Abstract
Abnormal trophoblast lineage proliferation and differentiation in early pregnancy have been associated with the pathogenesis of placenta diseases of pregnancy. However, there is still a gap in understanding the molecular mechanisms of early placental development due to the limited primary trophoblast cultures and fidelity of immortalized trophoblast lines. Trophoblasts stem (TS) cells, an in vitro model of trophectoderm that can differentiate into syncytiotrophoblasts and extravillous trophoblasts, can be an attractive tool for early pregnancy research. TS cells are well established in mouse but not in humans due to insufficient knowledge of which trophoblast lineage-specific transcription factors are involved in human trophectoderm (TE) proliferation and differentiation. Here, we applied induced pluripotent stem cell technique to investigate the human trophoblast lineage-specific transcription factors. We established human induced trophoblast progenitor (iTP) cells by direct reprogramming the fibroblasts with a pool of mouse trophoblast lineage-specific transcription factors consisting of CDX2, EOMES, and ELF5. The human iTP cells exhibit epithelial morphology and can be maintained in vitro for more than 2 months. Gene expression profile of these cells was tightly clustered with human trophectoderms but not with human neuron progenitor cells, mesenchymal stem cells, or endoderm cells. These cells are capable of differentiating into cells with an invasive capacity, suggesting extravillous trophoblasts. They also form multi-nucleated cells which secrete human chorionic gonadotropin and estradiol, suggesting syncytiotrophoblasts. Our results provide the evidence that transcription factors CDX2 and EOMES may play critical roles in human iTP cell generation.
Keywords: CDX2, EOMES, transcription factors, human induced trophoblast progenitor cells
1. Introduction
It is well accepted that aberrant trophoblast proliferation and differentiation are two of the major causes of placenta-associated diseases, but the pathogenesis of these diseases are still largely unknown. The molecular mechanisms of human trophoblast lineage proliferation and differentiation are difficult to study due to the existing ethical (use of human embryos) and practical (use of < 6-week placenta) issues. Trophoblast stem (TS) cells, which represent trophectoderm (TE) in vivo, can be a useful tool for the study of trophoblast lineage proliferation and differentiation in vitro [1]. However, unlike mouse TS cells which are well established and extensively studied, established human TS cell line does not exist. Numerous studies have been attempted to use human embryonic stem (ES) cells or 1st trimester placenta (8–12 week) to generate human TS cells [2; 3; 4; 5; 6; 7; 8; 9; 10]. Other studies have focused on analyzing transcriptomes between human inner cell mass (ICM) and TE or differentiation of human ES cells into trophoblasts over time in order to identify the transcription factors involved in human trophoblast lineage commitment and differentiation [11; 12; 13; 14; 15; 16]. It has been shown that mouse TS cells and human TE share similar lineage transcription factors. However, applying similar culture conditions which are effective in mouse ES cells/blastocysts differentiation into TS cells does not work for human ES cells, indicating the existence of different transcription factor loops/pathways between humans and mice. Thus, there is an urgent need to identify human trophoblast lineage-specific transcription factors and generate viable human trophoblast cell lines to advance reproductive research.
Induced pluripotent stem (iPS) cell technique is the direct reprogramming of fibroblasts into various cell types via transduction with different groups of lineage-specific transcription factors [17]. iPS technique shows promise in clinical applications; for example, dopaminergic neurons, cardiac cells, and hematopoietic cells have been successfully generated directly from fibroblasts using this technique [18; 19; 20]. iPS technique has also been proven to be a useful tool to investigate the biofunction of transcription factors; over-expression of POU5F1 in mouse TS cells can lead to generation of mouse ES cells, suggesting POU5F1 as a critical transcription factor in ES cells [21]. A similar study identified three transcription factors as a group of critical loop for induction of human cardiomycytes [19]. Therefore, it is rational to use this strategy to examine the transcription factors required for establishing human trophoblast cells directly from the fibroblasts. In this study, we transduced the well-documented mouse trophoblast lineage-specific transcription factors: caudal-type homeobox transcription factor 2 (CDX2) [22; 23; 24], eomesodermin (EOMES) [25; 26], and E74-like factor 5 (ELF5) [27; 28; 29], which are known to maintain mouse TS cells pluripotency and lineage specificity as a loop. CDX2, EOMES, and ELF5 are also found expressed in human 1st trimester placental trophoblast [2; 10]. Additionally, we included two oncogenes: MYC avian myelocytomatosis viral oncogene homolog c (C-MYC) and Kruppel-like factor 4 (KLF4), which are essential for cell proliferation and transformation [17]. The iTP cells generated in this study represent a useful tool for the study human trophoblast lineage-specific transcription factor biofunction.
