Abstract
In mammals, odor detection in the nose is mediated by a diverse family of olfactory receptors (ORs), which are used combinatorially to detect different odorants and encode their identities. The OR family can be divided into subfamilies whose members are highly related and are likely to recognize structurally related odorants. To gain further insight into the mechanisms underlying odor detection, we analyzed the mouse OR gene family. Exhaustive searches of a mouse genome database identified 913 intact OR genes and 296 OR pseudogenes. These genes were localized to 51 different loci on 17 chromosomes. Sequence comparisons showed that the mouse OR family contains 241 subfamilies. Subfamily sizes vary extensively, suggesting that some classes of odorants may be more easily detected or discriminated than others. Determination of subfamilies that contain ORs with identified ligands allowed tentative functional predictions for 19 subfamilies. Analysis of the chromosomal locations of members of each subfamily showed that many OR gene loci encode only one or a few subfamilies. Furthermore, most subfamilies are encoded by a single locus, suggesting that different loci may encode receptors for different types of odorant structural features. Comparison of human and mouse OR subfamilies showed that the two species have many, but not all, subfamilies in common. However, mouse subfamilies are usually larger than their human counterparts. This finding suggests that humans and mice recognize many of the same odorant structural motifs, but mice may be superior in odor sensitivity and discrimination.
Volatile odorants are detected by a large family of olfactory (odorant) receptors (ORs), which are found on olfactory sensory neurons lining the nasal cavity (1-3). Related families of ORs have been found in numerous vertebrate species, but their size ranges from an estimated 100 receptors in fish (4) to ≈1,000 in mice (5, 6). ORs are members of the seven-transmembrane domain, G protein-coupled receptor superfamily, but they share sequence motifs not found in other superfamily members (1). Aside from these motifs, ORs are exceptionally diverse in protein sequence. The immense size of the OR family and its diversity are consistent with an ability to detect a vast array of odorants with varied structures.
ORs are used in a combinatorial manner to detect odorants and encode their identities. Individual ORs can recognize multiple odorants (7-16) and single odorants are detected by multiple ORs (9, 12), but, importantly, different odorants are detected, and thus encoded, by different combinations of ORs (9). If each odorant were encoded by only three ORs, this combinatorial scheme could generate nearly one billion different odor codes. Because odorants with nearly identical structures can be recognized by partially overlapping sets of ORs, the odorants have different receptor codes and can be distinguished (9).
The OR family can be divided into subfamilies on the basis of sequence relationships (1). Current evidence suggests that members of the same subfamily recognize the same type of odorant structure, or structural motif, but that different members of the same subfamily may recognize different variants of that structure (9, 12). This characteristic of the OR family, like the combinatorial use of ORs, is likely to be important in the discrimination of closely related odorants. Interestingly, OR genes are found at many chromosomal locations, but highly related OR genes are often found in the same region (5, 6, 17-21). This finding raises the possibility that different loci might encode different OR subfamilies and thereby be involved in the detection of different types of odorants.
Studies of the mouse genome have provided considerable information on the mouse OR family (5, 6). However, its subfamily structure and the chromosomal distribution of different subfamilies are still unknown. In addition, whereas close relatives of many human ORs can be found in mice (5, 6), relationships between human and mouse subfamilies remain to be defined.
To investigate these issues, we analyzed the mouse OR family and compared it with that of human. Here, we describe the composition of the mouse OR gene family, the chromosomal position of each OR gene, the subfamily structure of the mouse OR family, the chromosomal organization of individual OR subfamilies, and a comparison of OR subfamilies in human and mouse.
Methods
Database Searches. To identify mouse OR genes, we used tblastn to search for genes encoding ORs in the Celera mouse genome database (www.celeradiscoverysystem.com). As queries in these searches, we used the region stretching from the end of transmembrane domain 3 (TM3) to the beginning of TM6 (TM3-TM6) in 10 divergent mouse ORs, as well as amino acid motifs commonly found in mammalian ORs (MAYDRYVAIC, MALDRYVAIC, MAFDRYVAIC, and KAFSTCASH). The TM3-TM6 DNA sequences identified in these searches were translated by using orf finder [National Center for Biotechnology Information (NCBI)]. The translated sequences were aligned by using clustal w 1.83, and a new divergent subset of ORs was chosen for additional searches of the database. Multiple searches were performed until no new sequences were obtained. Over 6,000 sequences were analyzed from which a set of 1,567 unique TM3-TM6 sequences was identified.
When an assembled version of the database became available, it was searched with representatives of the 1,567 partial sequences. DNA in the region of each match was then translated by using orf finder. Encoded proteins were classified as ORs if they contained four motifs (or variants thereof) common to ORs in specific locations (GN; MAYDRYVAIC, KAFSTCASH, and PMLNPFIY). Proteins that only partially satisfied these criteria were used as queries in blastp searches of the NCBI protein database, and only those whose closest matches were ORs were judged to be ORs. Sequences encoding highly pseudogenized OR genes and OR gene fragments were discarded.
