Abstract
The excitatory neurotransmitter, glutamate, activates N-methyl-d-aspartate (NMDA) receptors to induce long-lasting synaptic changes through alterations in gene expression. It is believed that these long-lasting changes contribute to learning and memory, drug tolerance, and ischemic preconditioning. To identify NMDA-induced late-response genes, we used a powerful gene-identification method, differential analysis of primary cDNA library expression (DAzLE), and cDNA microarray from primary cortical neurons. We report here that a variety of genes, which we have named plasticity-induced genes (PLINGs), are up-regulated with differential expression patterns after NMDA receptor activation, indicating that there is a broad and dynamic range of long-lasting neuronal responses that occur through NMDA receptor activation. Our results provide a molecular dissection of the activity-dependent long-lasting neuronal responses induced by NMDA receptor activation.
Neuronal plasticity and development of the CNS depends, in part, on neuronal activity (1). Short-term cellular changes, lasting minutes to hours, are due to rapid membrane ionic conductance changes and associated protein phosphorylation events (2, 3). In contrast, long-term plasticity requires the synthesis of new mRNA and proteins (4).
Insights into the genomic mechanisms that underlie long-term plasticity by using differential cloning techniques have been used to identify mRNAs that are rapidly induced by excitatory activity (5, 6). Most of these studies have focused on the identification of immediate-early gene changes (7, 8), whereas little attention has been paid to the late-response genes. It is now well established that induction of immediate-early genes in response to neuronal activity is responsible for setting the stage for long-term changes in synaptic function. However, the genes that are ultimately responsible for long-term changes in neuronal function are poorly characterized.
In the brain, neuronal plasticity is mediated largely by the activation of the N-methyl-d-aspartate (NMDA) glutamate receptor through increases in intracellular calcium (9, 10). A dichotomy of NMDA receptor signaling exists with excessive stimulation, leading to neuronal damage that occurs during stroke and chronic neurodegenerative diseases, whereas normal bursts of excitatory activity result in synaptic transmission and the expression of molecular substrates of long-term plasticity, growth, and survival (11). The activation of NMDA receptors in glutamatergic neurons induces long-lasting synaptic changes through multiple downstream signaling molecules, and these signaling pathways can express late cellular responses through changes in gene expression (12, 13). NMDA receptor-mediated neuronal activity is essential for activity-dependent modification of synaptic connections and refinement of functional circuits; it is critical for several forms of synaptic plasticity that underlie learning and memory (14). NMDA receptor stimulation is also important for long-term changes that lead to neuronal survival and resistance to toxic insults in neurodegenerative disease (15).
Investigation and characterization of the neuronal transcriptional profile after NMDA receptor activation will provide a better understanding of the processes underlying long-term changes in response to neuronal activity and NMDA receptor activation. Using differential analysis of primary cDNA library expression (DAzLE), an extremely sensitive method of differential gene-expression analysis to identify differentially regulated genes from neurons (41), coupled with a microarray (16), we report the identification and characterization of NMDA-induced late-response plasticity-induced genes (PLINGs).
Materials and Methods
Animals, Cell Culture, and Treatment. Primary cortical cell cultures were prepared from gestational day 15 fetal Sprague-Dawley rats as described (17). Experiments were performed at day in vitro 14. Under these conditions, neurons represent 70-90% of the culture. Mature neurons were washed with Tris-buffered control salt solution (CSS) containing 120 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 25 mM Tris·HCl (pH 7.4), and 15 mM d-glucose. NMDA (50 μM) and glycine (10 μM) in CSS solution was applied to the cells for 5 min, then the cells were washed and replaced with minimum essential medium containing 5% horse serum and incubated for 6 h in the incubator. Sham treatment control was performed as above except for a 5-min treatment with only CSS solution. All experimental procedures were in accordance with the National Institutes of Health Guidelines and were approved by our institutional animal care and use committees.
cDNA Library Construction. Total RNA was extracted from neurons with TRIzol reagent (GIBCO/BRL), and poly(A)+ RNA was purified with oligo(dT) cellulose chromatography as described in the manufacturer's instructions. The cDNA library from the mRNA of NMDA-treated neurons was constructed with the SuperScript Plasmid System (GIBCO/BRL) following the manufacturer's protocol. In brief, RNA was NotI-poly(dT)-primed, and cDNA fractions between 1 and 12 kb were pooled for cloning. cDNA was directionally cloned with 5′ NotI and 3′ SalI adapter in the pSPORT1 vector and introduced into Escherichia coli. The unamplified library contained 2 × 106 transformants.