2. Materials and methods
2.1. Generation of human induced trophoblast progenitor (iTP) cells
Human lenti-virus constructs for CDX2, ELF5, C-MYC, KLF4 (Open Biosystems) and EOMES (home-made) were used to generate iTP cells from human fetal fibroblasts (IMR90, ATCC). 1.25×105 fibroblasts were transducted for 24 hours in a mixture of 5 viral genes with fibroblast medium: DMEM supplemented with 10% fetal bovine serum(FBS), 1% L-glutamine, 1% non-essential amino acids (NEAA), 1% penicillin, and 1% streptomyocin. After 96 hours, cells were passaged onto in-activated CF-1 mouse embryonic fibroblast (Millipore) in fibroblast medium which was replaced with human iTP medium (mouse TS medium [1] modified as follows): RPMI1640 supplemented with 20% FBS, 1% L-Glutamine, 1% sodium pyruvate, 0.5% penicillin, 0.5% streptomyocin, 0.1mM beta mercaptoethanol (2-ME), 5ng/ml Activin A, 2ng/ml Transforming Growth Factor beta-1 (TGFb-1), 1μg/ml heparin, and 25ng/ml Fibroblast Growth Factor-4 (FGF-4). iTP colonies were manually picked approximately 25–30 days after viral transduction and mechanically passaged every 5 to 7 days. Two iTP colonies which passaged over 2 months were chosen for further analysis.
2.2. Differentiation of iTP cells
iTP cells were cultured in human iTP medium (-FGF4, -Activin A, -TGFβ1 and -heparin) in the presence of 1.0 μM cAMP (Sigma) for 7 days. The media was refreshed every other day and cells were analyzed on day 7.
2.3. ELISA assay
Supernatant of day-7 differentiated iTP cell culture was collected to detect hCG and estradiol using ELISA assays (R&D), following manufacturer's instructions. The spectral readouts were performed at 490nm–560nm and each sample was analyzed in duplicates.
2.4. Invasion assay
2×104 iTP cells were seeded above transwell chambers with or without Matrigel-coating (BD Biosciences.). Fibroblasts (IMR-90) were used as a control. The medium used in upper well was: RPMI1640 with 0.1% BSA, 1% sodium pyruvate, 1% L-Glutamine, and 0.1mM 2-ME while in bottom well: RPMI1640 with 20% FBS, 1% sodium pyruvate, 1% L-Glutamine, and 0.1mM 2-ME. 20–22 hours later, cells were stained with 0.09% Crystal violet solution. Non-invading cells on the upside of the transwell were carefully removed by cotton swab. Cells on the lower surface of the membrane were then counting the number of invaded cells (5 fields) and photographed (Nikon software).
2.5. qRT-PCR (quantitative real-time PCR)
Total RNA was isolated with PicoPure RNA isolation kit (Molecular Devices) and first strand cDNA synthesis was performed with Superscript II (invitrogen). qRT-PCR was performed using SYBR green PCR master Mix (Applied Biosystems). Gene names and the primers were summarized in table S1.
