Abstract
Many membrane-active peptides, such as cationic cell-penetrating peptides (CPPs) and antimicrobial peptides (AMPs), conduct their biological functions by interacting with the cell membrane. The interactions of charged residues with lipids and water facilitate membrane insertion, translocation or disruption of these highly hydrophobic species. In this mini-review we will summarize high-resolution structural and dynamic findings towards the understanding of the structure-activity relationship of lipid membrane-bound CPPs and AMPs, as examples of the current development of solid-state NMR (SSNMR) techniques for studying membrane peptides. We will present the most recent atomic-resolution structure of the guanidinium-phosphate complex, as constrained from experimentally measured site-specific distances. These SSNMR results will be valuable specifically for understanding the intracellular translocation pathway of CPPs and antimicrobial mechanism of AMPs, and more generally broaden our insight into how cationic macromolecules interact with and cross the lipid membrane.
Keywords: Cationic membrane peptide, Cell-penetrating peptides, Antimicrobial peptides, Guanidinium-phosphate complex, Solid-state NMR
Introduction
A large number of functionally important proteins and peptides carry out their functions in the cellular membrane, among which cationic cell-penetrating peptides (CPPs) and antimicrobial peptides (AMPs) have recently attracted much attention. Cellular internalization of bioactive agents, such as therapeutics, imaging agents and reporter molecules, garners significant biomedical and pharmaceutical interest because these molecules must bind to intracellular targets to function. However, the impermeable nature of the plasma membrane inhibits cellular entry of hydrophilic molecules that are relatively large. CPPs, a category of small peptides usually rich in arginine and lysine residues, have remarkable cellular uptake ability to deliver electrostatically or covalently bound bioactive agents into cells in culture and preclinical models in vivo (Zhang and Smith 2005; Schug and Lindner 2005; Fischer et al. 2005). Another important category of cationic membrane peptides is AMPs. They are important components of the innate immune system of many animals and plants (Zalsloff et al, 2002). They kill a broad spectrum of microbes including bacteria and fungi by destroying their cell membranes (Matsuzaki 1999). This rapid and general mechanism of action makes AMPs appealing alternatives to conventional antibiotics, most of which are rendered ineffective due to the increased incidence of antibiotic-resistant microbes.
CPPs and AMPs share common characteristics. Structurally, they have a highly charged character, a relatively small size (< 35 amino acids) and are lipid-bound. Functionally, the similar membrane insertion dictates the non-invasive cellular internalization potential of CPPs as drug transporters and is responsible for the antimicrobial efficiency and selectivity of AMPs. The mechanism addressing how these cationic peptides overcome the low-dielectric hydrophobic interior of the cell membrane and insert into the lipid bilayer remains unclear, limiting their therapeutic applications and presenting challenges to the design of more efficient alternatives (Chan et al. 2006; von Heijne 2006). This question has remained largely unanswered due to difficulty of characterizing the structures of these complex membrane systems at the atomic level. CPPs and AMPs are similar in structural motif of amino acid composition, but differ in biological activities within the lipid membrane. This begs the question, what is the underlying mechanism that distinguishes the two structurally similar but functionally different species?
To address these questions, SSNMR presents the most facile techniques, because it can probe the high-resolution structures of insoluble and noncrystalline membrane proteins that are difficult to analyze by traditional X-ray crystallography and solution NMR techniques (McDermott 2009; Tycko 2011; Hong et al. 2012b). Use of isotopic labeling, magic angle spinning (MAS) and multi-pulse techniques have made high-resolution NMR feasible in the solid state (De Paëpe 2012; Hong 1999; Rienstra et al. 2000). In recent years, it has gained much importance as a versatile tool to elucidate the structure, dynamics and functions of membrane-bound proteins, protein complexes, sedimented proteins, amyloid fibrils and more (Hong et al. 2012b; Tycko 2011; Bertini et al. 2011; Renault et al. 2011; Hong and Su 2011).
In the past few years, we have extensively investigated many CPPs and AMPs to elucidate the structural basis of how these cationic macromolecules interact with membrane lipids and water (Hong and Su 2011; Hong 2006)(Table 1). The two CPPs studied in our research, TAT and penetratin, are the first two discovered and also most frequently applied CPPs (Hong and Su 2011). TAT peptide contains the protein transduction domain (PTD) of the trans-activator of transcription (Tat protein) of HIV-1 virus (Green and Loewenstein 1988; Frankel and Pabo 1988), and penetratin is derived from the Drosophila homeodomain protein (Derossi et al. 1994). Two examples of AMPs in our recent studies include Protegrin-1 (PG-1) (Tang and Hong 2009) and human neutrophil peptide-1 (HNP-1) (Zhang et al. 2010b; Zhang et al. 2010a). PG-1 is representative of many β-sheet AMPs in its disulfide-linked structure and Arg-rich sequence. HNP-1 belongs to the α-defensin family of antimicrobial peptides and is the mediator of the host innate immune response. It is larger in size than PG-1 and contains different conformational domains.
Table 1.
Examples of cationic membrane-active peptides showing guanidinium-phosphate interactions. Cationic residues (Arg and Lys) are underlined in peptide sequences. Refer to the main text for detailed references. All conformations were determined in phospholipid bilayers at physiological temperature. r(Cζ-P) refers to the atomic distance between guanidinium Cζ of the specified arginine and the phosphorus atom of the lipid headgroup.
| Category | Peptide | Origins | Sequence | Conformation | r(Cζ-P) |
|---|---|---|---|---|---|
| CPP | TAT | the basic domain of Tat protein of HIV-1 virus | GRKKRRQRRRPPQ | random coil | R8: 4.1 Å |
| penetratin | the 3rd helix of Antennapedia Homeodomain | RQIKIWFQNRRMKWKK | coil-like | R10: 4.3 Å | |
| AMP | PG-1 | porcine leukocytes | RGGRLCYCRRRFCVCVGR | β-hairpin | R11: 4.0 Å |
| HNP-1 | human α-defensin | ACYCRIPACIAGERRY GTCIYQGRLWAFCC |
β-strand-rich | R25: 4.0 Å |
In this review, we will first address the crucial interactions of cationic residues, Arg and Lys, with lipids and water, which is widely recognized to play important roles in membrane protein function. Experimental evidences verifying the existence of these important interactions will be present. We’ll then have two separate sections of CPPs and AMPs as examples to demonstrate the indispensible role of solid-state NMR for elucidating the structure and dynamics of membrane-bound macromolecules. In each section, we will mainly focus on elucidating how these SSNMR findings give specific insights about the corresponding mechanism of action. Finally, the structural basis distinguishing these two categories of membrane-active macromolecules will be discussed.