DAzLE. Bacteria containing the cDNA library were applied to nylon filters (≈2,000 colonies per plate) on agar plates with ampicillin and incubated at 37°C. The colonies were transferred onto two filters, lysed, and neutralized. The transferred DNA was cross-linked with a UV-cross-linker and stored at 4°C in 2× SSC. mRNAs were purified from NMDA-treated (mRNA1) and sham (CSS solution)-treated (mRNA2) neurons; first-strand cDNAs were synthesized by reverse transcriptase with dT15V as a primer, and double-strand cDNAs were synthesized by DNA polymerase I (see Fig. 1). After heating at 70°C for 10 min and quenching on ice for 5 min, DNA probes were made by random primed DNA labeling with digoxigenin-dUTP (Roche Diagnostics) according to the manufacturer's instructions. Differential expression of genes was scrutinized by hybridizing two probes that originated from control and NMDA-treated neurons. The membrane filters were hybridized at 42°C overnight in a DIG Easy Hyb solution (Roche Diagnostics). The hybridized membrane filters were washed at room temperature for 30 min with 2× SSC containing 0.2% SDS twice and at 65°C for at least 15 min with 0.1× SSC containing 0.2% SDS. The digoxigenin-labeled cDNA probes were used several times until the hybridization signal diminished in intensity. The bacterial colonies that showed higher intensity on x-ray film from the NMDA-treated neuronal probe than sham-treated neurons were picked, cultured in LB broth containing ampicillin, and preserved at -80°C in 50% glycerol.
Fig. 1.
NMDA-induced PLING identification by DAzLE. DAzLE was used to identify genes induced at 6 h after NMDA receptor activation in rat cortical neurons. Differentially expressed transcripts were screened from nonamplified cDNA libraries by a modified differential hybridization method using poly(dA/dT)-tailless probes. The positive clones from primary screening were amplified individually by PCR and arrayed onto nylon membranes. The differential gene expressions were confirmed by microarray analysis, reverse Northern blot analysis, and Northern blot analysis.
Reverse Northern Blotting Procedure. Plasmid DNAs from the positive bacterial clones were isolated, denatured and spotted on a positively charged Nylon membrane (Amersham Pharmacia) with a 96-well vacuum manifold. The spotted DNA was cross-linked to the membrane with a UV-cross-linker (Amersham Pharmacia). 32P-labeled first-strand cDNA was prepared by reverse transcription of total RNA. Thirty micrograms of total RNA was mixed with 4 μg of dT15V and incubated 10 min at 70°C and cooled on ice for 5 min. The mixture was added with 50 mM Tris·HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 0.5 mM dATP, 0.5 mM dGTP, 0.5 mM dTTP, 0.02 mM dCTP, 100 μCi dCTP, and 200 units of SuperScript RT II (GIBCO/BRL) in a final 25-μl reaction solution. The reaction mixture was incubated at 42°C for 1 h and at 42°C for 30 min after addition of 200 units of SuperScript RT II. The membrane was hybridized and washed as described above. The signal intensity of each spot was measured by imagequant software (Amersham Pharmacia), followed normalization and comparison between control and NMDA-treated neurons. Each image was overlaid with grids so that signal intensities of individual spots could be assessed. The cDNAs displaying differential expression that were significantly different (P < 0.05) between two samples were selected and sequenced for further analysis.
Microarray Construction and Analysis. The NMDA-induced, gene-enriched microarray was constructed by arraying PCR-amplified cDNA clones at high density on a nylon membrane. Bacterial clones (1,152) were selected from differential screening. The plasmids were purified from 96-well bacterial cultures (Edge BioSystem, Gaithersburg, MD) and the cDNA inserts were amplified by PCR. Each PCR product was verified by agarose gel electrophoresis and each product was printed onto nylon membrane by an array robot (see Fig. 1). Thirty micrograms of total RNA was used to label cDNA probes by reverse transcription for hybridizing to the microarrays. 33P-labeled cDNAs from CSS- and NMDA-treated cortical neurons were used as the reference probe and the sample probe, respectively in all hybridizations. Ten micrograms of polydeoxyadenylic acid and 20 μg of human CoT1 DNA (Invitrogen) were added to a DIG easy hybridization solution (Roche Diagnostics) and the microarray membrane was prehybridized at 42°C for 1 h before the probe was added directly to the prehybridization solution. Hybridizations, washes, and image scans were performed as described (18). Hierarchical clustering algorithms were applied to all the genes after normalization by using software programs (genesis and ibmt-tug). Genes were selected as differentially expressed clones if their expression level deviated from that of CSS-treated neurons by a factor of 2.5 in at least five of the samples from NMDA-treated neurons or if the standard deviation for the set of five values of z-ratios (19) determined in the analysis of the time course of gene expression exceeded 0.8. Genes fitting these criteria were sequenced, and 5′ sequence tags were generated.
