Abstract
Rationale: Asthma is a heterogeneous lung disorder characterized by airway inflammation and airway dysfunction, manifesting as hyperresponsiveness and obstruction. Glutathione S-transferase M1 (GSTM1) is a multifunctional phase II enzyme and regulator of stress-activated cellular signaling relevant to asthma pathobiology. A common homozygous deletion polymorphism of the GSTM1 gene eliminates enzyme activity.
Objectives: To determine the effect of GSTM1 on airway inflammation and reactivity in adults with established atopic asthma in vivo.
Methods: Nineteen GSTM1 wild-type and eighteen GSTM1-null individuals with mild atopic asthma underwent methacholine and inhaled allergen challenges, and endobronchial allergen provocations through a bronchoscope.
Measurements and Main Results: The influx of inflammatory cells, panels of cytokines and chemokines linked to asthmatic inflammation, F2-isoprostanes (markers of oxidative stress), and IgE were measured in bronchoalveolar lavage fluid at baseline and 24 hours after allergen instillation. Individuals with asthma with the GSTM1 wild-type genotype had greater baseline and allergen-provoked airway neutrophilia and concentrations of myeloperoxidase than GSTM1-null patients. In contrast, the eosinophilic inflammation was unaffected by GSTM1. The allergen-stimulated generation of acute-stress and proneutrophilic mediators, tumor necrosis factor-α, CXCL-8, IL-1β, and IL-6, was also greater in the GSTM1 wild-type patients. Moreover, post-allergen airway concentrations of IgE and neutrophil-generated mediators, matrix metalloproteinase-9, B-cell activating factor, transforming growth factor-β1, and elastase were higher in GSTM1 wild-type individuals with asthma. Total airway IgE correlated with B-cell activating factor concentrations. In contrast, levels of F2-isoprostane were comparable in both groups. Finally, GSTM1 wild-type individuals with asthma required lower threshold concentrations of allergen to produce bronchoconstriction.
Conclusions: The functional GSTM1 genotype promotes neutrophilic airway inflammation in humans with atopic asthma in vivo.
Keywords: atopic asthma, GSTM1 polymorphism, inflammatory asthma phenotypes, neutrophilic airway inflammation
At a Glance Commentary
Scientific Knowledge on the Subject
The genetic regulation of distinct clinical and inflammatory asthma phenotypes is unclear. Glutathione S-transferase M1 (GSTM1) is a multifunctional enzyme expressed in airway cells that has been linked to asthma.
What This Study Adds to the Field
The wild-type GSTM1 genotype augments allergen-induced neutrophilic airway inflammation in humans with atopic asthma in vivo.
Asthma is a heterogeneous airway disorder characterized by inflammation, hyperresponsiveness, and obstruction. There is considerable interest in classifying asthma phenotypes. It has been suggested that the type of airway inflammation might define asthma phenotypes. One proposed classification uses eosinophilic, neutrophilic, and paucigranulocytic cellular inflammation to define three subtypes of asthma (1). The neutrophilic inflammation has recently drawn more attention because of its association with severe asthma and poor response to corticosteroids (2). The relationship between various asthmatic phenotypes and genotypes in humans is poorly understood.
Glutathione S-transferase M1 (GSTM1) is a phase II cytoprotective protein from the family of cytosolic GSTs (3). The GSTM1 gene is located on chromosome 1p13.3 and has two alleles: a wild-type allele, GSTM1*1, and a nonfunctional variant null allele, GSTM1*0 (3). Subjects homozygous for the GSTM1*0 express no GSTM1 protein (4). The frequency of the GSTM1 null genotype is ethnic-dependent and varies from 18 to 66% (5, 6). Approximately half of individuals of Northern European ancestry are homozygous for the GSTM1 null genotype (6). In normal lungs, GSTM1 is predominantly expressed in airway epithelial and smooth muscle cells (7). Functions of GSTM1 include xenobiotic detoxification (3) and regulation of critical stress-activated cellular signaling (8–10). GSTM1 may exert proinflammatory and cytotoxic effects due to increased catabolism of prostaglandins J2 and A2 (11), and enhanced formation of lipid peroxidation-protein adducts (12).
Most, although not all, epidemiological studies have suggested a decreased risk of asthma among individuals expressing GSTM1 (13–16). Indeed, GSTM1 mitigates airway inflammation due to prooxidant and proasthmatic environmental stressors such as ozone (17), tobacco smoke (5), and smog pollutants (18). However, to our knowledge, the effect of GSTM1 on allergen-provoked airway inflammation in humans with established atopic asthma has not been studied.
We hypothesized an inhibitory role of GSTM1 on allergen-induced oxidant stress and airway inflammation in humans with atopic asthma. The objective of our study was to determine the effect of the wild-type and null GSTM1 genotypes (GSTM1+ and GSTM1−, respectively) on allergen-induced oxidant stress, airway inflammation, and reactivity in adults with atopic asthma in vivo. Contrary to our hypothesis, GSTM1 did not affect the allergen-induced F2-isoprostane levels but augmented neutrophilic airway inflammation and altered the biochemical milieu in epithelial lining fluid that was linked to enhanced airway reactivity to specific allergens in patients with established atopic asthma in vivo.
Methods
Subjects
Data was derived from volunteers with atopic asthma who participated in a study exploring the role of natural-source d-α-tocopheryl in atopic asthma (19). Thirty-seven nonsmoking volunteers with a physician diagnosis of mild atopic asthma according to National Asthma Education and Prevention Program guidelines (20) were recruited from the Nashville area. Patients had positive skin tests with aeroallergens, methacholine challenge, and inhaled allergen provocation. Asthma medications were not allowed for at least 48 hours prior to procedures. All women had negative urine human chorionic gonadotropin tests. The study was approved by the Vanderbilt University Committee for the Protection of Human Subjects, and the use of instilled allergens was approved by the U.S. Food and Drug Administration.
Protocol
Patients underwent fiberoptic bronchoscopy with baseline (before allergen challenge) bronchoalveolar lavage (BAL) in the lingua of the left upper lobe followed by segmental allergen challenge in a subsegment of the right middle lobe. The allergen-challenged subsegment was lavaged 24 hours later. Methacholine challenge followed by an inhaled allergen provocation the next day was performed at least 3 weeks after the bronchoscopy studies.