Peptide-lipid and peptide-water interactions
An ultimate goal of membrane protein studies is to understand the underlying molecular mechanisms of their biological functions. One important facet is to address the functional contributions of intermolecular interactions, at an atomic or molecular level. Compared with the relatively well established biophysical and biochemical tools for analyzing structures and interactions of globular proteins, the study of membrane proteins is still relatively nascent. This is due to the extraordinary complexity of the membrane environment for most structural analysis methods. For example, the translocation efficiency and kinetics of CPPs and the antimicrobial potency of AMPs have been extensively studied, however, the atomic-level structural basis of how they interact with lipids to conduct their biological functions remains mysterious and unclear. Here we will show the discovery of mechanistically essential peptide-lipid and peptide-water interactions in our recent SSNMR studies.
Arg guanidinium-phosphate and Lys ammonium-phosphate interactions
Charge interactions between cationic residues and lipids are essential to the biological function of many membrane peptides and proteins, for example, cellular translocation of CPPs, membrane disruption of AMPs, and gating of voltage-sensing K+ channels (Freites et al. 2005; Tang and Hong 2009; Schmidt et al. 2009). The highly basic Arg guanidinium and Lys ammonium groups remain protonated under physiological pH conditions and thus can function as hydrogen bond (H-bond) donors in various protein-protein and protein-lipid interactions. The guanidinium-phosphate salt bridge is involved in many biological functions. As early as 1991, this interaction was proposed by Frankel and coworkers to explain site-specific binding to RNA (Calnan et al. 1991). As a highly stable cation in aqueous solutions, the guanidinium group accounts for most of Arg’s noncovalent bonding with anionic groups such as phosphates, sulfates and carboxylates (Sanchez-Quesada et al. 1996; Haack et al. 1999; Fernandez-Carneado et al. 2005). In lipid membranes, the guanidinium group in protein side chain can form a branched moiety by interacting with water and lipid molecules. Mutation of Arg residues (i.e. deletion or replacement) in cationic membrane peptides is usually found to cause significant reduction of their biological activity. Wender and coworkers showed oligomers of arginine containing dimethylguanidinium head groups have 95% lower translocation efficiency than the wild type, indicating the importance of this bidentate H- bond donor (Szyk et al. 2006). Thus, charged residues clearly play a crucial role in the biological functions of cationic membrane peptides. However, no high-resolution structure of this distinctive and mechanistically essential complex had been experimentally obtained until recently.
In recent years, SSNMR measurements of our several CPPs and AMPs have identified Arg guanidinium-phosphate and Lys ammonium-phosphate interactions by measuring site-specific distances and dynamics, as summarized in Table 1. The site-specific distance were measured using a SSNMR technique called rotational-echo double-resonance (REDOR) (Gullion and Schaefer 1989; Jaroniec et al. 2001). It uses rotor-synchronized π-pulses to recouple dipolar coupling, which encodes the distance between two nuclear spins. Short site–specific 13C-31P distance (~ 4 Å) between the distal end carbon (arginine Cξ and lysine Cε of the cationic residues and the phosphate of lipid headgroups provided the most direct molecular-level evidence to validate the existence of H-bonding interaction, since such a short distance is the minimum gap allowed between Arg/Lys and phosphate groups to avoid steric conflict (Su et al. 2009; Su et al. 2010b).
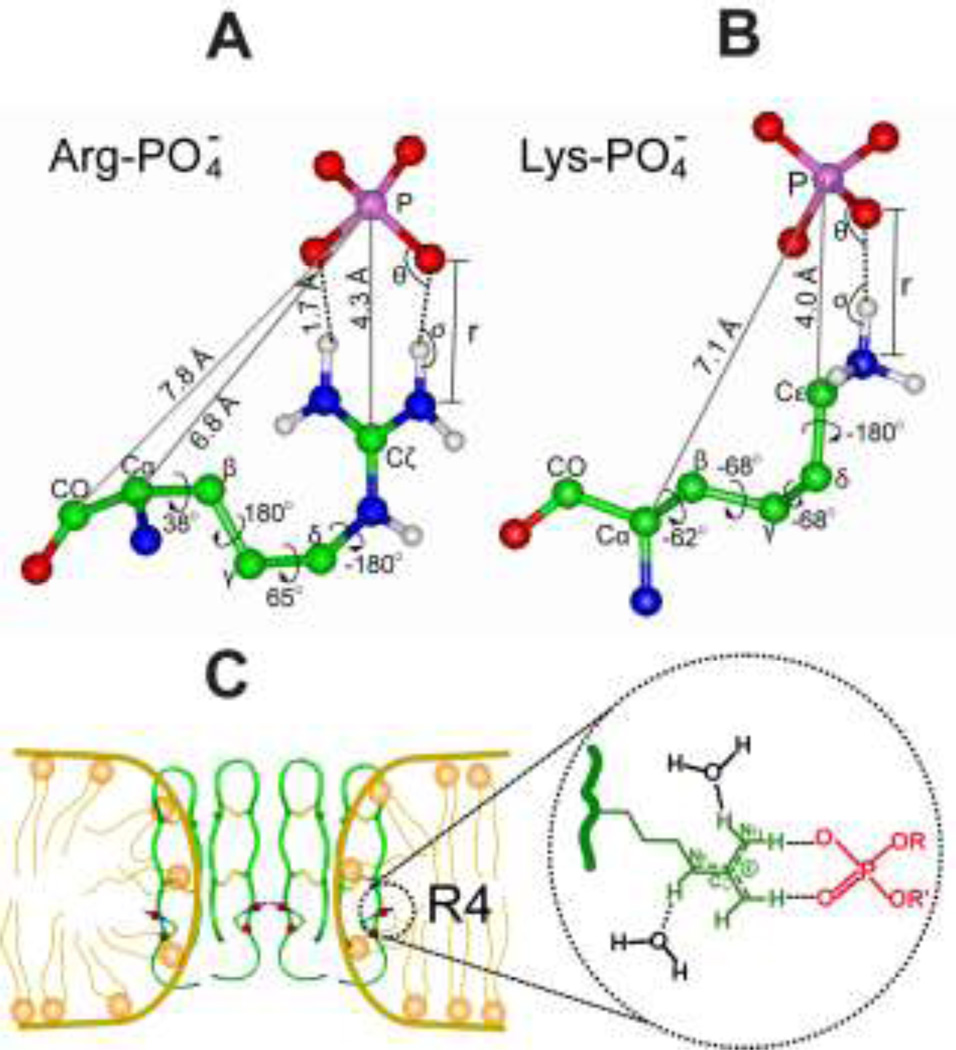
Fig. 1 shows the Arg-/Lys- lipid interactions for penetratin and PG-1, as examples for CPPs and AMPs, respectively (Su et al. 2010b; Li et al. 2010; Su et al. 2009; Tang et al. 2007). In the 13C-31P REDOR measurement of the membrane-bound penetratin (Su et al. 2009), Arg10 showed a short Cξ-P distances of 4.3 Å indicating the presence of guanidinium-phosphate complexation (Fig. 1A). Another cationic residue, Lys13 showed a similarly short (4.0 Å) distance between its side chain Cε and the lipid 31P (Fig. 1B), indicating Lys-lipid interaction. These 13C-31P distances, together with geometry parameters from previous studies, lead to the highest resolution structure of Arg-lipid