitro translation of FRB or FKBP assay, TNT Quick Coupled Translation/Transcription system (Promega) was used. pcDNA3 plasmid encoding FRB-GFP or FKBP-GFP was mixed with rabbit reticulocyte lysate (RRL) supplemented with [35S] L-methionine and recombinant proteins as indicated. In vitro translation was performed in the presence or absence of 1 μM Rapamycin. The products at different time points were collected and resolved on SDS-PAGE. The gel was dried and viewed by phosphor imaging screen (HE healthcare) and the band intensity was quantified using ImageQuant 5.2.
Supplementary Material
HIGHLIGHTS.
Proteotoxic stress reduces global protein synthesis by influencing elongation
Proteotoxic stress induces ribosome pausing on mRNAs in the first 50 codons
Molecular chaperones facilitate translation elongation by binding to nascent chains
Ribosomes fine-tune elongation rate in response to proteotoxic stress
Acknowledgments
We’d like to thank Qian lab members for helpful discussion and Drs. Patsy Brannon and William Brown for critical reading of the manuscript. We also thank Dr. Yewdell (NIH) for providing adenoviruses expressing Hsc70(WT) and Hsc70(K71M), and Cornell University Life Sciences Core Laboratory Center for performing deep sequencing. This work was supported by grants to S.-B.Q. from National Institutes of Health (1 DP2 OD006449-01), Ellison Medical Foundation (AG-NS-0605-09), and DOD Exploration-Hypothesis Development Award (TS10078).
Footnotes
ACCESSION NUMBERS
Sequencing data were deposited in the SRA database with the accession number SRA061778.
Supplemental information includes Extended Experimental Procedures and 7 figures and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- del Alamo M, Hogan DJ, Pechmann S, Albanese V, Brown PO, Frydman J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- Fredrick K, Ibba M. How the sequence of a gene can tune its translation. Cell. 2010;141:227–229. doi: 10.1016/j.cell.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Goldberg AL, Dice JF. Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem. 1974;43:835–869. doi: 10.1146/annurev.bi.43.070174.004155. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, De Los Rios P. The mechanism of Hsp70 chaperones: (entropic) pulling the models together. Trends Biochem Sci. 2007;32:372–380. doi: 10.1016/j.tibs.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Gray NK, Hentze MW. Regulation of protein synthesis by mRNA structure. Mol Biol Rep. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen WJ, Cowan NJ, Welch WJ. Prefoldin-nascent chain complexes in the folding of cytoskeletal proteins. J Cell Biol. 1999;145:265–277. doi: 10.1083/jcb.145.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal H, Conz C, Otto H, Wolfle T, Fitzke E, Mayer MP, Rospert S. The chaperone network connected to human ribosome-associated complex. Mol Cell Biol. 2011;31:1160–1173. doi: 10.1128/MCB.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RE, Johnson AE. Protein translocation: is Hsp70 pulling my chain? Curr Biol. 1999;9:R779–782. doi: 10.1016/S0960-9822(00)80012-3. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm JD, Schreiber SL, Crabtree GR. Dimerization as a regulatory mechanism in signal transduction. Annu Rev Immunol. 1998;16:569–592. doi: 10.1146/annurev.immunol.16.1.569. [DOI] [PubMed] [Google Scholar]
- Kolb VA, Makeyev EV, Spirin AS. Co-translational folding of an eukaryotic multidomain protein in a prokaryotic translation system. J Biol Chem. 2000;275:16597–16601. doi: 10.1074/jbc.M002030200. [DOI] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- Lavner Y, Kotlar D. Codon bias as a factor in regulating expression via translation rate in the human genome. Gene. 2005;345:127–138. doi: 10.1016/j.gene.2004.11.035. [DOI] [PubMed] [Google Scholar]
- Leu JI, Pimkina J, Pandey P, Murphy ME, George DL. HSP70 inhibition by the small-molecule 2-phenylethynesulfonamide impairs protein clearance pathways in tumor cells. Mol Cancer Res. 2011;9:936–947. doi: 10.1158/1541-7786.MCR-11-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Massey AJ, Williamson DS, Browne H, Murray JB, Dokurno P, Shaw T, Macias AT, Daniels Z, Geoffroy S, Dopson M, et al. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother Pharmacol. 2010;66:535–545. doi: 10.1007/s00280-009-1194-3. [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Tam S, Kaganovich D, Frydman J. Protein quality control: chaperones culling corrupt conformations. Nat Cell Biol. 2005;7:736–741. doi: 10.1038/ncb0805-736. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- Newmyer SL, Schmid SL. Dominant-interfering Hsc70 mutants disrupt multiple stages of the clathrin-coated vesicle cycle in vivo. J Cell Biol. 2001;152:607–620. doi: 10.1083/jcb.152.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PJ, McConkey EH. Evidence for control of protein synthesis in HeLa cells via the elongation rate. J Cell Physiol. 1980;104:269–281. doi: 10.1002/jcp.1041040302. [DOI] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian SB, Waldron L, Choudhary N, Klevit RE, Chazin WJ, Patterson C. Engineering a ubiquitin ligase reveals conformational flexibility required for ubiquitin transfer. J Biol Chem. 2009;284:26797–26802. doi: 10.1074/jbc.M109.032334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian SB, Zhang X, Sun J, Bennink JR, Yewdell JW, Patterson C. mTORC1 links protein quality and quantity control by sensing chaperone availability. J Biol Chem. 2010;285:27385–27395. doi: 10.1074/jbc.M110.120295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotondrafara AM, Hentze MW. An efficient factor-depleted mammalian in vitro translation system. Nat Protoc. 2011;6:563–571. doi: 10.1038/nprot.2011.314. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Sun J, Conn CS, Han Y, Yeung V, Qian SB. PI3K-mTORC1 attenuates stress response by inhibiting cap-independent Hsp70 translation. J Biol Chem. 2011;286:6791–6800. doi: 10.1074/jbc.M110.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter EW, Kao CM, Berenfeld L, Botstein D, Petsko GA, Gray JV. Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J Biol Chem. 2002;277:44817–44825. doi: 10.1074/jbc.M204686200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Qian SB. Chaperone-mediated hierarchical control in targeting misfolded proteins to aggresomes. Mol Biol Cell. 2011;22:3277–3288. doi: 10.1091/mbc.E11-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.