Experimental Procedures
The skin prick test was performed with diluent and histamine controls and a set of standardized allergenic extracts (Dermatophagoides pteronyssinus, Dermatophagoides farinae, cat hair, Bermuda grass, Kentucky bluegrass, fescue meadow grass, orchard grass, redtop grass, ryegrass, sweet vernal grass, timothy grass, and short ragweed) (Greer Laboratories, Lenoir, NC) (21). The methacholine (Methapharm Inc., Brantford, Ontario, Canada) challenge was completed using a Salter dosimeter (Salter Labs, Arvin, CA) and Flow Screen spirometer (VIASYS Healthcare GmbH, Hoechberg, Germany) (22). Allergen inhalation challenge was done according to Cockcroft and colleagues (23). The maximal fall in FEV1 between 0 and 3 hours after allergen inhalation was defined as the early airway response, and the greatest FEV1 fall at 3 to 8 hours was the late airway response. Allergen aerosols were generated using a Wright nebulizer connected to a two-way Hans Rudolph valve mouthpiece (Roxon, Montreal, Quebec, Canada) attached to a wall oxygen source at 50 psi (23). Bronchoscopy studies were done in subjects who fasted overnight. Midazolam 1 to 2 mg and fentanyl 50 to 100 μg were administered intravenously. Airway anesthesia was achieved with topical lidocaine (≤7 mg/kg). BAL was performed using three 50-ml aliquots of sterile normal saline. Segmental allergen challenge was done with two incremental doses of the allergen to which the volunteers had a positive skin test, each dose in a 5-ml aliquot of normal saline. The first dose of the allergen was 10-fold higher than the concentration that caused a 5- to 8-mm wheal/20- to 30-mm erythema on intradermal test followed by the second dose of the same allergen at a concentration 100-fold higher than the 2+ skin reaction, and the bronchoscope was then removed (19).
Laboratory Methods
DNA was extracted from blood using the Wizard Genomic DNA Extraction Kit (Promega, Madison, WI). GSTM1 genotyping was performed by polymerase chain reaction using 5′-CTGCCCTACTTGATTGATGGS-3′ and 5′-CTGGATTGTAGCAGATCATGC-3′ primers to GSTM1 and 5′-CTGGATTGTAGCAGATCATGC-3′ and 5′-CAACTTCATCCACGTTCACC-3′ primers to the β-globin gene as loading controls, as described by Davies and colleagues (24). PCR amplification was performed in a thermal cycler (Techne TC-412; Techne, Burlington, NJ). After amplification, the PCR products were resolved by electrophoresis in a 2% agarose gel (Bio-Rad, Hercules, CA). The observed amplicon lengths were 273 bp for positive GSTM1 and no band for a null GSTM1.
Cytospin slides of BAL (CytoSpin 4, Thermo Scientific, Asheville, NC) were stained with Hema 3 (Fisher, Pittsburgh, PA). IL-1β, IL-4, IL-5, IL-8 (CXCL8), IL-12p70, IL-13, IFN-γ, and tumor necrosis factor-α (TNF-α) were measured by human Plex Multi-Spot assay (Meso Scale Discovery, Gaithersburg, MD). IL-6 and IgE were analyzed by Cytometric Bead Array (BD Biosciences, San Jose, CA). Commercial ELISA kits were employed to measure levels of myeloperoxidase, IL-17, IL-33, matrix metalloproteinase-9 (MMP-9), B-cell activating factor (BAFF), and CXCL1 (R&D Systems, Minneapolis, MN); transforming growth factor-β1 (TGF-β1) (Enzo Life Sciences, Farmingdale, NY); and elastase (eBioscience, San Diego, CA). F2-isoprostanes were quantified by stable isotope dilution method in conjunction with gas chromatography/negative ion chemical ionization/mass spectrometry (25).
Statistical Analysis
A two-tailed Mann Whitney test was used for comparison of GSTM1+ versus GSTM1− individuals with asthma, Wilcoxon signed-rank test for comparisons within the groups, and Spearman’s rank correlation coefficient for correlation analyses. Subgroup analyses were further conducted in subjects with dust mite, cat, and grass allergen extracts. Because only three subjects were challenged with ragweed, we excluded these three subjects from the subgroup analyses. Multivariable linear regression with subjects’ baseline oxidative stress or inflammatory levels as covariate was performed. The model was further adjusted for allergen type, gender, and race. Normality of residues of linear models was diagnosed, and transformation of the dependent variables was done to correct normal residuals if needed. Data was expressed as median (interquartile range) or mean ± standard deviation. Significance was accepted when P < 0.05. R-software version 2.11.1 was used for data analyses.
Results
Table 1 displays the demographic and clinical characteristics of the volunteers and types of aeroallergens used for provocations. All individuals with asthma had normal baseline spirometry, and the vast majority of them did not use daily asthma medications. Temporary discontinuation of asthma drugs before challenges caused no symptoms’ exacerbation or spirometric changes. Patients continued the same asthma regimen throughout the study.
TABLE 1.
CHARACTERISTICS OF VOLUNTEERS AND ALLERGENS USED FOR ASTHMA PROVOCATIONS (EACH SUBJECT UNDERWENT INHALED AND ENDOBRONCHIAL CHALLENGE USING THE SAME ALLERGEN)
| GSTM1+ | GSTM1− | |
| Male/female | 5/14 | 7/11 |
| Age | 29.8 ± 7.5 (20–43) | 28 ± 6.1 (20–42) |
| Race | ||
| Northern European ancestry | 15 | 18 |
| African American ancestry | 4 | 0 |
| Family history of atopy | 19 | 18 |
| Baseline spirometry | ||
| FVC, L | 4.1 ± 0.4 (3.38–4.7) | 4.9 ± 1.0 (3.5–6.5) |
| FVC, % predicted | 98.7 ± 9.9 (81–120) | 111.9 ± 17.9 (90–121) |
| FEV1, L | 3.15 ± 0.52 (2.6–3.9) | 3.65 ± 0.64 (2.84–4.7) |
| FEV1, % predicted | 84.6 ± 12.3 (65–111) | 94.9 ± 12.5 (77–122) |
| Treatment (regular or as needed) | ||
| Albuterol inhaler | 19 | 18 |
| Long-acting bronchodilator | 2 | 2 |
| Inhaled corticosteroids | 3 | 2 |
| Nasal corticosteroids | 2 | 1 |
| Montelukast | 1 | 4 |
| Antihistamines | 6 | 9 |
| Allergen used for challenge | ||
| Dust mite | 6 | 4 |
| Cat | 7 | 10 |
| Ragweed | 2 | 1 |
| Bermuda grass | 1 | 1 |
| Bluegrass | 1 | 1 |
| Rye grass | 1 | 1 |
| Timothy grass | 1 | 0 |
Data are presented as count or mean ± SD (range).