and Lys-lipid interactions so far. In contrast to the short distances observed for cationic residues, a much longer distance of 6.9 Å from the neutral side chain of Ile3 Cγ/δ to lipid 31P was observed using double-quantum filtered selective REDOR (Su et al. 2009), indicating that neutral residues do not contribute to peptide-lipid interactions. The different roles of cationic and neutral residues suggest that the nature of peptide-lipid contact is charge-charge interaction. Interestingly, the guanidinium-phosphate interaction is not only revealed by the short Cξ-P distance (4.1 Å) but also by the fact that the guanidinium group exhibits higher rigidity than the rest of the Arg side chains in the study of lipid-bound TAT (Su et al. 2010b). The dipolar order parameter (SXH) indicates the motional amplitude of X-H bond and can be measured using the 2D dipolar-chemical-shift correlation (DIPSHIFT) technique. The order parameters of TAT Arg8 decrease from Nα (SNH = 0.20) to Cδ (SCH = 0.08) but then increase to NηSNH, indicating two relatively rigid ends sandwiching a mobile middle. Thus, the Arg sidechain end experiences stabilizing interactions in the membrane, consistent with the formation of N - H⋯O - P H-bonds.
Fig. 1.
(A) Arginine–phosphate and (B) Lysine–phosphate complexes stabilized by N-H⋯O-P H-bonds in POPC-POPG-bound CPP penetratin. The structural models were built based on experimentally measured site-specific 13C-31P distances (solid line) (Su et al, 2011), sidechain dihedral angles from the penultimate rotamer library (Lovell et al, 2000) and N-H⋯O-P geometric parameters in previous studies (Zhang et al, 1999; de Dios and Oldfield, 1994). Geometry parameters of the N-H⋯OP H-bonds: θ =120°, σ =168° and r = 2.8 Å. (C) Guanidinium-phosphate and guanidinium-water interactions of POPE-POPG-bound PG-1.
Cationic AMPs such as PG-1 and HNP-1 have also shown H-bonding of the guanidinium ion with phosphates (Zhang et al. 2010b; Tang et al. 2008b) (Fig. 1C). 13C-31P REDOR experiments indicated that PG-1 Arg11 and HNP-1 Arg25 have short Cξ-P distance of 4.0 Å. The important role of the guanidinium-phosphate interaction was also revealed by the fact that dimethylation of PG-1 Arg residues to Argmm, which diminishes the ability of the guanidinium group to act as a H-bond donor, significantly weakened the antimicrobial ability of the peptide.
The fundamental role of guanidinium-phosphate interaction for activities of these cationic membrane peptides was also supported many molecular dynamics (MD) simulations (Herce and Garcia 2007; Freites et al. 2005; Yoo and Cui 2008; Vorobyov et al. 2008). For example, in MD simulation of TAT translocation across a DOPC bilayer (Herce and Garcia 2007), Arg-lipid interaction was found to be the driving force for the initial insertion, transient water-pore formation and attraction from the phosphates of the distal leaflet. The guanidinium-phosphate complex was also found and proposed to be a crucial interaction for the motion of the gating helix S4 of voltage-sensitive potassium channels (Kv) (Freites et al. 2005). Our SSNMR study of the Kv S4 helix in lipid bilayers has identified such an interaction at residue Arg8 (Doherty et al. 2010). Overall, the knowledge gained from these experimentally identified intermolecular interactions and from MD simulations furthered our understanding of protein-lipid interactions and their importance in membrane-active biological systems.
The thermodynamic principles behind membrane insertion of these cationic peptides are important to consider. On the one hand, the insertion of cationic residues into a membrane-water interfacial layer appears to be energetically unfavorable due to the Born repulsion (Wimley and White 1996). On the other hand, interactions between the highly charged residues and lipid phosphates and water can contribute to the compensation energy necessary to overcome the insertion energy barrier (Ziegler et al. 2003; Torchilin et al. 2001). The H-bond-stabilized complex is proposed to minimize the polarity of Arg-rich peptides via charge-charge neutralization, which reduces the free-energy barrier from three different aspects: electrostatic interaction between the cationic residues and anionic lipid headgroups, H-bonding within the Arg guanidinium–phosphate complex or the Lys ammonium-phosphate complex, and Arg-water and Lys-water H-bonding. All these interactions have been observed in our SSNMR experiments of CPPs and AMPs (Su et al. 2010b; Li et al. 2010; Su et al. 2009; Tang et al. 2007).