2.6. Immuno-histochemistry
Cells were fixed 10 minutes in 4.0% paraformaldehyde, permeablized with 0.3% Triton X-100 in PBS for 15mins, and then blocked in 0.1% triton X-100 with 5% BSA for 30 mins. Following primary antibody incubation for 2 hrs at room temperature (or 4°C overnight), cells were then incubated with secondary antibodies (Alexa Flours, invitrogen) for 45 mins and nuclei counterstained with Hochest33342 (Sigma). Imaging and analysis were performed using a Nikon 2000 inverted microscopy and Nuance imaging system (CRI). Primary antibodies used in this study were CK7 (1:100, Dako) and HLA-G (1:50, Abcam).
2.7. Gene expression microarray and analysis
To investigate the dynamic transcriptome changes during the transition from fibroblast to iTP cells, we compared total RNA from iTP cells with control fibroblasts using the human genome 4×44K gene expression chip (Agilent Technologies, CA) according to manufacturer's instructions and primary data were collected from an Agilent Scanner. Probe intensities were extracted using Agilent Feature Extraction Software (v9.5). The signals of each array were normalized by median centering of ratios with fibroblast as a reference. Approximately 25,700 probes could be detected and were included in the analysis. All the statistical calculations were performed on logarithmic values of signals to the base 2. Statistical significance of differentially-expressed genes between iTP1 or iTP2 versus fibroblast was evaluated by two-tailed one-sample t-tests. Global expression profiles of fibroblast and iTP cell lines were examined using hierarchical clustering analysis by Cluster 3.0 (http://www.falw.vu/~huik/cluster.htm).
To compare the transcriptomes of iTP (based on Agilent expression chip) with previously published TE database studies based on the Affimetrix Human Genome U133 Plus 2.0 (GSM706172, GSM706171, GSM706170, GSM706169, GSM706168), neuron progenitor cells (NPC) (GSM335938, GSM335941), endoderm cells Endo (GSM1032057) and mesenchymal stem cells (MSC)(GSM878100, GSM878101), we included the IMR90 cells in both Agilent (our data) and Affimetrix (GSM713542, GSM713543) expression microarrays in the comparison as background. We then used GeneSpring software (Agilent Technologies) to pick up the “present” gene expression in all checked cell type in both Agilent and Affimetrix arrays. Affimetrix raw data (.CEL file) and Agilent raw data (.TXT) was loaded into software, background subtracted, and the signals were normalized by MAS5 or percentile shift method, respectively. After transferring probe-based data into gene-based data by “none” in Affimetrix and “quantile” method in Agilent data, we did the cross-section of “present” gene in each cells as the “common” expressed gene list (n=3711). Then, log fold change of “common gene” in each cell type vs. IMR90 in their own array type were used to demonstrate the similarity among cell types by hierarchical clustering analysis using Cluster 3.0 software.
2.8. Statistical analysis
We used Student's t-test to compare relative expression of genes in iTP1, iTP2 and IMR90 (donor fibroblasts). P < 0.05 is taken as statistically significant.
3. Results
3.1. Characterization of human iTP cell lines
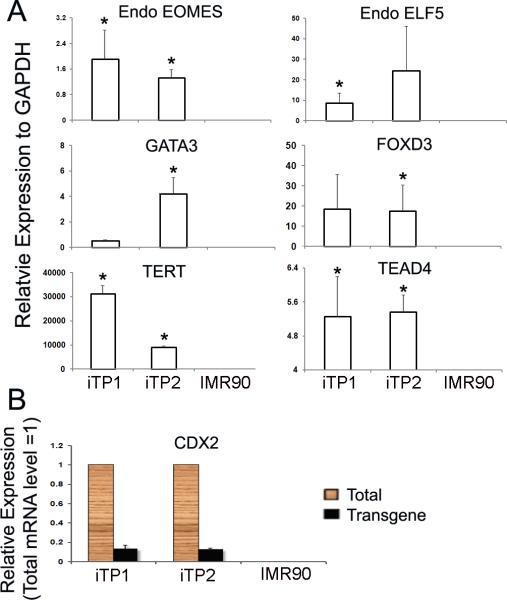
Human iTP cell lines were successfully established from human fetal fibroblasts (IMR90) by lenti-viral transduction with a pool of transcription factors (CDX2, EOMES, ELF5, KLF4, and c-MYC) [Fig.1A and B]. Four iTP cell lines showed similar growth and morphological characteristics and two (iTP1 and iTP2) were further characterized in detail. 25–30 days after initial transduction, the donor fibroblasts were reprogrammed into iTP cells which formed distinct colonies composed of small round cells with increased nuclear/cytoplasm ratio. The iTP cell lines maintained normal karotyping throughout the lenti-viral transduction and differentiation process [Fig.S1]. Quantitative real time PCR showed highly-expressed endogenous CDX2, EOMES, and ELF5, with expression of transducted viral transcripts CDX2 slightly detected as well [Fig.2A and B]. Compared with donor fibroblasts, iTP cell lines also showed significantly high expression of several mouse TS cell genes such as GATA binding protein 3 (GATA3) and tea domain family member 4 (TEAD4), as well as pluripotent stem cell genes such as forkhead box D3 (FOXD3) and telomerase reverse transcriptase (TERT) [Fig.2A]. The cells were also positively stained for CK7, a known trophoblast marker [30] [Fig.1C].