Northern Blot Analysis. Twenty micrograms of total RNA was resolved in 1% agarose gel and transferred onto nylon membrane (Hybond-N, Amersham Pharmacia). Each probe was labeled with [α-32P]dCTP by using Ready-To-Go (Amersham Pharmacia). The membrane was prehybridized and then hybridized at 55°C overnight. The membrane was washed with 1× SSC at 37°C and 0.5× SSC at 65°C, respectively. The membrane was exposed on phosphor screen, and signals were detected by using Cyclone Storage Phosphor System (Packard).
Maximal Electroconvulsive Seizures (MECSs). MECSs were induced in male Sprague-Dawley rats (30-40 d of age and 120-160 g). Current was passed transcranially (90 mA for 1.0 sec) by ear-clip electrodes. At this stimulation level, animals showed tonic/clonic seizures. Animals were allowed to survive 6 h(n = 4), 24 h (n = 6), 48 h (n = 3), and 72 h (n = 2). MK801 (0.6 mg/kg) or normal saline was injected i.p. 30 min before MECS. Rats were decapitated at appropriate times after MECS. Brains were removed and frozen in powdered dry ice. Sections were stored at -80°C until use.
Results
To begin to explore the long-term changes that occur in response to NMDA-glutamate receptor activation, we used DAzLE, an extremely sensitive method of differential gene-expression analysis (41), coupled with microarray analysis to identify late-response PLINGs. Late-response PLINGs were identified by comparing the expression profile of primary cortical neuronal cultures 6 h after a brief (5-min) stimulation with NMDA (50 μM) or with control buffer solution. We used a dose and length of stimulus of NMDA that induces sustained cAMP response element-binding protein phosphorylation and long-term changes in neuronal function that render cortical neurons resistant to subsequent toxic challenges (S.J.H., T.M.D., and V.L.D., unpublished work). The DAzLE method relies on screening nonamplified primary libraries with poly(A/T) tailless cDNAs. A full-length cDNA library from NMDA-stimulated cultures was constructed (Fig. 1). The construction of the cDNA library was followed by library screening with poly(A/T) tailless digoxigenin-labeled dUTP. Double-stranded cDNA probes reverse transcribed from mRNA samples of unstimulated (driver) neurons and NMDA-stimulated (tester) neurons were synthesized with A, C, G, anchored poly(T)16 to fix the size of the poly(A/T) tail of the cDNA. The use of poly(dA/T) tailless, double-stranded DNA as probes limits cross-hybridization among the 3′ ends of the sequences, so that rare transcripts are easily recovered as positive clones. Clones were picked by colony hybridization, and only those clones that were dramatically up-regulated 5- to 10-fold on visual inspection were picked. Individual clones (140,000) were screened by DAzLE, and 1,200 (0.86%) colonies that showed higher intensity by chemluminescence detection on x-ray film with the NMDA-treated neuronal probes were picked, cultured, and cataloged. These clones were subjected to PCR, and the PCR products were arrayed on nylon membranes (Fig. 1). These clones were then further screened for differential expression by reverse Northern blot analysis, and 661 clones are found to be differentially expressed after NMDA receptor stimulation of rat cortical neurons. The differentially expressed clones were then sequenced and identified and/or functionally characterized by comparison with the GenBank database. Northern blot analysis was used to confirm changes in mRNA expression after NMDA receptor stimulation (Fig. 1).