Different functional roles of Arg and Lys
Although both Arg and Lys in CPP sequences play principal roles in cellular uptake, the two residues contribute differently (Wender et al. 2008; Rothbard et al. 2005; Futaki et al. 2001; Wender et al. 2000; Sundlass and Raines 2011; Takechi et al. 2011). Polyarginine (Arg)n, (n=7 or 9), a synthetic CPP with n-consecutive Arg, was reported to enter cells more efficiently than polylysine (Lys)n (Mitchell et al. 2000). Wender et al. (Wender et al. 2000) showed that the D-arginine oligomer (Arg)9 exhibited 100-fold higher translocation efficiency than HIV Tat (49–57) and proposed that Arg contributes more than Lys to the cellular uptake. In addition to these differences as CPPs, TAT and R9 can actively bind to RNA while K9 cannot (Calnan et al. 1991). For AMPs, the Lys-for-Arg mutation also significantly weakens antimicrobial activities. For example, cryptdin-4 (Crp4), a potent bactericidal peptide of the α-defensin family, disrupts the lipid membrane by altering the membrane curvature. A recent study by Schmidt et al (Schmidt et al. 2012) showed that complete Lys-for-Arg mutation of Crp4 dramatically decreased its antimicrobial potency. However, none of these studies have provided molecular evidence of why Arg and Lys play distinct functional roles.
Although the NMR-observed 13C-31P distances indicate strong charge-charge interactions for both Arg10 and Lys13 of penetratin to phosphates of gel-phase lipid membranes, dynamic measurements at physiological temperature showed that Arg10Nη had a much larger order parameter (SNH=0.3) than the SNH of Lys13Nε which was estimated to be much less than 0.1. This indicates that Lys-phosphate interactions are less stable than Arg-phosphate interactions at physiological temperature. Moreover, Arg10 adopts rigid β-strand conformation at high temperature while Lys13 adopts a relatively mobile β-turn state, which further supports the stronger Arg-lipid interaction. The weaker stability of the Lys-phosphate interaction is reasonable because of the fast rotation of side chain NH3, which prohibits the formation of stable H-bonding. In comparison, the guanidinium group can form up to four H-bonds with two phosphate groups, creating the so-called guanidinium-phosphate bidentate complex. Therefore, the Lys-phosphate interaction should provide less compensation energy for membrane insertion. Overall, the different stabilities of Arg-lipid and Lys-lipid interactions at physiological temperature explains why lysine substitution of CPPs and AMPs weakens membrane insertion of these peptides. This agrees well with the finding that the inefficient CPP, (Lys)n, is not able to insert into the membrane whereas Arg-rich TAT and (Arg)9 can cross the lipid membrane (Ben-Tal et al. 1996).
Arg-water interaction
The hydration of proteins plays an essential role in the folding, stability, dynamics, and function of proteins, as well as in ligand binding and recognition. So, the investigation of water–protein interactions is necessary for understanding many biological processes. Charged residues such as arginine and lysine are readily exchanged and charge-stabilized with water by forming H-bonds. Thus, charged residues in membrane proteins are hot spots for interaction with water (Freites et al. 2005). In recent years, several effective SSNMR techniques have been developed to probe protein-water interactions in the lipid membrane environment.
A two-dimensional (2D) 1H spin diffusion SSNMR method was introduced to semi-quantitatively determine the distances from the mobile lipid chains and water to a rigid membrane-associated protein (Huster et al. 2002). Magnetization initiating from the mobile 1H magnetization sources such as H2O and lipid CH2 groups is allowed to transfer to nearby protein residues. Protein with different depths of insertion will manifest different spin diffusion rates. This technique has been successfully applied to investigate the protein-water interactions in a number of membrane proteins including the influenza A virus M2 protein (Luo and Hong 2010), the chimeric potassium channel KcsA-Kv1.3 (Ader et al. 2008), TAT (Su et al. 2010b) and various AMPs such as tachyplesin I (TP-1) (Doherty et al. 2006a), PG-1 (Mani et al. 2006) and a charge-reduced PG-1 mutant IB484 (Su et al. 2011). In our TAT study, all polar residues containing labile protons exhibited strong water cross-peaks in the 2D 13C-detected 1H spin diffusion spectrum, indicating stabilization of the charged residues by peptide-water interactions (Su et al. 2010b). Together with the observed guanidinium-phosphate interaction, we proposed that TAT minimizes it hydrophobicity by interacting with lipid phosphate groups and water molecules.
Hetero-nuclear correlation (HETCOR) NMR experiments with 1H homo-nuclear decoupling have been used to investigate Arg-water interactions in membrane-bound PG-1 (Li et al. 2010; Li and Hong 2011). Arg4 guanidinium-water correlation was clearly detected in the 1H−15N Hartman−Hahn (HH)-CP HETCOR spectra, indicating water solvation of this hydrophobically embedded Arg residue (Fig. 1C). This observtion of the arginine-water interactions for residues inside the lipid membrane was consistent with MD simulation results (Dorairaj and Allen 2007; Li et al. 2008).
To facilitate the detection of water cross peaks in 13C, 15N-labeled membrane peptides without perdeuteration in the HETCOR experiments, a 13C and/or 15N dipolar-edited MELODI−HETCOR experiment was introduced to avoid overlap of protein 1H signals, primarily Hα and HN, with the water signal (Li et al. 2010; Yao et al. 2001). Protein-water interactions can also be manifested from the chemical shift variations of water. Very recently, the histidine-water interaction in the influenza M2 proton channel was experimentally observed from the correlations between the imidazole ring and water at pH ranging from 4.5 to 8.5 in 1H-15N HETCOR experiments (Hong et al. 2012a), indicating the presence of strong H-bonding between the histidine side chain and water in the influenza M2 protein.
Cell-penetrating peptides
Since the discovery of cellular uptake of Tat protein in 1988 (Green and Loewenstein 1988; Frankel and Pabo 1988), TAT peptide and many other protein-derived or chemically synthesized CPPs, such as penetratin, transportan and polyarginines, have been widely characterized using biophysical and biochemical techniques, because of great potential for biological research and therapeutic applications. Several CPPs have already entered the clinical phase of animal or human testing (Schwarze et al. 1999; Khafagya et al. 2009; Gratton et al. 2003; Dietz et al. 2008). Despite the extensive biological characterization and applications, the mechanism of translocation still remains unclear because of lack of structure (Christiaens et al. 2002; Persson et al. 2001). By utilizing various solid-state NMR techniques, we have investigated molecular interactions of TAT and penetratin with lipid membranes, which provided valuable information about the relation between their Arg-rich structural properties and membrane translocation (Su et al. 2010b; Su et al. 2009; Su et al. 2008b; Su et al. 2008a; Su and Hong 2011).