Fig1.
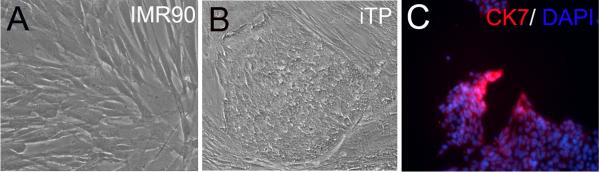
Induced trophoblast progenitor (iTP) cells established from human fibroblasts. (A) Human fetal fibroblasts (IMR90). (B) iTP1 cell line generated from IMR90 after transduction with 5 viral genes (CDX2, EOMES, ELF5, C-MYC and KLF4). (C) Expression of CK7 in iTP cells. The original picture is 200×.
Fig2.
iTP cells express stemness and trophoblast lineage transcripts. (A) Endogenous EOMES (P < 0.05) and ELF5 were highly-expressed in the iTP cells. iTP cells expressed pluripotent stem cell-associated transcripts: FOXD3 and TERT (P < 0.05) and mouse TS cell-associated transcripts: TEAD4 (P < 0.05) and GATA3. The expression level relative to fibroblast = 1. (B) CDX2 was highly-expressed while viral CDX2 is slightly detectable in iTP cells.
3.2. Epigenetic signature of iTP cells
Since ELF5 is unmethylated in mouse TS cells, we sought to investigate the epigenetic regulation of ELF5 in iTP cells. Our results show that ELF5 has similar hypermethylation patterns in donor fibroblast IMR90 and iTP cells [Fig.S2].
3.3. Transcriptome changes from transition of fibroblasts to iTP cells
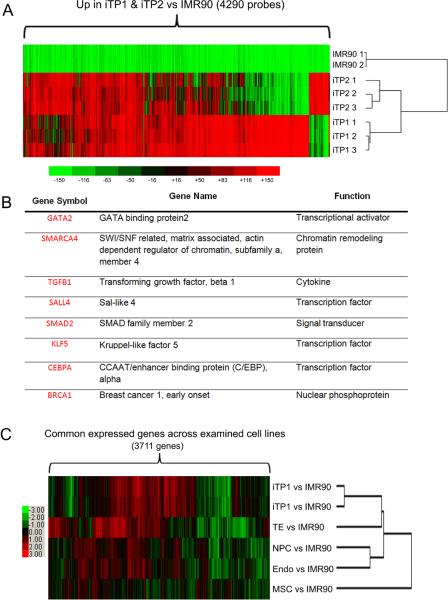
We performed global gene expression analysis by microarray and identified significant differentially-expressed genes between fibroblasts and iTP cells. Cluster analysis revealed that our two iTP cell lines formed the same cluster with each other while were a sub-cluster from donor fibroblasts [Fig.3A]. Selected highly-expressed trophoblast lineage genes are listed [Fig.3B]. Our transcriptome analysis shows that human iTP cells is more tightly clustered with human TE than NPC, MSC, or Endo cells [Fig.3C], indicating iTP cells are in the trophoblast lineage.