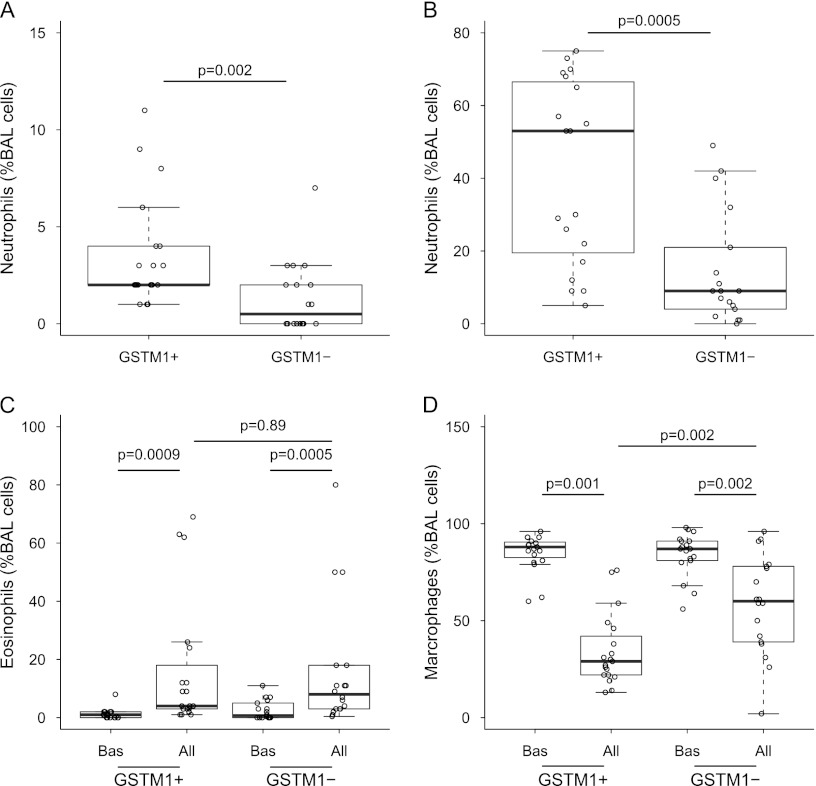
With the purpose of testing the hypothesis that GSTM1 genotype affects allergen-induced airway inflammation in individuals with atopic asthma, we analyzed cellular composition of BAL cells recovered at baseline and after allergen challenge in vivo. Baseline percentages of neutrophils in BAL were higher in GSTM1+ than in GSTM1− individuals with asthma (3.6 ± 2.8 vs. 1.3 ± 1.8, P = 0.002, GSTM1+ vs. GSTM1−) (Figure 1A). Allergen augmented the percentages of BAL neutrophils in both groups, but the increase was greater in GSTM1+ individuals with asthma (41.9 ± 25.2 vs. 14.5 ± 15.5, P = 0.0005, GSTM1+ vs. GSTM1−) (Figure 1B). Similar findings of neutrophil percentages were also found in subjects with dust mite and cat allergen extract, but not in grass allergen extract. Multivariable regression also indicated a significant difference in percentages of neutrophils after allergen challenge after adjusting for the type of allergen. In contrast, GSTM1 did not affect airway eosinophilia, either at baseline or after allergen (Figure 1C). Airway neutrophilia in GSTM1+ individuals with asthma was associated with higher post-allergen BAL concentrations of myeloperoxidase and elastase, markers of neutrophil activation (Table 2 and Figure 2A, respectively). These data suggest the capability of GSTM1 to modulate the inflammatory phenotype associated with atopic asthma but failed to demonstrate inhibition of the influx of inflammatory cells due to allergen in GSTM1+ individuals with asthma.
Figure 1.
Combined box and dot plots showing the effect of the GSTM1 genotype on neutrophils, eosinophils, and alveolar macrophages in BAL fluid. (A, B) Percentages of neutrophils at baseline (A) and after allergen provocation (B). (C, D) Baseline and allergen-provoked percentages of eosinophils (C) and alveolar macrophages (D). All = allergen challenge; BAL = bronchoalveolar lavage; Bas = baseline; GSTM1 = glutathione S-transferase M1.
TABLE 2.
EFFECT OF THE GSTM1 GENOTYPE ON MEDIATORS’ CONCENTRATIONS IN BAL AT BASELINE AND 24 HOURS AFTER ALLERGEN INSTILLATION
| Mediator | GSTM1+ | P Bas vs. All | GSTM1− | P Bas vs. All | P GSTM1+ vs. GSTM1− | |
| IL-4 | Bas | 0.12 (0.017-0.24) | 0.17 | 0.055 (0.0–0.1) | 0.0008 | 0.17 |
| All | 0.17 (0.07–0.32) | 0.15 (0.05–0.28) | 0.55 | |||
| IL-5 | Bas | 0.1 (0.03–0.16) | 0.0002 | 0.06 (0.02–0.16) | 0.0005 | 0.4 |
| All | 1.42 (0.5–3.6) | 0.4 (0.2–6.57) | 0.13 | |||
| IL-13 | Bas | 0.01 (0.0–0.6) | 0.01 | 0 (0.0–0.38) | 0.02 | 0.48 |
| All | 1.33 (0.2–5.5) | 0.43 (0.0–0.82) | 0.08 | |||
| IFN-γ | Bas | 0.49 (0.14–0.7) | 0.01 | 0.33 (0.16–0.43) | 0.29 | 0.23 |
| All | 0.6 (0.21–1.7) | 0.39 (0.3–0.66) | 0.46 | |||
| IL-12 | Bas | 0.12 (0.0–0.2) | 0.007 | 0.1 (0.0–0.19) | 0.8 | 0.9 |
| All | 0.2 (0.05–0.64) | 0.19 (0.0–0.2) | 0.08 | |||
| IL-17 | Bas | 21.9 (20.4–25.9) | 0.27 | 21.8 (20.1–24.8) | 0.25 | 0.96 |
| All | 21.13 (18.2–25.2) | 20.6 (19.4–22.9) | 0.8 | |||
| IL-33 | Bas | 6.28 (2.9–16.5) | 0.49 | 6.86 (3.88–9.5) | 0.62 | 0.97 |
| All | 3.7 (2.5–13.5) | 6.12 (3.8–8.5) | 0.57 | |||
| CXCL1 | Bas | 536.5 (402–856.7) | 0.27 | 441.7 (287–610.7) | 0.06 | 0.14 |
| All | 399.4 (230.6–670) | 339 (189.6–474) | 0.31 | |||
| F2-IsoPs | Bas | 0.86 (0.57–1.26) | 0.0002 | 1.16 (0.84–1.5) | 0.04 | 0.15 |
| All | 1.36 (1.07–2.75) | 1.4 (0.92–2.35) | 0.56 | |||
| MPO | Bas | 26.1 (11.56–26.2) | 0.0004 | 14.9 (5.1–26.2) | 0.001 | 0.14 |
| All | 300.3 (116–552) | 98.9 (27.2–186.5) | 0.002 | |||
Definition of abbreviations: All = allergen; BAL = bronchoalveolar lavage; Bas = baseline; F2-IsoPs = F2-isoprostanes; MPO = myeloperoxidase.
Concentrations of all mediators are expressed in pg/ml BAL. Concentrations are median (interquartile range).