Dynamic structure of membrane-bound CPPs: mobility makes opportunity
For the first time, we have determined the conformation of TAT and penetratin in hydrated lipid bilayers using SSNMR (Su et al. 2010b; Su et al. 2008a). 13C and 15N chemical shifts at physiological temperature indicate penetratin adopts a mobile β-turn rich structure, while TAT shows a random coil conformation. The latter is also supported by very low backbone order parameters (SCH = 0.14–0.20), suggesting similarly large mobility as lipids. More recently, the highly dynamic conformations of TAT and penetratin have been detected in live cells by Raman microscopy (Ye et al. 2010) and CD spectroscopy (Guo et al. 2012). Using a novel one-side paramagnetic relaxation enhancement (PRE) method, 1H spin diffusion and 13C-31P REDOR experiments, we found that both peptides insert into the membrane-water interface of the lipid bilayer. Thus, TAT arguably provides the first clear documented case of a membrane-bound random coil peptide (Su et al. 2010b; Su et al. 2009; Su et al. 2008b).
CPP translocation mechanism
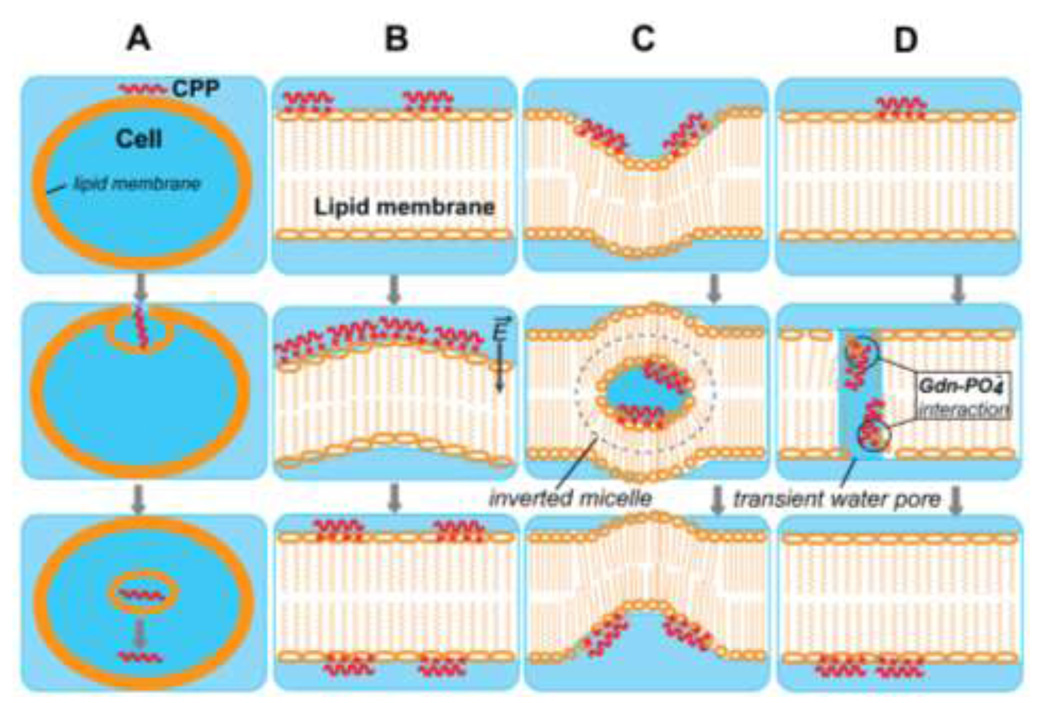
CPPs are non-invasive and highly efficient intracellular transporters, however, their therapeutic application and development of cell uptake efficiency enhancement are limited by an insufficient understanding of the translocation mechanism. Highly cationic CPPs are extremely unlikely to cross the hydrophobic lipid bilayer by simple passive diffusion without a free-energy penalty. Debate about the various models centers on whether membrane translocation of these peptides proceeds through an inverted-micelle-mediated route, through endocytosis or through direct membrane penetration (Weissig and D'Souz 2012; Madani et al. 2011) (Fig. 2). The endocytosis pathway is proposed based on the fact that CPP uptake in cells was found to adopt an energy-dependent mechanism (Richard et al. 2003) (Fig. 2A). However, cellular uptake is not completely prohibited at low temperatures or in the presence of endocytosis inhibitors (Richard et al. 2003; Letoha et al. 2003), suggesting a non-energy consuming route. The electroporation model (Fig. 2B) suggests that at a certain high concentration (P/L>1/40), the cationic peptides induce an electric field through the membrane, which changes membrane curvature and causes electroporation-like membrane permeabilization (Binder and Lindblom 2003). However, our one-side PRE results show that penetratin peptides are located on both leaflets of the bilayer at both low (P/L=1:40) and high (P/L=1:20) peptide concentrations (Su et al. 2008b). The inverted-micelle model invokes the formation of transient vesicles that trap CPPs and escort them across the bilayer (Derossi et al. 1998) (Fig. 2C). However, this model is ruled out by static 31P NMR spectra which preclude any highly mobile isotropic vesicles in lipid membranes in the presence of either penetratin or TAT (Su et al. 2010b; Su et al. 2008b). Recently, the finding of strong peptide-lipid interactions supports direct translocation of the peptides across the membrane (Fig. 2D) (Herce and Garcia 2007; Szyk et al. 2006; Su et al. 2010b). In addition, many studies also propose cellular uptake can follow multiple routes depending on experimental conditions, specific CPP sequence and the cargo structure (von Heijne 2006).
Fig. 2.
Schematic models of CPP translocation across the cellular membrane. (A) Endocytosis model. (B) Electroporation model. (C) Inverted micelle model. (D) Direct penetration model.