Fig3.
Transcriptome profiling of iTP cells. (A) Heat map of upregulated 4290 probes with 2-fold change or more (P < 0.05) in replicates of iTP1 or iTP2 vs. fibroblasts (IMR90). iTP1 cells and iTP2 cells are tightly clustered when compared with fibroblasts. (B) Eight TS cell-related genes are highly-expressed in iTP cells. (C) Heat map of 3721 transcripts which commonly exist across all examined cells showed that iTP1 cells and iTP2 cells clustered more tightly with human trophectoderms (TE) as compared with human neuron progenitor cells (NPC), mesenchymal stem cells (MSC) or endoderm cells (Endo).
3.4. Trophablast cells differentiation from iTP cells
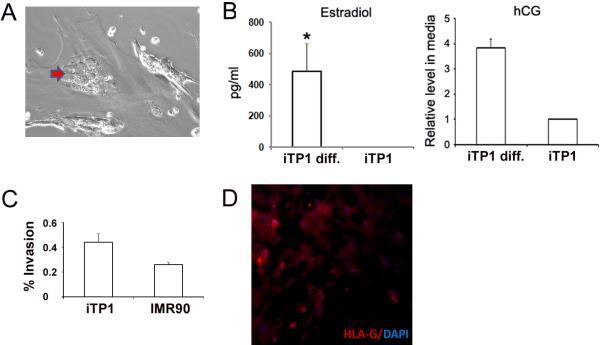
To determine whether iTP cells are functionally similar to human trophoblast progenitor cells, we evaluated the ability of iTP1 cells to differentiate into trophoblast cell subtypes: invasive trophoblasts and syncytiotrophoblasts. After 7 days in differentiation culture, multi-nucleated cells were formed [Fig.4A] and the secretion of estradiol and hCG, two placenta-specific hormones, could be detected in the supernatant [Fig.4B], suggesting formation of syncytiotrophoblasts. iTP1 cells were also grown on Matrigel-coated transwell (BD) to assess their invasive capacity [31]. We found a higher percentage of invasion in iTP1 cells vs. control fibroblasts (44.0% vs. 26.1% at 20 hr, n=3) [Fig.4C]. with invading cells staining positively for HLA-G (invasive trophoblast marker) [Fig.4D], suggesting the formation of extravillous trophoblasts.
Fig4.
iTP cells can differentiate into trophoblast subtypes. (A) Formation of multi-nucleated trophoblast cells (red arrow). (B) Estradiol and hCG levels detected from day-7 culture medium. (n=3). (C) iTP cells showed higher % of invasion compared with donor fibroblasts (n=3). (D) Invasive cells were stained with HLA-G (Red) with Hochest 33342 fluorescence as nuclear counterstaining (blue). The original picture is 200×.
4. Discussion
In this study, we generated human iTP cells to elucidate the trophoblast lineage-specific transcription factors required for human trophoblast lineage proliferation and differentiation. Here, we used 3 transcription factors (CDX2, EOMES, and ELF5) which were previously shown to form a transcription loop in mouse trophoblast lineage [32; 33]. CDX2 is critical for generating mouse TS cells; mouse ES cells could be transformed into TS cells solely by over-expressing CDX2 [23; 34] whereas CDX2 null blastocysts failed to implant due to aberrant trophoblast function [24]. Further, it has been shown that CDX2 null embryos were arrested during the early blastocyst stage, indicating that CDX2 also plays a critical role in TE formation [22]. Similarly, EOMES was also previously shown to be required for mouse TS cell line maintenance [25] and EOMES null blastocysts displayed defective trophoblast differentiation [26]. ELF5 is hypomethylated in both mouse ICM and TE cells [35] as well as TS cells, and is highly-expressed in mouse TS cells, suggesting ELF as a trophoblast lineage gate keeper [27]. In addition, previous studies have shown that mouse ES cells, in the presence of bFGF growth factor, could be transformed into epiblast stem cells, which is equivalent to human ES cells in terms of morphology, transcriptome, epigenetic, and pluripotent biofunction [36; 37]. Therefore, we applied this same strategy to culture human iTP cell line in mouse TS medium. To our knowledge, this is the first time that CDX2 and EOMES have been shown to play important biofunctional roles in human trophoblast lineage.