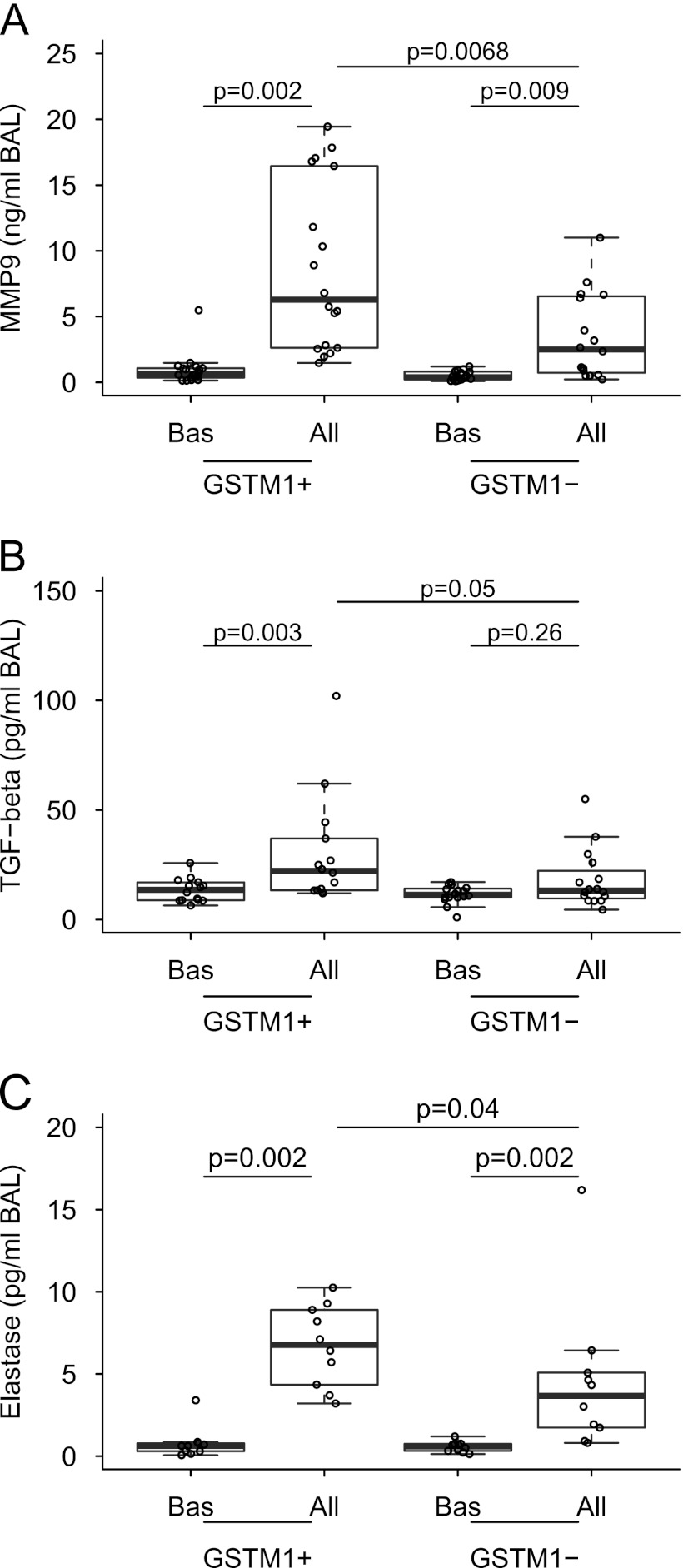
Figure 2.
Combined box and dot plots presenting BAL concentrations of (A) MMP-9, (B) TGF-β, and (C) elastase at baseline and after allergen challenge in individuals with asthma with GSTM1+ and GSTM1− genotypes. All = allergen challenge; BAL = bronchoalveolar lavage; Bas = baseline; GSTM1 = glutathione S-transferase M1; MMP9 = matrix metalloproteinase-9; TGF = transforming growth factor.
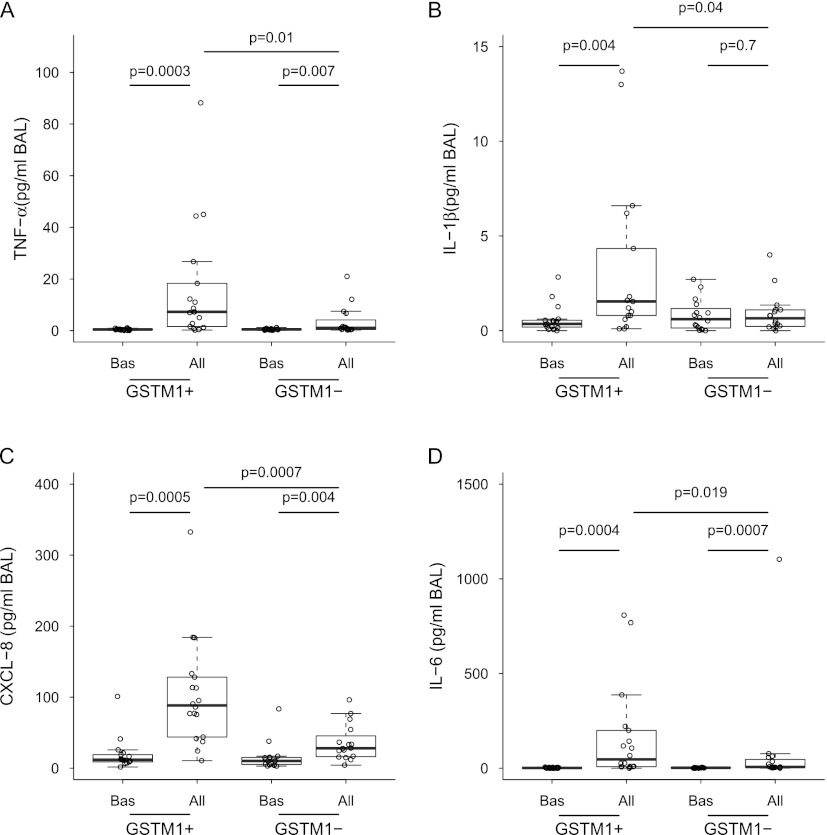
After finding different inflammatory patterns in individuals with asthma with divergent GSTM1 genotypes, we sought to answer the question of whether airway neutrophilia in GSTM1+ patients was associated with up-regulation of proneutrophilic factors in the lung. Indeed, although baseline concentrations of all measured mediators were comparable between groups (Figures 2–4 and Table 2), allergen challenge uncovered distinct mediator patterns related to the GSTM1 genotypes. In particular, the generation of CXCL8 and IL-1β as well as other acute-stress cytokines (TNF-α and IL-6) was greater in GSTM1+ individuals with asthma (Figure 3). In contrast, baseline and post-allergen concentrations of IL-17, a lymphocyte-originated proneutrophilic factor (26), and IL-33, a chromatin-associated nuclear cytokine from the IL-1 family (27), did not vary between GSTM1+ and GSTM1− patients (Table 2). CXCL1 levels were not increased by allergen at 24 hours (Table 2). The allergen-induced airway neutrophilia weakly correlated with concentrations of TNF-α in BAL (Spearman r = 0.4, P = 0.02). Concentrations of Th1 cytokines, IFN-γ and IL-12p70, and Th2 cytokines, IL-4, IL-5, and IL-13, did not differ between GSTM1+ and GSTM1− patients although statistically significant augmentation of IFN-γ and IL-12p70 after allergen occurred only in GSTM1+ individuals with asthma (Table 2). These results suggest that the GSTM1 genotype is involved in regulation of a specific molecular endotype, likely in airway epithelial cells, that results in up-regulation of the neutrophilic phenotype in atopic asthma in humans.