The chromosomal locations of ORs were established from their positions within Celera's annotated scaffold assemblies. Adjacent OR genes were assigned to different loci if their coding regions were >1 Mb apart (19). This analysis also allowed us to exclude allelic variants of OR genes and to identify with certainty a nonredundant set of OR genes (as of September 2002).
Phylogenetic Analysis. OR sequence alignments were used to generate consensus neighbor-joining phylogenetic trees (clustal w 1.83) after 1,000 rounds of bootstrapping. Nucleotide and protein sequence identities were determined by using the DISTANCES function of the Genetics Computer Group (wisconsin package, GCG). The uncorrected distance matrix was then used to assign sequences to subfamilies. Each member of the subfamily showed 60% or greater sequence identity to all other members of the same subfamily and displayed a strong phylogenetic grouping (clade) as judged from bootstrap values that were generally >50%.
Results
Composition of the Mouse OR Gene Family. In initial studies, we searched the Celera mouse genome database for genes encoding proteins related to known ORs (see Methods). After exhaustive searches of the database, we aligned OR coding regions and excluded replica sequences. By examining the exact contig positions of genes that were ≥99% identical, we also excluded likely allelic variants.
These experiments identified 1,209 mouse OR genes. Of these, 913 genes (76%) have uninterrupted open reading frames and are therefore able to encode functional ORs. The remaining 296 genes (24%) are pseudogenes that have frame shifts, stop codons, insertions, or deletions in the coding region. Sequence identity among the intact OR coding regions is 34-99%, emphasizing the extreme diversity of the OR gene family.
To assess the efficacy of our search strategy, we asked whether the ORs we had identified included 53 mouse OR sequences in the NCBI database that were not derived from the Celera database. We found 52 of 53 sequences in our set, suggesting that our search was highly effective.
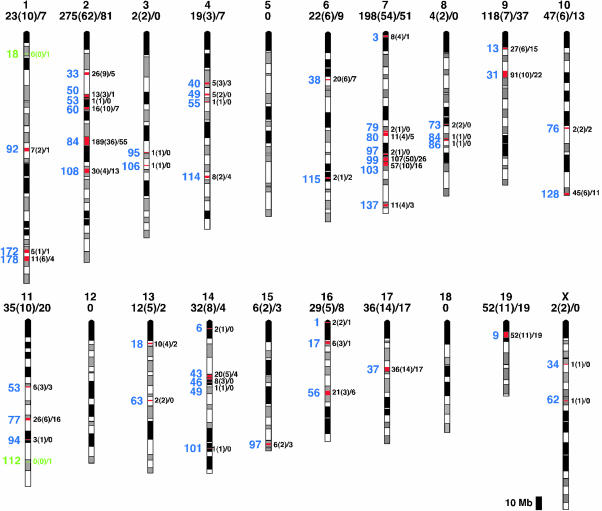
Chromosomal Organization of Mouse OR Genes. We next determined the chromosomal locations of the mouse OR genes (Fig. 1 and Table 1). We used the criterion that adjacent OR genes are at a different locus if their coding regions are >1 Mb apart (19). We were able to map the locations of 1,190 of the 1,209 OR genes. These studies defined 51 different chromosomal loci distributed over 17 mouse chromosomes. OR genes were assigned to all mouse chromosomes except chromosomes 5, 12, and 18, and the Y chromosome. We defined the position of each OR gene locus in terms of its distance in Mb from the centromeric end of the chromosome.
Fig. 1.
Chromosome locations of mouse OR genes. Mouse OR genes were mapped to 51 loci on 17 chromosomes. OR loci are indicated in red, except for those with no intact OR genes, which are shown in green. The position of each locus (blue) is shown on the left in Mb from the centromere (top). The number of intact OR genes and OR pseudogenes and the number of subfamilies encoded are indicated on the right for each locus and above for each chromosome [intact genes (subfamilies)/pseudogenes].
Table 1. Composition of mouse OR gene loci.