This study shows that our iTP cells express high levels of endogenous CDX2 and EOMES, which is consistent with data obtained from mouse TS cells. Since the role of ELF5 in human placental trophoblast is unclear [2; 10], we examined ELF5 DNA methylation in our iTP cells and found similar hypermethylation pattern with donor fibroblasts, with endogenous ELF5 expressed. Our data and others suggest that, although ELF5 plays an important role in trophoblast lineage specificity in mouse, its role in human trophoblast lineage requires further investigation.
Besides the three trophoblast lineage-specific transcription factors mentioned above, TEAD4 and GATA3 were also highly expressed in our iTP cells. TEAD4 is the earliest known trophoblast lineage transcription factor whose expression coincides with TE and inner cell mass (ICM) separation. TEAD4 null embryos cannot form blastocoel cavities but their ICM is able to establish ES cells [38; 39]. It has also been shown that GATA3 is expressed in TE at mouse blastocyst stage and its expression pattern is parallel with that of CDX2 [40; 41]. Our study also shows that these iTP cells are tightly clustered with human TE. Taken together, these data indicate that iTP cells are of trophoblast lineage. Furthermore, FOXD3 and TERT are highly-expressed in our iTP cells. Reports have confirmed that FOXD3 is required for trophoblast progenitor cells self-renewal and multi-differentiation [42], whereas TERT is required for trophoblast progenitor cell self-renewal [43]. These data further suggest that our iTP cells have retained the progenitor status.
To investigate the biofunctions of our iTP cells, we performed differentiation studies and found multi-nucleated cells and hormone secretion, indicating syncytiotrophoblast formation. Further studies revealed the more invasive potential of iTP cells when compared with donor fibroblasts. Although these iTP cells have slightly viral transcript CDX2 expression, the differentiation capacity was not affected. Taken together, our data are consistent with previous findings on human trophoblast progenitor cells [3; 10; 44].
We conclude that CDX2 and EOMES play critical biofunctional roles in human iTP cells, whereas the role of ELF5 requires further investigation. This study can be further applied to generate reporter cell lines (i.e. CDX2 reporter cell line) for efficient screening of human trophoblast lineage-specific transcription factor loops/pathways as well as therapeutic application to study the pathogenesis of placenta-associated diseases. In addition, we exhibit a proof-of-concept that human iTP cells can be directly trans-differentiated from fibroblasts; this approach provides a new alternative to establish human trophoblast progenitor in vitro, instead of using human embryonic stem cells and 1st trimester placenta.
Supplementary Material
*Highlights
-
➢
CDX2 and EOMES play critical roles in human induced trophoblast progenitors(iTP).
-
➢
iTP cells directly transformed from fibroblasts.
-
➢
Differentiation of iTP cells into extravillous trophoblasts and syncytiotrophoblasts.