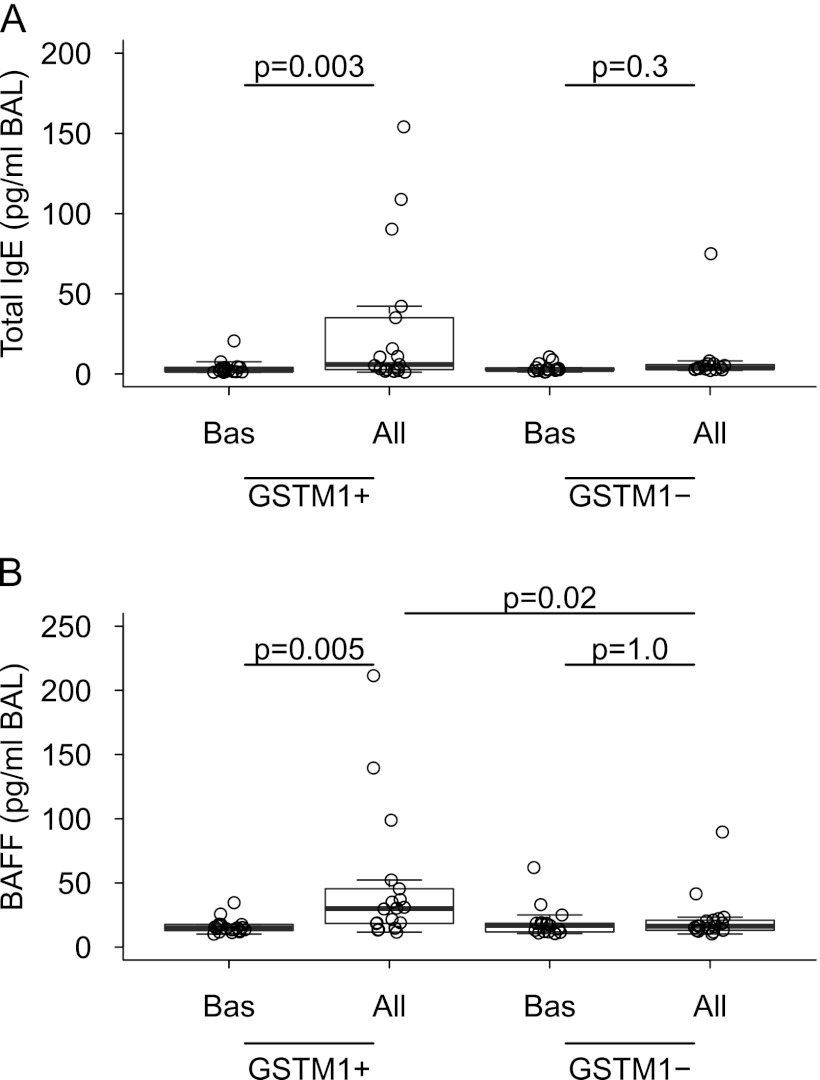
Figure 4.
Combined box and dot plots showing BAL concentrations of (A) total IgE and (B) BAFF at baseline and after allergen challenge in individuals with asthma with GSTM1+ and GSTM1− genotypes. All = allergen challenge; BAFF = B-cell activating factor; BAL = bronchoalveolar lavage; Bas = baseline; GSTM1 = glutathione S-transferase M1.
Figure 3.
Combined box and dot plots demonstrating BAL levels of (A) TNF-α, (B) IL-1β, (C) CXCL8, and (D) IL-6 at baseline and after allergen challenge in individuals with asthma with GSTM1+ and GSTM1− genotypes. All = allergen challenge; BAL = bronchoalveolar lavage; Bas = baseline; GSTM1 = glutathione S-transferase M1; TNF-α = tumor necrosis factor-α.
Neutrophils are capable of generating a vast spectrum of mediators capable of enhancing, modulating, and resolving inflammatory responses (28). To demonstrate whether neutrophilic inflammation associated with the GSTM1+ genotype modulates the inflammatory milieu in asthmatic airways, we analyzed selected mediators that have been previously described in the neutrophilic asthma. Indeed, allergens provoked significantly greater concentrations of the metalloproteinase MMP-9 (29); TGF-β, a pluripotent cytokine involved in inflammation and tissue repair and remodeling (30); and elastase (31) in GSTM1+ than in GSTM1− individuals with asthma (Figure 2). The finding that allergens provoked a significant increase of total BAL IgE levels exclusively in GSTM1+ individuals with asthma (Figure 4) prompted us to determine production of BAFF, a neutrophil-produced T cell–independent factor participating in activation and differentiation of B cells (32). We found augmented concentrations of allergen-induced BAFF in BAL fluid in the GSTM1+ individuals. It is noteworthy that the IgE and BAFF levels correlated with each other (Spearman r = 0.54, P = 0.001) (Figure 4). These data expand the current knowledge concerning biochemical consequences of neutrophilic inflammation and link them to new genomic information in humans with atopic asthma.
With the aim of testing the hypothesis that GSTM1 regulates oxidative stress in atopic asthmatic lungs, we measured BAL concentrations of F2-isoprostanes, specific markers of oxidative stress in vivo (25). We found no effect of the GSTM1 genotype on the generation of F2-isoprostanes at baseline and after allergen challenge in asthmatic airways (Table 2). These data suggest that GSTM1 regulates airway inflammation independently of the oxidative stress in humans with atopic asthma in vivo.
Finally, we sought to determine consequences of the GSTM1-dependent regulation of inflammation on airway function in individuals with atopic asthma. We found that GSTM1+ individuals with asthma were more reactive to specific allergens as suggested by lower threshold doses necessary to produce a 20% fall in FEV1 (102.8 ± 133.5 vs. 39.3 ± 42.9, dilution fold of the allergen stock solution, P = 0.04, GSTM1+ vs. GSTM1−). The late asthmatic reaction produced by inhaled allergen occurred in 11 of 23 GSTM1+ and 7 of 23 GSTM1− individuals with asthma (P = 0.2 by Fisher’s exact test). The lower threshold to inhaled methacholine in GSTM1+ compared with GSTM1− barely missed statistical significance (2.0 ± 2.2 vs. 4.5 ± 5.3 mg/ml, P = 0.06, GSTM1+ vs. GSTM1−). Nevertheless, these data suggest that airway hyperresponsiveness in human asthma could at least in part depend on the inflammatory and biochemical events regulated by GSTM1.
Discussion
Our study demonstrates greater neutrophilic airway inflammation at baseline and after allergen provocation in GSTM1+ than in GSTM1− individuals with asthma in vivo. The allergen-stimulated generation of acute-stress and proneutrophilic factors, CXCL-8, IL-1β, TNF-α, and IL-6 was also enhanced in the GSTM1+ patients. The post-allergen levels of total IgE in BAL fluid increased only in GSTM1+ individuals with asthma and correlated with concentrations of B-cell activating factor that could be released by neutrophils (32). Furthermore, levels of selective neutrophil-generated mediators, MPP-9, TGF-β, and elastase were also higher in the GSTM1+ individuals with asthma. On the other hand, GSTM1 did not modulate the generation of F2-isoprostanes in BAL fluid. Finally, GSTM1+ individuals with asthma required lower threshold concentrations of specific allergen to provoke bronchoconstriction. These data imply that GSTM1 regulates the inflammatory airway phenotype produced by allergens in individuals with atopic asthma in vivo. To our knowledge, these findings are novel.