| Locus* | No. of intact genes | No. of pseudogenes | % pseudogenes | No. of subfamilies |
|---|---|---|---|---|
| 1-18 | 0 | 1 | 100 | 0 |
| 1-92 | 7 | 1 | 13 | 2 |
| 1-172 | 5 | 1 | 17 | 1 |
| 1-178 | 11 | 4 | 27 | 6 |
| 2-33 | 26 | 5 | 16 | 9 |
| 2-50 | 13 | 1 | 7 | 3 |
| 2-53 | 1 | 0 | 0 | 1 |
| 2-60 | 16 | 7 | 30 | 10 |
| 2-84 | 189 | 55 | 23 | 36 |
| 2-108 | 30 | 13 | 30 | 4 |
| 3-95 | 1 | 0 | 0 | 1 |
| 3-106 | 1 | 0 | 0 | 1 |
| 4-40 | 5 | 3 | 38 | 3 |
| 4-49 | 5 | 0 | 0 | 2 |
| 4-55 | 1 | 0 | 0 | 1 |
| 4-114 | 8 | 4 | 33 | 2 |
| 6-38 | 20 | 7 | 26 | 6 |
| 6-115 | 2 | 2 | 50 | 1 |
| 7-3 | 8 | 1 | 11 | 4 |
| 7-79 | 2 | 0 | 0 | 1 |
| 7-80 | 11 | 5 | 31 | 4 |
| 7-97 | 2 | 0 | 0 | 1 |
| 7-99 | 107 | 26 | 20 | 50 |
| 7-103 | 57 | 16 | 22 | 10 |
| 7-137 | 11 | 3 | 21 | 4 |
| 8-73 | 2 | 0 | 0 | 2 |
| 8-84 | 1 | 0 | 0 | 1 |
| 8-86 | 1 | 0 | 0 | 1 |
| 9-13 | 27 | 15 | 36 | 6 |
| 9-31 | 91 | 22 | 19 | 10 |
| 10-76 | 2 | 2 | 50 | 2 |
| 10-128 | 45 | 11 | 20 | 6 |
| 11-53 | 6 | 3 | 33 | 3 |
| 11-77 | 26 | 16 | 38 | 6 |
| 11-94 | 3 | 0 | 0 | 1 |
| 11-112 | 0 | 1 | 100 | 0 |
| 13-18 | 10 | 2 | 17 | 4 |
| 13-63 | 2 | 0 | 0 | 2 |
| 14-6 | 2 | 0 | 0 | 1 |
| 14-43 | 20 | 4 | 17 | 5 |
| 14-46 | 8 | 0 | 0 | 3 |
| 14-49 | 1 | 0 | 0 | 1 |
| 14-101 | 1 | 0 | 0 | 1 |
| 15-97 | 6 | 3 | 33 | 2 |
| 16-1 | 2 | 1 | 33 | 2 |
| 16-17 | 6 | 1 | 14 | 3 |
| 16-56 | 21 | 6 | 22 | 3 |
| 17-37 | 36 | 17 | 32 | 14 |
| 19-9 | 52 | 19 | 27 | 11 |
| X-34 | 1 | 0 | 0 | 1 |
| X-62 | 1 | 0 | 0 | 1 |
Locus is shown as chromosome-distance of locus in Mb from centromere.
We found extensive variation in the number of OR genes at individual OR loci (1-244 genes) as well as on different chromosomes (2-356 OR genes)(Table 1 and Fig. 1). The percentage of pseudogenes also varies among loci (7-100%) (Fig. 1 and Table 1). Of 51 OR loci, 2 have only pseudogene(s), indicating that 49 chromosomal loci are capable of encoding one or more ORs. Of these 49 “functional” loci, 10 have only one intact OR gene.
Subfamily Structure of the Mouse OR Family. To gain insight into the functional organization of the mouse OR family, we determined its subfamily structure. The criterion we used for defining subfamilies was that all members of a subfamily are ≥60% identical to all other members in amino acid sequence (22). The 60% identity cutoff was selected on the basis of functional studies of ORs, which indicate that ORs that are 60% or more identical in protein sequence tend to recognize odorants with related structures (refs. 9 and 12 and K. Nara, P.A.G., and L.B.B., unpublished results). For example, mouse ORs S1 and S3, which are 63% identical, recognize n-aliphatic acids with 7-9 carbon atoms and n-aliphatic alcohols with 5-7 carbon atoms, respectively (9).
These studies showed that the mouse OR family is divisible into 241 subfamilies (for the composition and accession numbers of individual subfamilies, see Table 5, which is published as supporting information on the PNAS web site). Ninety-four of the subfamilies contain only a single OR (Table 2). The other 147 subfamilies range in size from 2 to 34 members. Although most subfamilies have 1-5 members, 23 subfamilies contain >10 ORs. The differing sizes of the subfamilies could conceivably result in variation in the ability to detect or discriminate different types of odorants.
Table 2. Sizes of OR subfamilies.
| Number of ORs | Number of subfamilies* |
|---|---|
| 1 | 94 |
| 2 | 40 |
| 3 | 32 |
| 4 | 20 |
| 5 | 12 |
| 6 | 8 |
| 7 | 6 |
| 8 | 3 |
| 9 | 3 |
| 10-15 | 15 |
| 16-34 | 8 |
Number of subfamilies that contain the number of ORs on left.