DNA microarray hybridization was used to measure the temporal changes in mRNA levels of 1,152 genes at five times, ranging from 1 to 24 h after NMDA receptor stimulation (Fig. 2 A and B). The cDNA made from total RNA from each sample was labeled with [33P]dCTP, microarray membrane filters were hybridized, and the expression level of each gene was analyzed and segregated based on its temporal profile of gene expression by using hierarchical cluster analysis into six gene-expression patterns (Fig. 2C). Six hundred sixty-one of the 1,152 arrayed genes are found to be consistently up-regulated between 1 and 24 h after NMDA receptor stimulation consistent with the reverse Northern blot analysis. Because the initial DAzLE screen was focused on identifying NMDA-induced genes, we only focused our analysis on up-regulated genes. Of the genes, ≈5% are induced at 1 h (group 1) and their expression levels return to base line by 12 h (Fig. 2C). These genes probably represent early-response genes. Another group of genes (group 2) are induced at 1 h and remained up-regulated through the entire 24-h period and represent 18% of the up-regulated genes. Group 3 comprises 19% of the up-regulated genes and represents a set of genes whose expression gradually increases from 1 to 6 h and remains elevated for the remainder of the 24-h period. Fourteen percent of the genes (group 4) are up-regulated between 1 and 6 h after NMDA receptor stimulation, and their expression returns to control levels at 12 h and 24% of the genes (group 5) are down-regulated initially and then up-regulated at 12 h (Fig. 2C). Twenty percent of the genes (group 6) have fluctuating patterns of gene expression and are up- or down-regulated at various times after NMDA receptor stimulation (data not shown).
Fig. 2.
Microarray analysis of NMDA-induced PLINGs. (A) Microarray of cherry-picked PLINGs at 1, 3, 6, 12, and 24 h after 5 min of 50 μM NMDA treatment. Fold change in expression levels relative to the zero time point is displayed in red (increased expression) or green (decreased expression). (B) Hierarchical cluster analysis of the temporal gene-expression profile of NMDA-induced PLINGs. (C) NMDA-induced PLINGs were divided into different groups based on their expression profile. Expression curves were constructed for immediate-response genes (⋄, group 1), constant-response genes (▪, group 2), gradual-response genes (▴, group 3), and intermediate-response genes (×, group 4, and ○, group 5).
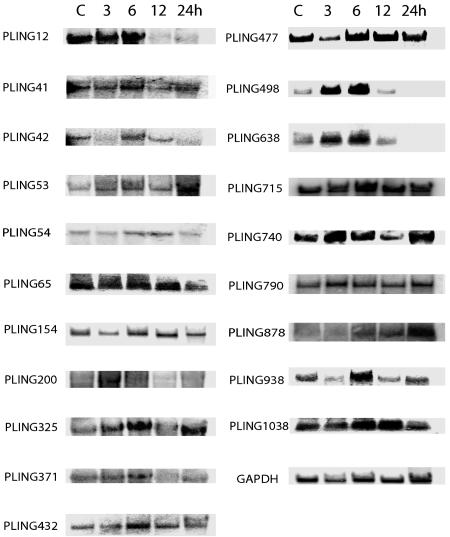
Northern blot analysis was used to further characterize the differentially expressed transcripts. We analyzed the expression pattern of 20 representative genes from the different expression pattern groups and confirm the microarray expression analysis results (Fig. 3). Of the genes selected for Northern blot analysis, ≈90% show remarkably similar expression patterns, as identified by microarray analysis (data not shown).
Fig. 3.
Northern blot analysis of NMDA-induced PLINGs. Selected PLINGs were used to confirm the temporal gene-expression changes after NMDA receptor activation by Northern blot analysis. Identical amounts of total RNA from neurons 6 h after control buffer (CSS) and 3, 6, 12, and 24 h after 5 min of 50 μM NMDA treatment were used for Northern blot analysis. GAPDH was used as control.
The functional breakdown of genes induced by NMDA receptor stimulation is shown in Fig. 4. There is a broad spectrum of functional categories of NMDA-induced genes (see Tables 1 and 2, which are published as supporting information on the PNAS web site, for a full list of genes and categories). DAzLE is extremely sensitive and identifies very rare transcripts; and, consistent with the power of the DAzLE technique, ≈51% of the NMDA-induced genes are genes of unknown function (Fig. 4). Several putative zinc-finger transcription factors are induced as late-response genes after NMDA receptor stimulation and may function in the secondary and tertiary transcriptional responses after NMDA receptor stimulation. Other known transcription factors, such as nuclear factor IA, ETS-related transcription factor ETV5, and upstream stimulatory factor 1, are up-regulated as late-response genes after NMDA receptor stimulation. Several genes involved in DNA synthesis, transcriptional regulation, protein synthesis, and RNA processing and stability are up-regulated after NMDA receptor activation. Many protein kinases and phosphatases are up-regulated after NMDA receptor stimulation. These molecules might be involved in the signaling mechanisms responsible for regulating or inducing LTP, gene expression, and synaptic plasticity.
Fig. 4.