Regardless of whether endocytosis is involved or not, CPPs need to penetrate either the plasma or the endosomal membrane to reach the intracellular compartments, therefore the structural information of CPPs crossing a lipid bilayer from SSNMR studies is valuable for critically evaluating the different models. The direct translocation process stresses the fundamental role of guanidinium-phosphate ion pairs, which have been observed in our TAT and penetratin studies. In the first stage of translocation, CPPs bind to the lipid membrane surface. The high affinity of CPP for the membrane surface results from electrostatic interaction and was confirmed in many thermodynamic and MD studies (Jing et al. 2012; Mitchell et al. 2000; Torchilin et al. 2001; Ziegler et al. 2003). Subsequenctly, the H-bonding interactions of Arg guanidinium and lipid phosphate facilitate the insertion of the surface-bound peptides. The experimentally observed complex makes CPPs more membrane-soluble by minimizing the exposure of the charged groups to the hydrophobic environment. H-bonding of the guanidinium group with lipid phosphate and water also provides sufficient compensation energy to overcome the Born repulsion from the membrane-water interface. The membrane-water interfacial insertion of TAT and penetratin has been identified in our studies. The movement of CPPs inside the lipid membrane may be facilitated by the transient water pore formation as proposed by Herce and Garcia in their MD simulation of TAT translocation (Herce and Garcia 2007) and indicated by the strong guanidinium-water interaction we observed (Su et al. 2010b). Meanwhile, peptide concentration difference at various membrane depths is caused by the accumulation of peptide near the outer membrane surface. In addition, CPPs also experience charge-charge interaction from the distal phosphate layer. Possibly driven by these two factors, the phosphate- or water-neutralized peptides could move in the intracellular direction and finally accomplish the translocation. Overall, the guanidinium-phosphate interactions stabilize the CPP peptides in lipids and facilitate the insertion, while the plastic conformation and high mobility further promote the translocation.
Antimicrobial Peptides
To date, hundreds of AMPs have been isolated from a variety of organisms ranging from microbes to plant and animals. Their biological and chemical properties have been extensively characterized in the past twenty years. In general, they have a high content of cationic residues, adopt amphipathic conformation, and carry out antimicrobial actions by permeablizing the microbial cell membrane. However, how these highly charged peptides insert into the lipid membrane seems puzzling. This question can hardly be answered without knowledge of the structure and dynamics of AMPs in the lipid membrane. Here, we will use PG-1 and its mutants as examples to summarize SSNMR findings of the structure, topology and dynamics of the family of β-hairpin AMPs, and most importantly to show how this structural information can be correlated to antimicrobial potency and selectivity.
Structure and topology of AMPs in lipid bilayers
Over the past few years, we used SSNMR to study many AMPs, including retrocyclin-2, tachyplesin-I, HNP-1, PG-1 and its mutants (Tang and Hong 2009; Doherty et al. 2006a, b; Buffy et al. 2004; Hong and Su 2011; Su et al. 2010a). In contrast to unstructured CPPs, AMPs normally adopt well-defined secondary structures to conduct their antimicrobial activity in lipid membranes. As a representative cationic cysteine-rich AMP, PG-1 adopts a β-hairpin conformation due to its two intramolecular disulfide bonds (Fahrner et al. 1996). The knowledge of structural behaviors of AMPs in hydrated lipid membrane, such as insertion depth and aggregation of peptides, is more valuable. For example, PG-1 in DPC micelles exists as anti-parallel NCCN dimers (Roumestand et al. 1998), while, our intermolecular distance measurments show that PG-1 peptides pack as parallel NCCN dimers in POPC and POPE-POPG lipid bilayers (Mani et al. 2006). Differenct lipid compositions of the membrane bilayer cuase distinct PG-1 oligomerization states and insertion depths. Our SSNMR data show PG-1 peptides form a toroidal β-barrel pore in a transmembrane fashion in bacteria-mimetic anioic lipids (i.e. POPE/POPG) but stay on the membrane surface as β-sheet aggregates in eukaryotic cell-mimetic cholesterol-rich membranes (i.e. POPC/cholesterol) (Mani et al. 2006).
Antimicrobial mechanisms
AMPs selectively kill bacteria mainly by disrupting cytoplasmic membrane integrity and sometimes by altering the membrane potential. The first step towards membrane disruption is the high affinity of the cationic peptides on the anionic lipid membrane surface due to electrostatic interactions. Many studies have shown that the charge density affects the membrane-binding ability and in turn the antimicrobial activity (Balali-Mood et al. 2003; Dathe et al. 2001). In addition to charge, structural properties such as conformation, hydrophobicity and amphipathicity of the peptide also contribute to the antimicrobial potency and mechanism (Yeaman and Yount 2003).