Prediction of Subfamily Functions. The subfamily structure of the mouse OR family provides a template for assigning potential functions to groups of ORs. For example, mouse OR73 and OR74 belong to the same subfamily, and both recognize aromatic aldehydes (12). One may therefore predict that this subfamily, which includes five ORs, detects aromatic aldehydes. In an initial attempt to gain insight into the functional organization of the mouse OR family, we examined OR subfamilies that contain the 22 mouse ORs for which odor ligands have been identified (7-10, 12-15).
This analysis showed that the 22 mouse ORs with identified ligands belong to a total of 19 different subfamilies, which altogether contain 96 ORs (Table 3). On the basis of this information, hypothetical functional assignments can be made for these subfamilies. The tentative functional assignments do not predict the precise odorants recognized, but rather general classes of odorant structures that may be recognized. Of the subfamilies given assignments in this manner, 13 subfamilies (59 ORs) are predicted to recognize aliphatic odorants and the remaining 6 subfamilies (37 ORs) are predicted to recognize odorants with other types of structural motifs.
Table 3. Subfamilies containing ORs with known ligands.
| OR* | Subfamily† | Locus | Ligand | Structure‡ |
|---|---|---|---|---|
| MOR23 (267-13) | 34 (5) | 1-172 | Lyral | 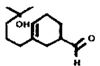 |
| ID3 (136-6) | 151 (14) | 2-33 | (±)-Carvone | 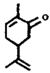 |
| IG7 (276-1) | 66 (3) | 2-50 | (±)-Limonene | |
| OR73 (174-9) | 78 (5) | 2-84 | Eugenol | 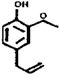 |
| OR74 (174-4) | 78 (5) | 2-84 | Ethyl vanillin | 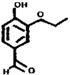 |
| OR912-93 (175-1) | 108 (5) | 2-84 | n-Aliphatic ketones | |
| S25 (204-32) | 118 (10) | 7-103 | n-Aliphatic alcohols | |
| S46 (32-4) | 208 (6) | 7-99 | n-Aliphatic acids | |
| S85 (13-6) | 222 (5) | 7-99 | n-Aliphatic acids | |
| S86 (8-2) | 224 (3) | 7-99 | n-Aliphatic acids | |
| S18 (31-2) | 204 (4) | 7-99 | n-Aliphatic acids/alcohols | |
| S19 (33-1) | 207 (2) | 7-99 | n-Aliphatic acids/alcohols | |
| S41 (22-2) | 203 (4) | 7-99 | n-Aliphatic acids/alcohols | |
| S51 (40-1) | 192 (2) | 7-99 | n-Aliphatic acids/alcohols | |
| S83 (40-4) | 193 (4) | 7-99 | n-Aliphatic acids/alcohols | |
| S6 (42-3) | 241 (3) | 7-99 | n-Aliphatic dicarboxylic acids | |
| S50 (42-1) | 241 (3) | 7-99 | n-Aliphatic dicarboxylic acids | |
| 17 (103-15) | 13 (2) | 7-103 | Heptanal | |
| M71 (171-2) | 111 (9) | 9-31 | Acetophenone | 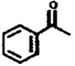 |
| S1 (106-1) | 16 (9) | 14-43 | n-Aliphatic acids | |
| S3 (106-13P) | 16 (9) | 14-43 | n-Aliphatic alcohols | |
| IC6 (118-1) | 4 (1) | 14-49 | (−)-Citronellal |
MOR designations of Zhang and Firestein (5) are in parentheses.
Subfamily designation is followed by number of ORs in subfamily in parentheses. Some ORs here belong to the same subfamilies.
For ORs that detect n-aliphatic acids and alcohols, only one example of each odorant type is shown.
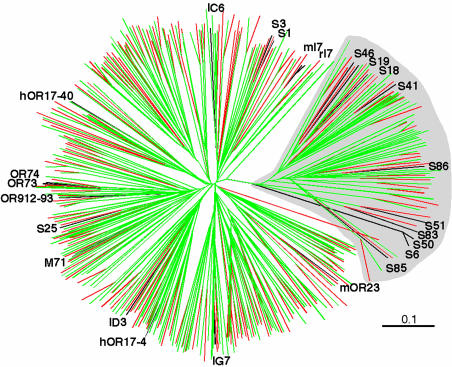
To further explore the functional organization of the mouse OR family, we constructed a phylogenetic tree using one OR sequence from each of the 241 subfamilies (as well as human OR sequences, as will be discussed below). We then examined the positions of ORs with identified ligands in the tree (Fig. 2). Strikingly, 9 of 13 subfamilies that contain receptors for n-aliphatic acids/alcohols are located in one distinct branch of the tree. Interestingly, all 9 of these subfamilies are encoded at a single locus [chromosome 7 (99 Mb)]. This distinctive branch was observed previously (5, 19, 23) and was proposed to contain a divergent set of “fish-like” or “class I” ORs (22, 24, 25). The two other subfamilies that contain ORs for n-aliphatic alcohols and/or acids are found in two other branches of the tree. Subfamilies that contain ORs for other types of odorant structures are scattered among the other branches.