Functional categorization of NMDA-induced PLINGs. PLINGs were divided into functional categories. The number next to each category indicates the percentage of the genes in that class of the total number of genes.
Several mammalian homologs of human, Drosophila, and Caenorhabditis elegans genes, such as TP53BP1, hucep-11, DnaJ, pp21, FLJ20279, KIAA0261, ZNF286, MGC19556, AL133206, ENL, RNA polymerase II, EIF4G1, nexin 17, Cbx3, Su(fu), peanut-like 2 homolog, CG1874-PA, C42C1.9, Fox-1, are up-regulated after NMDA receptor stimulation. TP53BP1 is known to play an important role in genomic stability as a transducer of the DNA damage signal to p53 and other tumor suppressor proteins (20). It is clear that NMDA receptor activation can induce DNA damage in neurons (21). The induction of TP53BP1 may be a protective mechanism against genomic damage after NMDA receptor activation. Peanut-like 2 homolog (H5/PNUTL2/CDCrel2b) is known to be required for cytokinesis in eukaryotic cells (22), and its splice variant is localized to mitochondria and has a pro-apoptotic function (23). The other homologs such as the Suppressor of Fused [Su(fu)] protein, the heterochromatin protein (Cbx3), and Fox-1 may regulate mRNA transcription after NMDA receptor activation (24-26).
Several genes that are involved in cellular oxidation-reduction, energy and cellular metabolism, and cell survival are up-regulated after NMDA receptor stimulation. These gene products include mitochondrial ATP synthase, creatine kinase, and adenylate kinase, key enzymes in the energy-generating system of the brain. We also observe increases in lactate dehydrogenase and phosphoglycerate kinase, genes that are involved in glycolysis. These gene products may play a role in cellular recovery after NMDA-induced ATP and creatine phosphate depletion (27, 28). Many mitochondrial genes, several cellular membrane ion transporters, mitochondrial transporters, and channels are also induced after NMDA receptor stimulation. Genes involved in maintaining intracellular calcium homeostasis, such as hippocalcin and lipocortin 1, are also increased after NMDA stimulation.
One of the largest functional groups of up-regulated genes after NMDA treatment are a group of genes involved in intracellular signaling. Many of the signaling proteins induced by NMDA are genes involved in intracellular structure and vesicular trafficking. A broad number of genes with putative cytoskeletal function are induced after NMDA receptor activation and these might be involved in structural reorganization of neurons following synaptic plasticity. Unexpectedly, many proteins involved in protein degradation, ubiquitination and protein turnover are induced following NMDA receptor stimulation. These proteins may protect neurons from oxidative stress following NMDA receptor stimulation, but they may also be involved in remodeling synapses and receptor structure following synaptic plasticity (29). The synaptic remodeling mediated by proteases in neurons might be initiated by calpain that is activated after NMDA receptor activation as an early response (30) and the changes in gene transcription of the proteases possibly leads to synaptic remodeling as a late response.
To further characterize the NMDA receptor induced genes identified by DAzLE, we monitored the expression pattern of the DAzLE identified gene sets in vivo by using the MECS paradigm (31). MECS produces extremely robust and long-lived potentiation of synaptic contacts in the hippocampus and blocks spatial learning. Furthermore, it induces many of the same genes as long-term potentiation. Thus, MECS is considered a model of long-term plasticity relevant to learning and memory. MECS was induced in male Sprague-Dawley rats and animals were euthanized at 6, 24, 48, and 72 h after MECS. Up-regulated genes following MECS are provided in Table 3, and the functional categories of genes are illustrated in Fig. 6, which is published as supporting information on the PNAS web site. Genes were selected as up-regulated if their gene expression was induced at least three times during the different MECS experiments. A total of 418 genes are induced after MECS and 161 genes are induced by both NMDA receptor stimulation of cortical culture and MECS in rat cortex (Fig. 5). The functional categories of genes induced by both NMDA receptor stimulation and MECS indicates the similarity between the two experimental paradigms (Fig. 5). Many of the genes that are induced by both NMDA receptor stimulation and MECS are possibly involved in long-term changes in synaptic plasticity.
Fig. 5.
MECS-induced PLINGs. MECSs were induced in male Sprague-Dawley rats, and total RNA was collected from rat cortex after 24 h. The z-ratios of PLINGs were compared between MECS- and NMDA-treated groups. The numbers on top of each bar indicate the z-ratio of each NMDA-induced gene. The genes with unknown function represent 60% of genes induced by both NMDA receptor stimulation and MECS, and four of the genes are shown.