Our results showing coexistence of neutrophilic inflammation with IL-1β, TNF-α, and CXCL8 concur with previous findings linking neutrophilic airway inflammation with these cytokines and nuclear factor-κB in persistent and allergen-induced experimental asthma in humans (33, 34). The most likely lung cells regulated by GSTM1 are bronchial epithelial and smooth muscle cells that express GSTM1 (7) and are capable of generating proneutrophilic cytokines after exposure to allergens (35). We did not find regulation by allergens of other proneutrophilic cytokines previously associated with asthma, including IL-17, a T cell-dependent cytokine (26); IL-33, a chromatin-associated nuclear cytokine from the IL-1 family of cytokines (27); and CXCL1 (36).
The role of GSTM1 in asthma has been mainly attributed to the antioxidant properties of the enzyme. However, our data showing similar generation of F2-isoprostanes irrespective of the GSTM1 genotype do not support a substantial antioxidant role of GSTM1 in atopic asthmatic airways. In fact, GSTM1 provides only 0.1% of GST activity in normal lungs (37). Currently, it is unclear why airway inflammation in the GSTM1+ individuals with atopic asthma is deviated toward the neutrophilic phenotype. Genes in the IL-1 and TNF-α/nuclear factor-κB pathways are regulated by the mitogen-activated protein kinase signaling cascade that can be inhibited by GSTM1 independently of its glutathione-conjugating or antioxidant activity (8–10). However, the GSTM1-dependent restraining mechanism could be disrupted in the atopic asthmatic airways as a result of down-regulation of the transcription factor nuclear factor (erythroid-derived 2)-like 2 (38, 39) controlling expression of GSTM1 (40). Airway epithelial cells could be particularly affected as they express GSTM1 (7) and are capable of generating robust proneutrophilic signals after allergen stimulation (35). A tight regulation of stress-signaling pathways would suggest the presence of an alternative inhibitory mechanism in GSTM1− individuals. Interestingly, exposure of the GSTM1-deleted mice to 1,2-dichloro-4-nitrobenzene (specific GSTM1 substrate) caused up-regulation of genes previously associated with the murine asthma, including Mmp9, Chi313, Trem3, and Epx, encoding matrix metallopeptidase 9, chitinase 3-like 3, activating-receptor on macrophages, and eosinophil peroxidase, respectively (41).
As expected, the different cellular phenotypes in GSTM1+ and GSTM1− individuals with asthma were associated with dissimilar biochemical milieus in the epithelial lining fluid. Indeed, neutrophils have the capacity to produce a myriad of mediators of innate and adaptive immunity (28). We found increased concentrations of MMP-9, BAFF, TGF-β, and elastase in GSTM1+ compared with GSTM1− patients. MMP-9 is a major metalloproteinase associated with neutrophilic inflammation in atopic asthma (29). Up-regulation of BAFF by allergen and its correlation with post-allergen total IgE in BAL, predominantly in GSTM1+ patients, is a novel observation that will need future investigation. BAFF is a critical regulator of B cells’ survival and responsiveness that can be produced by neutrophils (32). TGF-β is a powerful pluripotent cytokine linked to modulation and resolution of inflammation and remodeling in asthma (30). Elastase is an important protease enzyme causing lung damage that has been linked to neutrophilic asthma in humans (31, 42).
Our study also suggests modulation of airway responsiveness by GSTM1 in individuals with atopic asthma. Though the difference in methacholine hyperresponsiveness marginally missed statistical significance, the inhaled dose of allergen required to cause a similar fall in FEV1 in the two groups was lower in GSTM1+ individuals with asthma. This implies that the allergen sensitivity of the GSTM1+ group is greater.
The promoting role of GSTM1 on neutrophilic airway inflammation in our homogenous population of individuals with atopic asthma contradicts previous studies showing a protective role of the wild-type GSTM1 genotype on ozone- and endotoxin-induced airway neutrophilia (17, 43, 44) or diesel exhaust particles–enhanced response of nasal mucosa to allergen (18). However, it is important to emphasize that asthmatic airway epithelial cells are intrinsically different from normal or nasal epithelial cells; thus, biochemical interactions involving GSTM1 could be incomparable in these anatomically comparable but functionally dissimilar cells (45, 46). Although most functions of GSTM1 are considered protective, there are precedents for opposite actions of the enzyme. For example, there are studies showing that in atopic individuals the null, not wild-type, GSTM1 genotype decreases risk for asthma (14). Regardless, additional extensive studies will be necessary to explain the pathomechanism and clinical significance of our seemingly paradoxical findings.
Commercial allergens contain small amounts of endotoxin that may affect airway neutrophilia in individuals with asthma undergoing allergen challenges in vivo (47). We employed only standardized allergens provided by the same vendor, and importantly, identical lots of allergens were used in individuals with asthma with both GSTM1 genotypes. Although we do not believe that endotoxin contaminating allergens was responsible for the discrepancies found in our study, at present one cannot rule out the possibility that GSTM1 acts differently in individuals with atopic asthma. In fact, a recent study demonstrated a diminished response to endotoxin in individuals with atopic asthma compared with individuals without asthma (48). In this investigation, the absolute number of inflammatory cells recruited to the airways was lower in individuals with asthma, and moreover, levels of IL-1β, IL-18, and cell-surface expression of Toll-like receptors 4 and 2 were up-regulated only in normal subjects (48). Thus, a distinct inflammatory signaling could be associated with an altered modulatory action of GSTM1 in response to endotoxin in atopic asthmatic airways cells in vivo.
Major weaknesses of our investigation include a relatively small population of individuals with atopic asthma of predominantly Northern European ancestry. Thus, a larger study might uncover even sharper phenotypic contrasts related to the GSTM1 genotype. Future studies should also determine if GSTM1 similarly regulates airway response to allergens in individuals with atopic asthma from other ethnic groups. Patients with moderate or severe asthma were not enrolled because of the invasive nature of the experiments. Consequently, it remains to be established whether the GSTM1 genotype is associated with the neutrophilic variant of the severe asthma phenotype. Finally, noneosinophilic asthma is characterized by poorer lung function (18), less favorable response to corticosteroids (2), and systemic inflammation (49). Consequently, the role of the GSTM1 genotype on treatment response to currently available antiinflammatory asthma drugs, especially corticosteroids, should also be sought.
Presently, the genetic and pathomechanistic underpinnings linked to the neutrophilic asthma phenotype is unclear. Our study identifies a new and potentially important area of investigation concerning asthma heterogeneity.
In summary, our study provides a convincing evidence for an endogenous modulatory role of GSTM1 on asthmatic responses in human atopic asthma. Particularly, the wild-type GSTM1 genotype appears to promote a pathophysiologically relevant neutrophilic asthma phenotype. However, the exact mechanism of the GSTM1-dependent regulation of allergen-provoked asthma exacerbation is yet to be fully elucidated. Our study provides a novel link between genetic polymorphism and asthma phenotype that could be important for personalized tailoring of asthma therapy in the future.
Supplementary Material
Acknowledgments
The authors thank Dr. James R. Sheller for helpful comments.
Footnotes
Supported by grants from the NIH (K23 HL080030, M01 RR-00095, and P30 ES000267).