Fig. 2.
Comparison of mouse and human ORs. An unrooted phylogenetic tree showing protein sequence relationships among representatives of all mouse (green) and human (red) OR subfamilies. One member of each subfamily was used (435 sequences) in addition to 25 ORs with known ligands (black). Mouse and human ORs are interspersed throughout the tree, indicating that the OR repertoires of the two species are similar. Many ORs that detect aliphatic odorants (S19-S86) are located in a distinct branch, which is shaded in gray.
OR Gene Loci and OR Subfamilies. Previous studies (5, 6, 17-21) indicate that highly related OR genes are often located at the same chromosomal locus. To investigate how frequently members of the same OR subfamily are encoded by the same locus, we determined the chromosomal locations of genes encoding the members of each of the 241 subfamilies (Table 4; see also Table 5).
Table 4. Chromosomal organization of OR genes and subfamilies.
| Intact OR genes/locus*
|
Subfamilies/locus†
|
Loci/subfamily‡
|
|||
|---|---|---|---|---|---|
| No. of OR genes | No. of loci | No. of subfamilies | No. of loci | No. of loci | No. of subfamilies |
| 0 | 2 | 1 | 16 | 1 | 225 |
| 1-10 | 30 | 2 | 8 | 2 | 15 |
| 11-20 | 7 | 3 | 6 | 3 | 1 |
| 21-30 | 5 | 4 | 5 | ||
| 36 | 1 | 5 | 1 | ||
| 45 | 1 | 6 | 5 | ||
| 52 | 1 | 9 | 1 | ||
| 57 | 1 | 10 | 3 | ||
| 91 | 1 | 11 | 1 | ||
| 107 | 1 | 14 | 1 | ||
| 189 | 1 | 36 | 1 | ||
| 50 | 1 | ||||
No. of OR gene loci with 0-189 intact OR genes.
No. of loci that encode members of 1-50 subfamilies.
No. of subfamilies whose members are encoded at one to three loci.
These studies showed that the vast majority of OR subfamilies are encoded by genes at a single chromosomal locus (Table 4). In addition to the 94 subfamilies with a single member, 131 subfamilies with multiple members are each encoded by genes at one locus. The remaining 7% of OR subfamilies (16/241 subfamilies) are encoded by genes at adjacent loci (5 subfamilies), or by genes that are either on different chromosomes or widely separated on the same chromosome (11 subfamilies). Consistent with these findings are reports stating that the closest relatives of most mouse ORs are encoded at the same locus (5, 6). The clustering of related OR genes emphasizes the importance of local gene duplication and divergence in the evolution of the OR family.
In these studies, we also determined the number of OR subfamilies encoded by each OR gene locus (Table 4). Of 51 OR gene loci, 49 encode at least 1 OR. The number of subfamilies encoded by individual loci ranges from 1 to 50. Of the 49 functional loci, 16 code for members of a single subfamily, 14 code for 2-3 subfamilies, and 19 code for 4-50 subfamilies. Thus, 61% of loci encode only one or a few subfamilies each. This finding is consistent with the idea that different OR loci may be involved in the detection of different types of odorants.
Comparison of Mouse and Human OR Subfamilies. These studies identified a total of 913 mouse ORs and 241 mouse OR subfamilies. In similar studies of the human OR family, we identified 339 ORs and 172 OR subfamilies (26). Thus, mice have ≈2.7 times as many ORs as humans, and 1.4 times as many OR subfamilies.
To investigate the extent to which the mouse and human OR repertoires resemble one another, we compared human and mouse OR subfamilies. We first constructed a phylogenetic tree using one member of each human and mouse subfamily (Fig. 2). Human and mouse ORs are interspersed throughout the tree. Of 24 major branches in the tree, 21 branches contain both human and mouse ORs, suggesting that the human and mouse OR repertoires are similar.
We next asked whether humans and mice have the same OR subfamilies. We first aligned all 339 human ORs and 913 mouse ORs, and then examined the percent protein sequence identities among all of the ORs. After analyzing all pairwise comparisons of ORs, interspecies matches of ≥60% were used to assign human and mouse ORs to corresponding subfamilies. In all but five cases, all members of the corresponding subfamilies were ≥60% identical. Corresponding subfamilies were generally encoded by syntenic chromosomal regions in the two species.
These studies showed that the majority of subfamilies found in human or mouse are common to both species. Of 241 mouse subfamilies, 157 (65%) are also present in human and of 172 human subfamilies, 150 (87%) are found in mouse. This result is consistent with previous reports (5, 6) that many human ORs have close relatives in the mouse. In most cases, there is a one-to-one correspondence between a human subfamily and a mouse subfamily, but, in several instances, an OR subfamily in one species corresponds to two or three subfamilies in the other species.