Discussion
We have used DAzLE coupled with microarray analysis to identify the late-response PLINGs that are induced by NMDA receptor stimulation of primary cortical neurons. This method combines a novel and extremely sensitive screening method of differentially expressed genes with custom microarray analysis (41). This method has advantages over conventional microarray analysis and differential hybridization methodologies (16, 32). DAzLE selects for abundant and very rare transcripts that are differentially expressed. The preselection of differentially expressed genes (i.e., cherry picking) followed by microarray analysis provides a powerful method for analysis of a large number of dynamically regulated genes. Although a number of relatively complete genomic sequences are available for a variety of organisms, these databases still suffer from gene identification and annotation deficiencies that have hindered comprehensive identification of mRNA (33). Furthermore, extensive microarray analysis would be required to identify a similar set of differentially expressed genes that are identified by DAzLE. DAzLE coupled with microarray analysis has allowed us to identify a large set of late-response PLINGs that may be important in neuronal function, survival and plasticity of the nervous system. DAzLE is a sensitive technique that allowed us to identify a large number of previously unknown genes. This technique can be applied to a variety of experiments that are designed to investigate the transcriptional response of a variety of genes in different cells and tissues. DAzLE with microarray can also be readily applied to different experimental designs such as pharmacological inhibition of different signaling cascades including receptor agonists or antagonists to investigate the molecular mechanisms of cellular responses. For instance, we recently identified genes that are regulated by mitogen-activated protein kinase- and NO-dependent signaling pathways using DAzLE (S.-J.H., T.M.D., and V.L.D., unpublished observations).
A number of previously characterized PLINGs were identified in our screen and include, TIS11, ARPP-21, neuronatin, clathrin, HSC70, NF-l, PAI, HMG, GFAP, vasopressin, mitogen-activated protein kinase, nexin 1, prosaposin, androgen-binding protein, and mitochondrial cytochrome c oxidase (34, 35). Because most prior screens for PLINGs have mainly focused on the identification of immediate early-response genes (35-37), our gene set has minimal overlap with previously identified PLINGs. In particular, our screen was designed to identify genes that are induced 6 h after NMDA receptor stimulation and, accordingly, we failed to identify many previously characterized immediate-early genes whose expression returns to baseline before 6 h (38, 39). Thus, our gene set represents a heretofore-uncharacterized picture of the response of neurons to NMDA receptor stimulation.
The results we have gathered from this study show that the late response of NMDA receptor activation leads to changes in many functional groups of genes. Many genes are dynamically regulated in response to brief episodes of NMDA receptor stimulation. A diverse expression profile of late-response genes exists after NMDA receptor stimulation. This temporal gene expression probably links up the NMDA receptor activation with long-term changes in neuronal function. The set of NMDA-induced genes presented here gives us information about synaptic plasticity and suggests many possibilities for future experimental work. Discovered were >150 previously unknown genes, and the expression of these unknown genes is regulated in specific temporal patterns during the response of neurons to NMDA receptor activation. Although the molecular machinery of synaptic plasticity and neuronal survival is well studied, it is possible that some of the genes of unknown function that showed dramatic up-regulation after NMDA receptor stimulation may yet emerge as regulators of these processes. The induction of several transcription factors, including both uncharacterized genes and genes like NFIA, DDX3, upstream stimulatory factor 1, and ETV5, could play important roles in long-term changes in neuronal function. Consistent with this notion, upstream stimulatory factor 1 was recently shown to be required for calcium-dependent transcription for the brain-derived neurotrophic factor promoter (40). Analysis of promoter sequences of NMDA-induced genes in conjunction with knowledge of the binding site of transcription factors regulated by NMDA receptor stimulation may help illuminate the molecular mechanisms of transcriptional regulation that mediate NMDA-induced gene expression.
A large number of genes were identified that were induced both by NMDA receptor stimulation and MECS. Many of these genes may be important in inducing and maintaining long-term changes in neuronal function that underlie synaptic plasticity. A variety of cytoskeletal and microtubule-related genes are induced and may play a role in the establishment of synaptic connections. Furthermore, several ion pumps, transporters, and intracellular signaling molecules were identified that may also play important roles in the neurons adaptation to its cellular environment.
The list of NMDA-induced genes outlined here presents not only a set of candidate PLINGs, but it also represents possible neuronal survival genes. We expect our NMDA gene set to be of particular use in identifying genes that are involved in neuronal plasticity and in neuronal survival. Functional follow-up of all these NMDA-regulated genes, however, awaits further studies, but gene-expression profiles such as the data presented here provide the framework for investigating genes that are important in these processes.