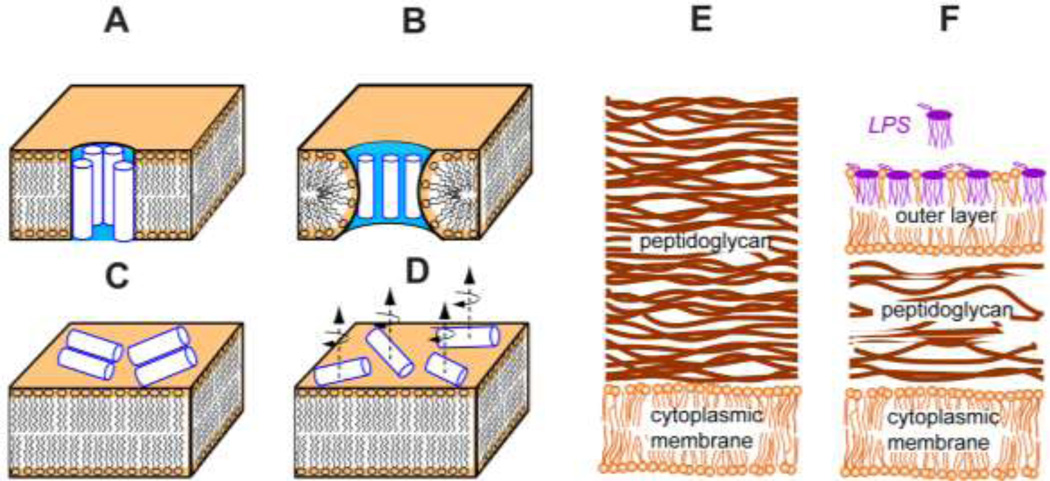
In recent years, several models have been suggested to rationalize the mechanism of action of AMPs. These include the barrel-stave model, the toroidal pore model, the carpet model, and the in-plane diffusion or partial-insertion model (Fig. 3A–D) (Bechinger 1999; Brogden 2005; Epand and Vogel 1999). These four models are distinguished by the peptide location in the membrane, peptide oligomeric structure, dynamics, and the membrane disorder. The barrel-stave model (Fig. 3A) involves a few peptides forming a barrel to span the lipid bilayer, and was proposed based on the studies of alamethicin using neutron scattering and single-channel conductance techniques (Baumann and Mueller 1974; Bechinger 1999). The toroidal pore model addresses amphipathic AMP interaction with lipids through a torus-shaped transmembrane channel (Fig. 3B). It was first proposed to explain the antimicrobial activity of magainin (Ludtke et al. 1996; Matsuzaki et al. 1994). Both the barrel-stave and the toroidal pore models posit a transmembrane pore but the latter involves significant orientational changes of lipid head groups from their normal lamellar bilayer orientations, such that the lipids merge the two leaflets of the membrane to form a pore. 13C-detected 1H spin diffusion experiments showed that POPE-POPG-bound PG-1 is in close contact (2 Å) with the lipid chain methyl groups, which represent the membrane center, strongly indicating that PG-1 adopts a transmembrane orientation. Together with the finding that some lipid headgroup are embedded in the hydrophobic region of the membrane (Tang et al. 2007), we propose that PG-1 forms a ‘toroidal pore’ to kill microbial cells (Mani et al. 2006; Tang et al. 2007). In the carpet model (Fig. 3C), AMPs aggregate on the membrane surface at a certain high peptide concentration, causing membrane thinning and eventual micellization (Shai and Oren 2001). In the in-plane diffusion or partial-insertion model (Bechinger 1999), surface-bound AMPs do not aggregate but induce local curvature of the lipid membrane (Fig. 3D). The fast diffusion of peptide triggers transient pores, which can cause sufficient disruption to lead to loss of cell viability. AMPs like TP-1, which is too short to span the bilayer to induce transmembrane pores and does not aggregate, adopt such an in-plane diffusion mechanism (Doherty et al. 2006a).
Fig. 3.
Membrane disruption models (A–D) of AMPs and Gram-positive (E) and Gram-negative (F) bacterial membranes. (A) Barrel-stave transmembrane model. (B) Toroidal pore model. (C) Carpet model. (D) In-plane diffusion or partial insertion model. AMP examples for each model are given in the text.
Mechanism of double antimicrobial selectivity
AMPs show double selectivity, i.e. different antimicrobial potency in bacterial and mammalian membranes and antimicrobial selectivity against gram-positive versus gram-negative bacteria. Different membrane compositions of bacterial and mammalian membranes help to make the peptides active towards microbial invaders and less harmful to the host cells. Bacterial membranes are rich in anionic phospholipids, while mammalian cell membranes consist of zwitterionic phospholipids (Rouser et al. 1968). The recognition of the former is realized by the preferred binding of positively charged peptides to negatively charged lipids. In addition, the higher anionic lipid content of bacterial membranes can also facilitate the insertion of cationic AMPs. In addition to anionic lipid content, cholesterol, a main component in mammalian cell membranes, is absent in bacterial cells. It is known cholesterol can rigidify the lipid membrane, protecting the host cell from AMP insertion.
These hypotheses have all been proven to be true by many others’ work as well as our SSNMR study of PG-1 in various lipid membranes. PG-1 is active against various bacteria and fungi and the MICs range from 0.3 – 3 µM but it is almost inactive in mammalian cells. SSNMR 31P lineshapes of lipid membranes containing PG-1 indicate that the peptide preferentially disrupts negatively charged membranes (Yamaguchi et al. 2002; Buffy et al. 2003). The membrane-selective disruption was suggested in studies of the insertion depth of PG-1: the bacteria-mimetic anionic POPE-POPG membrane allows peptide insertion, while the eukaryote-MIMIC POPC-cholesterol membrane prevents PG-1 insertion (Mani et al. 2006; Glukhov et al. 2005).
In addition to the selectivity between bacterial and mammalian cells, many AMPs show different antimicrobial potency in gram-positive versus gram-negative bacteria. The two types of bacteria are distinguished by the structural differences of their cell walls (Fig. 3E and F). Both kinds of bacteria have a cytoplasmic membrane layer composed of phospholipids, where most AMPs conduct their antimicrobial activity (Tang and Hong 2009). The difference is that the gram-negative bacterial membrane has an extra outer membrane, which is composed of lipopolysaccharide (LPS) in the outer leaflet and phospholipids (mostly PE) in the inner leaflet (Sperandeo et al. 2009). PG-1, active in both kinds of bacteria, and IB484, a charge-reduced PG-1 mutant that is only potent in gram-positive bacteria, have provided ideal model systems to understand the antimicrobial selectivity between the two types of bacteria (Chen et al. 2000). Our SSNMR results of the topological structure and interaction of the two peptides showed that both peptides insert into the center of the gram-positive-bacteria mimetic POPE-POPG membrane and cause lipid headgroup reorientation, consistent with the toroidal pores. However, PG-1 fully inserts into the gram-negative-bacteria mimetic ReLPS-DEPE membranes, whereas IB484 does not, indicating that the antimicrobial potency is modulated by insertion of the peptide in the outer membrane (Su et al. 2011). The bulky size of LPS makes the outer layer less permeable than the cytoplasmic membrane of gram-positive bacteria. Due to the largely reduced charge intensity, IB484 has relatively weak electrostatic attraction to the LPS layer, which prevents its further insertion into the cytoplasmic membrane of the gram-negative bacteria. Our comparative study of PG-1 and IB484 indicates that the charge density of AMPs is a crucial structural property determining their antimicrobial selectivity against gram-positive and gram-negative bacteria.
CPPs and AMPs: how different are they?
The comparison between CPPs and AMPs suggests the conformation and dynamics are the structural factors distinguishing the two categories of membrane peptides. Most AMPs adopt rigid amphipathic secondary structures, either α-helical or in β-sheet (White and Wimley 1999; Abbassi et al. 2008; Mangoni et al. 2000; Ulmschneider and Ulmschneider 2008; Thundimadathil et al. 2006; Steiner et al. 1981; Zasloff 1987). However, the two studied CPPs, TAT and penetratin, show a turn-rich conformation and random coil structrue in lipid bilayers, respectively, suggesting that the absence of intra- or intermolecular H-bonded conformation and high molecular mobility may be the hallmarks of CPPs differentiates them from AMPs.