Although the human and mouse OR repertoires are similar, the number of subfamilies unique to one species is larger for mouse than human. Whereas 22 subfamilies containing 29 ORs are found only in human, 84 subfamilies with a total of 177 ORs are present only in mouse. These “species-specific” subfamilies represent 13% of human subfamilies and 35% of mouse subfamilies. Differences in the complement of OR subfamilies in different species could result in interspecies differences in the detection of particular odorants.
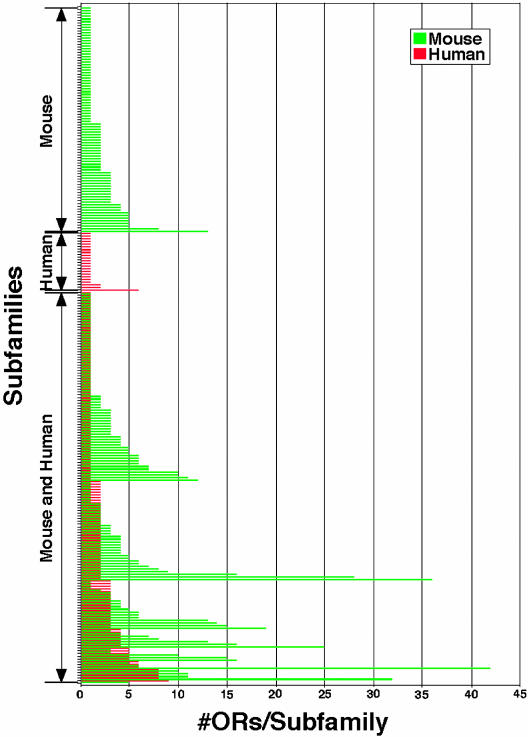
We next compared the sizes of the mouse and human subfamilies (Fig. 3). Of 150 human OR subfamilies also found in mouse (including some corresponding to more than one mouse subfamily), 52 subfamilies (35%) contain the same number of ORs in the two species. Most of these subfamilies (37 subfamilies) have only one member. However, many mouse subfamilies are larger than their human counterparts. Among 150 shared subfamilies, 81 (54%) have more members in mouse than in human. Some of these subfamilies are relatively large in both species (e.g., 8 receptors in human vs. 11 receptors in mouse) whereas others are quite different in size (e.g., 4 receptors in human vs. 25 receptors in mouse). In contrast, only 17 (11%) of the shared subfamilies are larger in human. Thus, although human and mouse share many OR subfamilies, most of these subfamilies have more members in mouse than human.
Fig. 3.
Sizes of OR subfamilies in human vs. mouse. Each human (red) and mouse (green) subfamily is indicated by a horizontal bar. The number of ORs per subfamily is shown on the x axis. The 150 subfamilies shared by the two species are indicated by pairs of red and green bars. Most of the shared subfamilies have more members in mouse than in human. Subfamilies unique to mouse or human, or found in both species, are indicated on the left.
Discussion
In these studies, we analyzed the composition of the mouse OR gene family and its chromosomal distribution. We then determined the subfamily structure of the mouse OR repertoire and the chromosomal organization of genes encoding individual subfamilies. Using these data, together with information on ORs with known odor ligands, we investigated potential links between the detection of different types of odorants, OR subfamilies, and OR gene loci. Finally, we conducted a detailed comparison of OR subfamilies in human and mouse.
These studies indicate that the mouse OR family is composed of ≈1,209 OR genes, 913 of which are likely to encode functional receptors in the nose. Earlier searches of the same database (5, 6) reported fewer intact mouse OR genes; this difference is probably due to our use of a later, more complete version of the database.
These studies showed that mouse OR genes can be found at 51 loci on 17 different chromosomes. Each locus was defined in terms of its megabase coordinates as well as its complement of intact OR genes and pseudogenes. Of 51 OR gene loci identified, 49 contain at least 1 intact OR gene and are therefore likely to be involved in odor detection. These results extend findings of two previous studies, one (5) that defined 27 mouse OR gene clusters with ≥5 OR genes, and another (6) that found 46 mouse OR gene loci but did not provide their exact locations.
The present studies indicate that the mouse OR family is composed of 241 subfamilies. Members of each subfamily are at least 60% identical in protein sequence. Previous studies (7-16) indicate that a single OR can detect multiple odorants that have related structures. In addition, ORs that are 60% or more identical in protein sequence can recognize structurally related odorants (refs. 9 and 12 and K. Nara, P.A.G., and L.B.B., unpublished results). This finding suggests that members of the same subfamily may recognize odorants that share a particular type of odorant structural feature and/or belong to the same class of odorants. Examples of different classes of odorants might be those classified as n-aliphatic odorants with straight carbon chains or odorants classified as aromatic aldehydes and contain both a benzene ring and an aldehyde group. Whereas members of the same subfamily might detect odorants of the same class, they might recognize different variations on the same structural theme. For example, two mouse ORs that belong to the same subfamily were previously found to both recognize n-aliphatic odorants, but one detected n-aliphatic alcohols with 5-7 carbons whereas the other recognized n-aliphatic acids with 7-9 carbons (9).