Supplementary Material
Acknowledgments
We thank Weza Cotman for manuscript preparation. This work was supported by U.S. Public Health Service Grants NS 37090, NS 40809, and DA 00266 and the National Alliance for Research on Schizophrenia and Depression Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NMDA, N-methyl-D-aspartate; PLING, plasticity-induced gene; MECS, maximal electroconvulsive seizure; DAzLE, differential analysis of primary cDNA library expression; CSS, control salt solution.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. CB724286-CB725070)
References
- 1.Cohen-Cory, S. (2002) Science 298, 770-776. [DOI] [PubMed] [Google Scholar]
- 2.Dolmetsch, R. E., Pajvani, U., Fife, K., Spotts, J. M. & Greenberg, M. E. (2001) Science 294, 333-339. [DOI] [PubMed] [Google Scholar]
- 3.Kornhauser, J. M., Cowan, C. W., Shaywitz, A. J., Dolmetsch, R. E., Griffith, E. C., Hu, L. S., Haddad, C., Xia, Z. & Greenberg, M. E. (2002) Neuron 34, 221-233. [DOI] [PubMed] [Google Scholar]
- 4.Steward, O. & Schuman, E. M. (2001) Annu. Rev. Neurosci. 24, 299-325. [DOI] [PubMed] [Google Scholar]
- 5.Nedivi, E., Hevroni, D., Naot, D., Israeli, D. & Citri, Y. (1993) Nature 363, 718-722. [DOI] [PubMed] [Google Scholar]
- 6.Lyford, G. L., Yamagata, K., Kaufmann, W. E., Barnes, C. A., Sanders, L. K., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Lanahan, A. A. & Worley, P. F. (1995) Neuron 14, 433-445. [DOI] [PubMed] [Google Scholar]
- 7.Brakeman, P. R., Lanahan, A. A., O'Brien, R., Roche, K., Barnes, C. A., Huganir, R. L. & Worley, P. F. (1997) Nature 386, 284-288. [DOI] [PubMed] [Google Scholar]
- 8.Qian, Z., Gilbert, M. E., Colicos, M. A., Kandel, E. R. & Kuhl, D. (1993) Nature 361, 453-457. [DOI] [PubMed] [Google Scholar]
- 9.Gnegy, M. E. (2000) Crit. Rev. Neurobiol. 14, 91-129. [PubMed] [Google Scholar]
- 10.Nishiyama, M., Hong, K., Mikoshiba, K., Poo, M. M. & Kato, K. (2000) Nature 408, 584-588. [DOI] [PubMed] [Google Scholar]
- 11.Skaper, S. D., Facci, L. & Strijbos, P. J. (2001) Ann. N.Y. Acad. Sci. 939, 11-22. [DOI] [PubMed] [Google Scholar]
- 12.Montarolo, P. G., Goelet, P., Castellucci, V. F., Morgan, J., Kandel, E. R. & Schacher, S. (1986) Science 234, 1249-1254. [DOI] [PubMed] [Google Scholar]
- 13.Husi, H., Ward, M. A., Choudhary, J. S., Blackstock, W. P. & Grant, S. G. (2000) Nat. Neurosci. 3, 661-669. [DOI] [PubMed] [Google Scholar]
- 14.Heynen, A. J., Quinlan, E. M., Bae, D. C. & Bear, M. F. (2000) Neuron 28, 527-536. [DOI] [PubMed] [Google Scholar]
- 15.Cull-Candy, S., Brickley, S. & Farrant, M. (2001) Curr. Opin. Neurobiol. 11, 327-335. [DOI] [PubMed] [Google Scholar]
- 16.Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. (1995) Science 270, 467-470. [DOI] [PubMed] [Google Scholar]
- 17.Dawson, V. L., Dawson, T. M., Bartley, D. A., Uhl, G. R. & Snyder, S. H. (1993) J. Neurosci. 13, 2651-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vawter, M. P., Barrett, T., Cheadle, C., Sokolov, B. P., Wood, W. H., III, Donovan, D. M., Webster, M., Freed, W. J. & Becker, K. G. (2001) Brain Res. Bull. 55, 641-650. [DOI] [PubMed] [Google Scholar]