Author Contributions: A.H. was responsible for recruitment of volunteers, performance of human experiments, and collection and interpretation of data, and aided in the preparation of the manuscript. S.R. performed genotyping and contributed to manuscript preparation. P.W. and N.C. contributed significantly to statistical analysis of data and preparation of the manuscript. W.H., R.A., and R.-h.D. contributed to BAL analysis, collection of data, and manuscript preparation. R.D. was the principal investigator of the study. He designed and coordinated the investigation, performed bronchoscopies, supervised all challenge procedures, and was responsible for interpretation of the results and manuscript preparation.
Originally Published in Press as DOI: 10.1164/rccm.201204-0786OC on November 29, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Wenzel SE. Asthma: defining of the persistent adult phenotype. Lancet 2006;368:804–813 [DOI] [PubMed] [Google Scholar]
- 2.McGrath KW, Icitovic N, Boushey HA, Lazarus SC, Sutherland ER, Chinchilli VM, Fahy JV. A large subgroup of mild-to-moderate asthma is persistently non-eosinophilic. Am J Respir Crit Care Med 2012;85:612–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol 2005;45:51–88 [DOI] [PubMed] [Google Scholar]
- 4.Brasch-Andersen C, Christiansen L, Tan Q, Haagerup A, Vestbo J, Kruse TA. Possible gene dosage effect of glutathione-S-transferases on atopic asthma: using real-time PCR for quantification of GSTM1 and GSTT1 gene copy numbers. Hum Mutat 2004;24:208–214 [DOI] [PubMed] [Google Scholar]
- 5.Gilliland FD, Li YF, Dubeau L, Berhane K, Avol E, McConnell R, Gauderman WJ, Peters JM. Effects of glutathione S-transferase M1, maternal smoking during pregnancy, and environmental tobacco smoke on asthma and wheezing in children. Am J Respir Crit Care Med 2002;166:457–463 [DOI] [PubMed] [Google Scholar]
- 6.Carlsten C, Sagoo GS, Frodsham AJ, Burke W, Higgins JP. Glutathione S-transferase M1 (GSTM1) polymorphism and lung cancer: a literature based systematic HuGE review and metaanalysis. Am J Epidemiol 2008;167:759–774 [DOI] [PubMed] [Google Scholar]
- 7.Anttila S, Hirvonen A, Vainio H, Husgafvel-Pursiainen K, Hayes JD, Ketterer B. Immunohistochemical localization of glutathione S-transferases in the lung. Cancer Res 1993;53:5643–5648 [PubMed] [Google Scholar]
- 8.Cho S-G, Lee YH, Park H-S, Ryoo K, Kang KW, Park J, Eom S-J, Kim MJ, Chang T-S, Choi S-Y, et al. Glutathione S-Transferase Mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. J Biol Chem 2001;16:12749–12755 [DOI] [PubMed] [Google Scholar]
- 9.Dorion S, Lambert H, Landry J. Activation of the p38 signaling pathway by heat shock involves the dissociation of glutathione S-transferase Mu from Ask1. J Biol Chem 2002;277:30792–30797 [DOI] [PubMed] [Google Scholar]
- 10.Ryoo K, Huh S-H, Lee YH, Yoon KW, Cho S-G, Choi E-J. Negative regulation of MEKKK1-induced signaling by glutathione S-transferase Mu. J Biol Chem 2004;279:43589–43594 [DOI] [PubMed] [Google Scholar]
- 11.Bogaards JJP, Venekamp JC, van Bladeren PJ. Stereoselective conjugation of prostaglandin A2 and prostaglandin J2 with glutathione, catalyzed by the human glutathione S-transferases A1-1, A2-2, M1a-1a, and P1. Chem Res Toxicol 1997;10:310–317 [DOI] [PubMed] [Google Scholar]
- 12.Rudd LP, Kabler SL, Morrow CS, Townsend AJ. Enhanced glutathione depletion, protein adduct formation, and cytotoxicity following exposure to 4-hydroxy-2-nonenal (HNE) in cells expressing human multidrug resistance protein-1 (MRP1) together with human glutathione S-transferase-M1 (GSTM1). Chem Biol Interact 2011;194:113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minelli C, Granell R, Newson R, Rose-Zerilli MJ, Torrent M, Ring SM, Newson R, Rose-Zerilli MJ, Torrent M, Ring SM, et al. Glutathione S-transferase genes and asthma phenotypes: a human epidemiology (HuGE) systemic review and meta-analysis including unpublished data. Int J Epidem 2010;39:539–562. [DOI] [PMC free article] [PubMed]
- 14.Mak JC, Ho SP, Leung HC, Cheung AH, Law BK, So LK, Chan JW, Chau CH, Lam WK, Ip MS, et al. Relationship between glutathione S-transferase gene polymorphism and enzyme activity in Hong Kong Chinese asthmatics. Clin Exp Allergy 2007;37:1150–1157 [DOI] [PubMed] [Google Scholar]
- 15.Kamada F, Mashimo Y, Inoue H, Shao C, Hirota T, Doi S, Kameda M, Fujiwara H, Fujita K, Enomoto T, et al. The GSTP1 gene is a susceptibility gene for childhood asthma and the GSTM1 gene is a modifier of the GSTP1 gene. Int Arch Allergy Immunol 2007;144:275–286 [DOI] [PubMed] [Google Scholar]
- 16.Tung K-Y, Tsai C-H, Lee YL. Microsomal epoxide hydrolase genotypes/diplotypes, traffic air pollution, and childhood asthma. Chest 2011;139:839–848 [DOI] [PubMed] [Google Scholar]
- 17.Alexis NE, Zhou HZ, Lay JC, Harris B, Hernandez ML, Lu T-S, Bromberg PA, Diaz-Sanchez D, Devlin RB, Kleeberger SR, et al. The glutathione-S-transferase Mu 1 null genotype modulates ozone-induced airway inflammation in human subjects. J Allergy Clin Immunol 2009;124:1222–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilliland FD, Li YE, Saxon A, Diaz-Sanchez D. Effect of glutathione- S-transferase M1 and P1 genotypes on xenobiotic enhancement of allergic responses: randomized, placebo-controlled crossover study. Lancet 2004;363:119–125 [DOI] [PubMed] [Google Scholar]
- 19.Hoskins A, Roberts JL, II, Milne G, Choi L, Dworski R. Natural-source d-α-tocopheryl acetate inhibits oxidant stress and modulates atopic asthma in humans in vivo. Allergy 2012;67:676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Asthma Education and Prevention Program. Expert panel report 3: guidelines for the diagnosis and management of asthma. Bethesda, MD: National Institute of Health, National Heart, Lung, and Blood Institute, 2007. Publication No. 08-5846.