If this view of odorant recognition by OR subfamilies is generally applicable, the identification of an odorant for one member of a subfamily should allow the tentative assignment of function to the entire subfamily, if only that the subfamily recognizes odorants that are in some (perhaps unknown) way related to the known ligand. If functions are known for two subfamily members, it may be possible to hone one's understanding to a particular odorant structural motif. As a first step in this direction, the present studies predict types of odorant structures that might be detected by 19 mouse subfamilies that contain a total of 96 ORs. However, given the current paucity of information on OR ligand specificities, it cannot yet be excluded that a subfamily (or even a single OR) might, in some cases, interact with different types of odorant structures.
These studies revealed that the vast majority of mouse subfamilies are encoded by genes at a single chromosomal locus. Moreover, many OR gene loci code for members of only one or a few subfamilies. These results are similar to those we obtained in complementary studies of the human OR family (26). These findings suggest that many OR loci may be involved in the recognition of a restricted range of odorant structures. Furthermore, different parts of the genome may be involved in the detection of different classes of odorants or odorant structural motifs. An odorant recognized by only one subfamily might involve only a single locus whereas an odorant detected by multiple subfamilies might involve a combination of loci.
Consistent with previous results (5, 6), these studies indicate that mice have ≈2.7 times as many ORs as humans. However, whereas humans have only 37% as many ORs as mice, they have 71% as many OR subfamilies. This finding suggests that the size of an OR family is not necessarily a good predictor of the diversity of odorant features a species can detect.
These studies further suggest that humans and mice are likely to recognize many of the same odorant structural motifs. Direct comparisons of human and mouse OR subfamilies revealed that the vast majority (87%) of human OR subfamilies have counterparts in the mouse repertoire. The majority (65%) of mouse subfamilies are also shared by human. This suggests a commonality between human and mouse in which the majority of odorant features detectable by one species may also be recognized by the other. Consistent with these findings are numerous studies showing that various compounds perceived by humans as having odors also induce neural activity in the olfactory system of mice (15, 27-31).
It is possible, however, that mice can detect and/or discriminate odorants more easily than humans. Of 150 subfamilies common to human and mouse, 81 are larger in mouse, 52 are the same size in the two species, and only 17 have more members in human than mouse. This finding is consistent with previous studies that noted that major branches of phylogenetic trees contain more mouse than human ORs (or OR “families”) (5, 6). If ORs that belong to the same subfamily do indeed recognize the same, or highly related, odorants, having more ORs in a subfamily could exert two effects on perception. First, it could result in a greater sensitivity of detection although the relative affinities of the ORs involved would also be important in this regard. Second, because highly related odorants are likely to be recognized by partially overlapping combinations of ORs (9, 12), having larger subfamilies should increase the likelihood of there being ORs that can distinguish between closely related odorants, thereby increasing the possibility that the odorants can be discriminated.
Although 150 OR subfamilies are shared by human and mouse, 22 subfamilies are present only in human and 84 subfamilies are found only in mouse. Because there can be functional redundancy among OR subfamilies (9),odorants detected by these “unique” subfamilies might also be recognized by other subfamilies that are shared by the two species, or they might be detected by different unique subfamilies in the two species. However, another, more intriguing possibility is that some of the unique subfamilies detect odorants that are sensed by only human or mouse. Obvious candidates would be chemicals of selective importance to one of the two species, such as pheromones.
Supplementary Material
Acknowledgments
We thank members of the Buck laboratory for helpful comments and suggestions. This project was supported by the Howard Hughes Medical Institute and by grants from the National Institutes of Health (National Institute on Deafness and Other Communication Disorders), the Department of Defense (Army Research Office), and Fundação de Amparo à Pesquisa do Estado de São Paulo (to B.M.).
Abbreviations: OR, olfactory receptor; TM, transmembrane domain.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY521456-AY521460).