Acknowledgements
The authors would like to thank Dr. Trixie Smith and Susan Ferguson for critically reading the manuscript and providing constructive criticism.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–5. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- [2].Hemberger M, Udayashankar R, Tesar P, Moore H, Burton GJ. ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum Mol Genet. 19:2456–67. doi: 10.1093/hmg/ddq128. [DOI] [PubMed] [Google Scholar]
- [3].Harun R, Ruban L, Matin M, Draper J, Jenkins NM, Liew GC, Andrews PW, Li TC, Laird SM, Moore HD. Cytotrophoblast stem cell lines derived from human embryonic stem cells and their capacity to mimic invasive implantation events. Hum Reprod. 2006;21:1349–58. doi: 10.1093/humrep/del017. [DOI] [PubMed] [Google Scholar]
- [4].Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–4. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- [5].Matin MM, Walsh JR, Gokhale PJ, Draper JS, Bahrami AR, Morton I, Moore HD, Andrews PW. Specific knockdown of Oct4 and beta2-microglobulin expression by RNA interference in human embryonic stem cells and embryonic carcinoma cells. Stem Cells. 2004;22:659–68. doi: 10.1634/stemcells.22-5-659. [DOI] [PubMed] [Google Scholar]
- [6].Gerami-Naini B, Dovzhenko OV, Durning M, Wegner FH, Thomson JA, Golos TG. Trophoblast differentiation in embryoid bodies derived from human embryonic stem cells. Endocrinology. 2004;145:1517–24. doi: 10.1210/en.2003-1241. [DOI] [PubMed] [Google Scholar]
- [7].Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci U S A. 2005;102:4783–8. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ezashi T, Telugu BP, Roberts RM. Model systems for studying trophoblast differentiation from human pluripotent stem cells. Cell Tissue Res. 349:809–24. doi: 10.1007/s00441-012-1371-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Das P, Ezashi T, Schulz LC, Westfall SD, Livingston KA, Roberts RM. Effects of fgf2 and oxygen in the bmp4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Res. 2007;1:61–74. doi: 10.1016/j.scr.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Genbacev O, Donne M, Kapidzic M, Gormley M, Lamb J, Gilmore J, Larocque N, Goldfien G, Zdravkovic T, McMaster MT, Fisher SJ. Establishment of human trophoblast progenitor cell lines from the chorion. Stem Cells. 29:1427–36. doi: 10.1002/stem.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bai Q, Assou S, Haouzi D, Ramirez JM, Monzo C, Becker F, Gerbal-Chaloin S, Hamamah S, De Vos J. Dissecting the first transcriptional divergence during human embryonic development. Stem Cell Rev. 8:150–62. doi: 10.1007/s12015-011-9301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chickarmane V, Peterson C. A computational model for understanding stem cell, trophectoderm and endoderm lineage determination. PLoS One. 2008;3:e3478. doi: 10.1371/journal.pone.0003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Marchand M, Horcajadas JA, Esteban FJ, McElroy SL, Fisher SJ, Giudice LC. Transcriptomic signature of trophoblast differentiation in a human embryonic stem cell model. Biol Reprod. 84:1258–71. doi: 10.1095/biolreprod.110.086413. [DOI] [PubMed] [Google Scholar]
- [14].Adjaye J, Huntriss J, Herwig R, BenKahla A, Brink TC, Wierling C, Hultschig C, Groth D, Yaspo ML, Picton HM, Gosden RG, Lehrach H. Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells. 2005;23:1514–25. doi: 10.1634/stemcells.2005-0113. [DOI] [PubMed] [Google Scholar]
- [15].Assou S, Boumela I, Haouzi D, Monzo C, Dechaud H, Kadoch IJ, Hamamah S. Transcriptome analysis during human trophectoderm specification suggests new roles of metabolic and epigenetic genes. PLoS One. 7:e39306. doi: 10.1371/journal.pone.0039306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Haouzi D, Dechaud H, Assou S, Monzo C, de Vos J, Hamamah S. Transcriptome analysis reveals dialogues between human trophectoderm and endometrial cells during the implantation period. Hum Reprod. 26:1440–9. doi: 10.1093/humrep/der075. [DOI] [PubMed] [Google Scholar]
- [17].Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- [18].Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 108:10343–8. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 142:375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 468:521–6. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- [21].Wu T, Wang H, He J, Kang L, Jiang Y, Liu J, Zhang Y, Kou Z, Liu L, Zhang X, Gao S. Reprogramming of trophoblast stem cells into pluripotent stem cells by Oct4. Stem Cells. 29:755–63. doi: 10.1002/stem.617. [DOI] [PubMed] [Google Scholar]