- 21.Bernstein IL, Li JT, Bernstein DI, Hamilton R, Spector SL, Tan R, Sicherer S, Golden DB, Khan DA, Nicklas RA, et al. Allergy diagnostic testing: an updated parameter. Ann Allergy Asthma Immunol 2008;100:S14–S24 [DOI] [PubMed] [Google Scholar]
- 22.American Thoracic Society Guidelines for methacholine and exercise challenge testing. Am J Respir Crit Care Med 2000;161:309–329 [DOI] [PubMed] [Google Scholar]
- 23.Cockcroft DW, Davis BE, Boulet L-P, Deschesnes F, Gauvreau GM, O’Byrne PM, Watson RM. The links between allergen skin test sensitivity, airway responsiveness and airway response to allergen. Allergy 2005;60:56–59 [DOI] [PubMed] [Google Scholar]
- 24.Davies MH, Elisa E, Acharya S, Cotton W, Faulder GC, Fryer AA, Strange RC. GSTM1 null polymorphism at the glutathione S-transferase M1 locus: phenotype and genotype studies in patients with primary biliary cirrhosis. Gut 1993;34:549–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts LJ, Morrow JD. Measurement of F2-isoprostanes as an index of oxidative stress in vivo. Free Radic Biol Med 2000;28:505–513 [DOI] [PubMed] [Google Scholar]
- 26.Wang Y-H, Wills-Karp M. The potential role of interleukin-17 in severe asthma. Curr Allergy Asthma Rep 2011;11:388–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith DE. IL-33: a tissue derived cytokine pathway involved in allergic inflammation and asthma. Clin Exp Allergy 2009;40:200–208 [DOI] [PubMed] [Google Scholar]
- 28.Mantovani A, Cassatella MA, Constantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Natl Rev 2011;11:519–531 [DOI] [PubMed] [Google Scholar]
- 29.Kelly EABK, Busse WW, Jarjour NN. Increased matrix metalloproteinase-9 in the airway after allergen challenge. Am J Respir Crit Care Med 2000;162:1157–1161 [DOI] [PubMed] [Google Scholar]
- 30.Salib RJ, Howarth PH. Transforming growth factor-β in allergic inflammatory diseases of the airways: friend or foe? Clin Exp Allergy 2009;39:1128–1135 [DOI] [PubMed] [Google Scholar]
- 31.Vignola AM, Bonanno A, Mirabella A, Riccobono L, Mirabella F, Profita M, Bellia V, Bousquet J, Bonsignore G. Increased levels of elastase and α1-antitrypsin in sputum of asthmatic patients. Am J Respir Crit Care Med 1998;157:505–511 [DOI] [PubMed] [Google Scholar]
- 32.Puga I, Cols M, Barra CM, He B, Cassis L, Gentile M, Comerma L, Chorny A, Shan M, Xu W, et al. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat Immunol 2012;13:170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol 2011;127:153–160 [DOI] [PubMed] [Google Scholar]
- 34.Nocker RET, Out TA, Weller FR, Mul EPJ, Jansen HM, van der Zee JS. Influx of neutrophils into the airway lumen at 4 h after segmental allergen challenge in asthma. Int Arch Allergy Immunol 1999;119:45–53 [DOI] [PubMed] [Google Scholar]
- 35.Österlund C, Grönlund H, Gafvelin G, Bucht A. Non-proteolytic aeroallergens from mites, cat and dog exert adjuvant-like activation of bronchial epithelial cells. Int Arch Allergy Immunol 2011;155:111–118 [DOI] [PubMed] [Google Scholar]
- 36.Fitzpatrick AM, Higgins M, Holguin F, Brown LAS, Teague WG. The molecular phenotype of severe asthma in children. J Allergy Clin Immunol 2011;125:851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bauer M, Herbarth O, Aust G, Henstler JG, Dotzauer A, Graebsh C, Schmuecking E. Expression patterns and novel splicing variants of glutathione-S-transferase isoenzymes of human lung and hepatocyte cell lines. Cell Tissue Res 2006;324:423–432 [DOI] [PubMed] [Google Scholar]
- 38.Dworski R, Han W, Blackwell TS, Hoskins A, Freeman ML. Vitamin E prevents NRF2 suppression by allergens in asthmatic alveolar macrophages in vivo. Free Radic Biol Med 2011;51:516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fitzpatrick AM, Stephenson ST, Hadley GR, Burwell L, Penugonda M, Simon DM, Hansen J, Jones DP, Brown LA. Thiol redox disturbances in children with severe asthma are associated with posttranslational modification of the transcription factor nuclear factor (erythroid-derived 2)-like 2. J Allergy Clin Immunol 2011;127:1604–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chanas SA, Jiang Q, McMahon M, McWalter GK, McLellan LI, Elcombe CR, Henderson CJ, Wolf CR, Moffat GJ, Itoh K, et al. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase GSTA1, GSTA2, GSTM1, GSTM2, GSTM3 and GSTM4 genes in the livers of male and female mice. Biochem J 2002;365:405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arakawa S, Maejima T, Kiyosawa N, Yamaguchi T, Shibaya Y, Aida Y, Kawai R, Fujimoto K, Manabe S, Takasaki W. Methemoglobinemia induced by 1,2-dichloro-4-nitrobenzene in mice with a disrupted glutathione S-transferase Mu 1 gene. Drug Metab Dispos 2010;38:1545–1552 [DOI] [PubMed] [Google Scholar]
- 42.Wood LG, Baines KJ, Fu J, Scott HA, Gibson PG. The neutrophilic inflammatory phenotype is associated with systemic inflammation in asthma. Chest 2012;142:86–96 [DOI] [PubMed] [Google Scholar]
- 43.Kim CS, Alexis NE, Rappold AG, Kehrl H, Hazucha MJ, Lay JC, Schmitt MT, Case M, Devlin RB, Peden DB, et al. Lung function and inflammatory responses in healthy young adults exposed to 0.06 ppm ozone for 6.6 hours. Am J Respir Crit Care Med 2011;183:1215–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dillon MA, Harris B, Hernandez ML, Zou B, Reed W, Bromberg PA, Devlin RB, Diaz-Sanchez D, Kleeberger S, Zhou H, et al. Enhancement of systemic and sputum granulocyte response to inhaled endotoxin in people with the GSTM1 null genotype. Occup Environ Med 2011;68:783–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braunstahl G-J, Fokkens WJ, Overbeek SE, KleinJan A, Hoogsteden HC, Prins JB. Mucosal and systemic inflammatory changes in allergic rhinitis and asthma: a comparison between upper and lower airways. Clin Exp Allergy 2003;33:579–587 [DOI] [PubMed] [Google Scholar]
- 46.Freishtat RJ, Watson AM, Benton AS, Iqbal SF, Pillai DK, Rose MC, Hoffman EP. Asthmatic airway epithelium is intrinsically inflammatory and mitotically dyssynchronous. Am J Respir Cell Mol Biol 2011;44:863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunt LW, Gleich GJ, Ohnishi T, Weiler DA, Mansfiels ES, Kita H, Sur S. Endotoxin contamination causes neutrophilia following pulmonary allergen challenge. Am J Respir Crit Care Med 1994;149:1471–1475 [DOI] [PubMed] [Google Scholar]
- 48.Hernandez ML, Herbs M, Lay JC, Alexis NE, Brickey WJ, Ting JPY, Zhou H, Peden DB. Atopic asthmatic patients have reduced airway inflammatory cell recruitment after inhaled endotoxin challenge compared with healthy volunteers. J Allergy Clin Immunol 2012;130:869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berry M, Morgan A, Shaw DE, Parker D, Green R, Brightling C, Bradding P, Wardlaw AJ, Pavord ID. Pathological features and inhaled corticosteroid response of eosinophilic and non-eosinophilic asthma. Thorax 2007;62:1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.