References
- 1.Buck, L. & Axel, R. (1991) Cell 65, 175-187. [DOI] [PubMed] [Google Scholar]
- 2.Buck, L. B. (2000) Cell 100, 611-618. [DOI] [PubMed] [Google Scholar]
- 3.Firestein, S. (2001) Nature 413, 211-218. [DOI] [PubMed] [Google Scholar]
- 4.Ngai, J., Dowling, M. M., Buck, L., Axel, R. & Chess, A. (1993) Cell 72, 657-666. [DOI] [PubMed] [Google Scholar]
- 5.Zhang, X. & Firestein, S. (2002) Nat. Neurosci. 5, 124-133. [DOI] [PubMed] [Google Scholar]
- 6.Young, J. M., Friedman, C., Williams, E. M., Ross, J. A., Tonnes-Priddy, L. & Trask, B. J. (2002) Hum. Mol. Genet. 11, 535-546. [DOI] [PubMed] [Google Scholar]
- 7.Zhao, H., Ivic, L., Otaki, J. M., Hashimoto, M., Mikoshiba, K. & Firestein, S. (1998) Science 279, 237-242. [DOI] [PubMed] [Google Scholar]
- 8.Krautwurst, D., Yau, K. W. & Reed, R. R. (1998) Cell 95, 917-926. [DOI] [PubMed] [Google Scholar]
- 9.Malnic, B., Hirono, J., Sato, T. & Buck, L. B. (1999) Cell 96, 713-723. [DOI] [PubMed] [Google Scholar]
- 10.Touhara, K., Sengoku, S., Inaki, K., Tsuboi, A., Hirono, J., Sato, T., Sakano, H. & Haga, T. (1999) Proc. Natl. Acad. Sci. USA 96, 4040-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wetzel, C. H., Oles, M., Wellerdieck, C., Kuczkowiak, M., Gisselmann, G. & Hatt, H. (1999) J. Neurosci. 19, 7426-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kajiya, K., Inaki, K., Tanaka, M., Haga, T., Kataoka, H. & Touhara, K. (2001) J. Neurosci. 21, 6018-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araneda, R. C., Kini, A. D. & Firestein, S. (2000) Nat. Neurosci. 3, 1248-1255. [DOI] [PubMed] [Google Scholar]
- 14.Gaillard, I., Rouquier, S., Pin, J. P., Mollard, P., Richard, S., Barnabe, C., Demaille, J. & Giorgi, D. (2002) Eur. J. Neurosci. 15, 409-418. [DOI] [PubMed] [Google Scholar]
- 15.Bozza, T., Feinstein, P., Zheng, C. & Mombaerts, P. (2002) J. Neurosci. 22, 3033-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spehr, M., Gisselmann, G., Poplawski, A., Riffell, J. A., Wetzel, C. H., Zimmer, R. K. & Hatt, H. (2003) Science 299, 2054-2058. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan, S. L., Adamson, M. C., Ressler, K. J., Kozak, C. A. & Buck, L. B. (1996) Proc. Natl. Acad. Sci. USA 93, 884-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouquier, S., Taviaux, S., Trask, B. J., Brand-Arpon, V., van den Engh, G., Demaille, J. & Giorgi, D. (1998) Nat. Genet. 18, 243-250. [DOI] [PubMed] [Google Scholar]
- 19.Glusman, G., Yanai, I., Rubin, I. & Lancet, D. (2001) Genome Res. 11, 685-702. [DOI] [PubMed] [Google Scholar]
- 20.Strotmann, J., Hoppe, R., Conzelmann, S., Feinstein, P., Mombaerts, P. & Breer, H. (1999) Gene 236, 281-291. [DOI] [PubMed] [Google Scholar]
- 21.Xie, S. Y., Feinstein, P. & Mombaerts, P. (2000) Mamm. Genome 11, 1070-1078. [DOI] [PubMed] [Google Scholar]
- 22.Glusman, G., Bahar, A., Sharon, D., Pilpel, Y., White, J. & Lancet, D. (2000) Mamm. Genome 11, 1016-1023. [DOI] [PubMed] [Google Scholar]
- 23.Zozulya, S., Echeverri, F. & Nguyen, T. (2001) Genome Biol. 2, research0018.1-0018.12. [DOI] [PMC free article] [PubMed]
- 24.Bulger, M., Bender, M. A., van Doorninck, J. H., Wertman, B., Farrell, C. M., Felsenfeld, G., Groudine, M. & Hardison, R. (2000) Proc. Natl. Acad. Sci. USA 97, 14560-14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulger, M., van Doorninck, J. H., Saitoh, N., Telling, A., Farrell, C., Bender, M. A., Felsenfeld, G., Axel, R., Groudine, M. & von Doorninck, J. H. (1999) Proc. Natl. Acad. Sci. USA 96, 5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malnic, B., Godfrey, P. A. & Buck, L. B. (2004) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 27.Sicard, G. (1986) Brain Res. 397, 405-408. [DOI] [PubMed] [Google Scholar]
- 28.Sato, T., Hirono, J., Tonoike, M. & Takebayashi, M. (1994) J. Neurophysiol. 72, 2980-2989. [DOI] [PubMed] [Google Scholar]
- 29.Bozza, T. C. & Kauer, J. S. (1998) J. Neurosci. 18, 4560-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inaki, K., Takahashi, Y. K., Nagayama, S. & Mori, K. (2002) Eur. J. Neurosci. 15, 1563-1574. [DOI] [PubMed] [Google Scholar]
- 31.Belluscio, L. & Katz, L. C. (2001) J. Neurosci. 21, 2113-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.