- [22].Bruce AW. What is the role of maternally provided Cdx2 mRNA in early mouse embryogenesis? Reprod Biomed Online. 22:512–5. doi: 10.1016/j.rbmo.2011.02.003. [DOI] [PubMed] [Google Scholar]
- [23].Tolkunova E, Cavaleri F, Eckardt S, Reinbold R, Christenson LK, Scholer HR, Tomilin A. The caudal-related protein cdx2 promotes trophoblast differentiation of mouse embryonic stem cells. Stem Cells. 2006;24:139–44. doi: 10.1634/stemcells.2005-0240. [DOI] [PubMed] [Google Scholar]
- [24].Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- [25].Kidder BL, Palmer S. Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res. 20:458–72. doi: 10.1101/gr.101469.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, Wilson V, Evans MJ. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–9. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- [27].Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10:1280–90. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Donnison M, Beaton A, Davey HW, Broadhurst R, L'Huillier P, Pfeffer PL. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development. 2005;132:2299–308. doi: 10.1242/dev.01819. [DOI] [PubMed] [Google Scholar]
- [29].Pearton DJ, Broadhurst R, Donnison M, Pfeffer PL. Elf5 regulation in the trophectoderm. Dev Biol. 360:343–50. doi: 10.1016/j.ydbio.2011.10.007. [DOI] [PubMed] [Google Scholar]
- [30].Maldonado-Estrada J, Menu E, Roques P, Barre-Sinoussi F, Chaouat G. Evaluation of Cytokeratin 7 as an accurate intracellular marker with which to assess the purity of human placental villous trophoblast cells by flow cytometry. J Immunol Methods. 2004;286:21–34. doi: 10.1016/j.jim.2003.03.001. [DOI] [PubMed] [Google Scholar]
- [31].Leach RE, Kilburn B, Wang J, Liu Z, Romero R, Armant DR. Heparin-binding EGF-like growth factor regulates human extravillous cytotrophoblast development during conversion to the invasive phenotype. Dev Biol. 2004;266:223–37. doi: 10.1016/j.ydbio.2003.09.026. [DOI] [PubMed] [Google Scholar]
- [32].Maltepe E, Bakardjiev AI, Fisher SJ. The placenta: transcriptional, epigenetic, and physiological integration during development. J Clin Invest. 120:1016–25. doi: 10.1172/JCI41211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Roper S, Hemberger M. Defining pathways that enforce cell lineage specification in early development and stem cells. Cell Cycle. 2009;8:1515–25. doi: 10.4161/cc.8.10.8381. [DOI] [PubMed] [Google Scholar]
- [34].Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–29. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- [35].Nakanishi MO, Hayakawa K, Nakabayashi K, Hata K, Shiota K, Tanaka S. Trophoblast-specific DNA methylation occurs after the segregation of the trophectoderm and inner cell mass in the mouse periimplantation embryo. Epigenetics. 7:173–82. doi: 10.4161/epi.7.2.18962. [DOI] [PubMed] [Google Scholar]
- [36].Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–5. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- [37].Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–9. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- [38].Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–36. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- [39].Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, Makita R, Kurihara H, Morin-Kensicki EM, Nojima H, Rossant J, Nakao K, Niwa H, Sasaki H. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- [40].Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem. 2009;284:28729–37. doi: 10.1074/jbc.M109.016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- [42].Tompers DM, Foreman RK, Wang Q, Kumanova M, Labosky PA. Foxd3 is required in the trophoblast progenitor cell lineage of the mouse embryo. Dev Biol. 2005;285:126–37. doi: 10.1016/j.ydbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- [43].Bayne S, Liu JP. Hormones and growth factors regulate telomerase activity in ageing and cancer. Mol Cell Endocrinol. 2005;240:11–22. doi: 10.1016/j.mce.2005.05.009. [DOI] [PubMed] [Google Scholar]
- [44].Udayashankar R, Baker D, Tuckerman E, Laird S, Li TC, Moore HD. Characterization of invasive trophoblasts generated from human embryonic stem cells. Hum Reprod. 26:398–406. doi: 10.1093/humrep/deq350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.