- 19.Cheadle, C., Vawter, M. P., Freed, W. J. & Becker, K. G. (2003) J. Mol. Diagn. 5, 73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, B., Matsuoka, S., Carpenter, P. B. & Elledge, S. J. (2002) Science 298, 1435-1438. [DOI] [PubMed] [Google Scholar]
- 21.Yu, S. W., Wang, H., Poitras, M. F., Coombs, C., Bowers, W. J., Federoff, H. J., Poirier, G. G., Dawson, T. M. & Dawson, V. L. (2002) Science 297, 259-263. [DOI] [PubMed] [Google Scholar]
- 22.Field, C. M. & Kellogg, D. (1999) Trends Cell Biol. 9, 387-394. [DOI] [PubMed] [Google Scholar]
- 23.Larisch, S., Yi, Y., Lotan, R., Kerner, H., Eimerl, S., Tony Parks, W., Gottfried, Y., Birkey Reffey, S., de Caestecker, M. P., Danielpour, D., et al. (2000) Nat. Cell Biol. 2, 915-921. [DOI] [PubMed] [Google Scholar]
- 24.Cheng, S. Y. & Bishop, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 5442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheutin, T., McNairn, A. J., Jenuwein, T., Gilbert, D. M., Singh, P. B. & Misteli, T. (2003) Science 299, 721-725. [DOI] [PubMed] [Google Scholar]
- 26.Jin, Y., Suzuki, H., Maegawa, S., Endo, H., Sugano, S., Hashimoto, K., Yasuda, K. & Inoue, K. (2003) EMBO J. 22, 905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pieper, A. A., Blackshaw, S., Clements, E. E., Brat, D. J., Krug, D. K., White, A. J., Pinto-Garcia, P., Favit, A., Conover, J. R., Snyder, S. H. & Verma, A. (2000) Proc. Natl. Acad. Sci. USA 97, 1845-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eliasson, M. J., Sampei, K., Mandir, A. S., Hurn, P. D., Traystman, R. J., Bao, J., Pieper, A., Wang, Z. Q., Dawson, T. M., Snyder, S. H. & Dawson, V. L. (1997) Nat. Med. 3, 1089-1095. [DOI] [PubMed] [Google Scholar]
- 29.Ehlers, M. D. (2003) Nat. Neurosci. 6, 231-242. [DOI] [PubMed] [Google Scholar]
- 30.Lynch, G. (1998) Neurobiol. Learn. Mem. 70, 82-100. [DOI] [PubMed] [Google Scholar]
- 31.Cole, A. J., Saffen, D. W., Baraban, J. M. & Worley, P. F. (1989) Nature 340, 474-476. [DOI] [PubMed] [Google Scholar]
- 32.Watson, J. B. & Margulies, J. E. (1993) Dev. Neurosci. 15, 77-86. [DOI] [PubMed] [Google Scholar]
- 33.Jenssen, T. K., Laegreid, A., Komorowski, J. & Hovig, E. (2001) Nat. Genet. 28, 21-28. [DOI] [PubMed] [Google Scholar]
- 34.Zagulska-Szymczak, S., Filipkowski, R. K. & Kaczmarek, L. (2001) Neurochem. Int. 38, 485-501. [DOI] [PubMed] [Google Scholar]
- 35.Hevroni, D., Rattner, A., Bundman, M., Lederfein, D., Gabarah, A., Mangelus, M., Silverman, M. A., Kedar, H., Naor, C., Kornuc, M., et al. (1998) J. Mol. Neurosci. 10, 75-98. [DOI] [PubMed] [Google Scholar]
- 36.Matsuo, R., Murayama, A., Saitoh, Y., Sakaki, Y. & Inokuchi, K. (2000) J. Neurochem. 74, 2239-2249. [DOI] [PubMed] [Google Scholar]
- 37.Lanahan, A. & Worley, P. (1998) Neurobiol. Learn. Mem. 70, 37-43. [DOI] [PubMed] [Google Scholar]
- 38.Platenik, J., Kuramoto, N. & Yoneda, Y. (2000) Life Sci. 67, 335-364. [DOI] [PubMed] [Google Scholar]
- 39.Dragunow, M. (1996) Behav. Genet. 26, 293-299. [DOI] [PubMed] [Google Scholar]
- 40.Chen, W. G., West, A. E., Tao, X., Corfas, G., Szentirmay, M. N., Sawadogo, M., Vinson, C. & Greenberg, M. E. (2003) J. Neurosci. 23, 2572-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, H., Gu, X., Dawson, V. L. & Dawson, T. M. (2004) Proc. Natl. Acad. Sci. USA 101, 647-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.