Due to the similar Arg-rich structural motif, CPPs and AMPs have strong Arg-lipid and Arg-water interactions, which stabilize these hydrophobic peptides by membrane neutralization and water solvation and thus facilitate the insertion. However, CPPs insert into the cellular membrane in a non-invasive manner, while AMPs function by disrupting the lipid membrane. For CPPs, a small guanidinium N-H order parameter (SNH = 0.30) was obtained for both penetratin Arg10 and TAT Arg8, indicating that Arg-lipid H-bonding is relatively weak and will not retard the mobility of CPPs in the lipid membrane. The dynamic structure may serve to prevent these Arg-rich CPPs from permanently residing in and damaging the lipid membrane, thus facilitating peptide across the lipid bilayer. For AMPs, as represented by PG-1, their backbone is largely immobilized due to intra- or intermolecular H-bonds and oligomerization. The high backbone rigidity of aggregated PG-1 in POPE-POPG membranes is indicated by SSNMR results of large dipolar order parameters (SXH=0.70–1.05) (Tang et al. 2008a). Thus, the stable transmembrane insertion and lipid interaction together with the formation of oligomerized structures such as β-barrels cause membrane disorder and pore formation, resulting in membrane disruption.
Conclusion and perspectives
We have carried out many comprehensive solid-state NMR studies to investigate the conformation, dynamics and depth of insertion of representative CPPs and AMPs in the lipid bilayer. The mechanistically crucial guanidinium-phosphate and guanidinium-water interactions have been experimentally identified. These atomic-level structural results have contributed significantly to uncover the underlying mechanisms of CPP translocation and AMP antimicrobial activity and gain insights into developing alternative compounds with more potent activites for pharmaceutical applications. While the lipid-bound studies provided plausible information on the equilibrium structures of these membrane peptides, to elucidate the insertion route of CPPs and AMPs, knowledge of in-situ translocation structures will be important (Ye et al. 2010). Thus, further understanding of the membrane insertion mechanism requires key intermediate structures and interactions, which are likely invisible in the equilibrium state, to be identified in vivo. One strategy is to trap transient stages in membrane insertion by rapid sample freezing on the 10−20 µs time scale (Hu et al. 2010). The low-intensity difficulty in the structural characterization due to the distribution of conformational intermediates may be overcome by using DNP-enhanced SSNMR (Zech et al. 2005).
Acknowledgement
The SSNMR research reviewed in this work was supported by grant GM066976 from National Institutes of Health (NIH) to M.H.
Abbreviations
- AMP
Antimicrobial peptide
- CPP
Cell-penetrating peptide
- CSA
Chemical shift anisotropy
- DARR
Dipolar-assisted rotational resonance
- DIPSHIFT
Dipolar-chemical-shift correlation
- DNP
Dynamic nuclear polarization
- HETCOR
Heteronuclear correlation
- HNP-1
Human neutrophil peptide-1
- INADEQUATE
Incredible natural abundance double quantum transfer experiment
- LPS
Lipopolysaccharide
- MAS
Magic angle spinning
- PE
Phosphatidylethanolamine
- PG
Phosphatidylglycerol
- PG-1
Protegrin-1
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol
- PRE
Paramagnetic relaxation enhancement
- REDOR
Rotational-echo double-resonance
References
- Abbassi F, Galanth C, Amiche M, Saito K, Piesse C, Zargarian L, Hani K, Nicolas P, Lequin O, Ladram A. Solution Structure and Model Membrane Interactions of Temporins-SH, Antimicrobial Peptides from Amphibian Skin. A NMR Spectroscopy and Differential Scanning Calorimetery Study. Biochemistry. 2008;47:10513–10525. doi: 10.1021/bi8006884. [DOI] [PubMed] [Google Scholar]
- Ader C, Schneider R, Seidel K, Etzkorn M, Becker S, Baldus M. Structural Rearrangements of Membrane Proteins Probed by Water-Edited Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2008;131(1):170–176. doi: 10.1021/ja806306e. [DOI] [PubMed] [Google Scholar]
- Balali-Mood K, Harroun TA, Bradshaw JP. Molecular Dynamics Simulations of a Mixed DOPC/DOPG Bilayer. Eur Phys J E. 2003;12:S135–S140. doi: 10.1140/epjed/e2003-01-031-3. [DOI] [PubMed] [Google Scholar]
- Baumann G, Mueller P. A molecular model of membrane excitability. J. Supramolecular Struc. 1974;2(5–6):538–557. doi: 10.1002/jss.400020504. [DOI] [PubMed] [Google Scholar]
- Bechinger B. The structure, dynamics, and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim Biophys Acta. 1999;1462:157–183. doi: 10.1016/s0005-2736(99)00205-9. [DOI] [PubMed] [Google Scholar]
- Ben-Tal N, Honig B, Peitzsch RM, Denisov G, McLaughlin S. Binding of small basic peptides to membranes containing acidic lipids: theoretical models and experimental results. Biophys J. 1996;71:561–575. doi: 10.1016/S0006-3495(96)79280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I, Luchinat C, Parigi G, Ravera E, Reif B, Turano P. Solid-state NMR of proteins sedimented by ultracentrifugation. Proc Natl Acad Sci U S A. 2011;108(26):10396–10399. doi: 10.1073/pnas.1103854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder H, Lindblom G. Charge-dependent translocation of the Trojan peptide penetratin across lipid membranes. Biophys J. 2003;85:982–995. doi: 10.1016/S0006-3495(03)74537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Buffy JJ, McCormick MJ, Wi S, Waring A, Lehrer RI, Hong M. Solid-State NMR Investigation of the Selective Perturbation of Lipid Bilayers by the Cyclic Antimicrobial Peptide RTD-1. Biochemistry. 2004;43:9800–9812. doi: 10.1021/bi036243w. [DOI] [PubMed] [Google Scholar]
- Buffy JJ, Waring AJ, Lehrer RI, Hong M. Immobilization and Aggregation of the Antimicrobial Peptide Protegrin-1 in Lipid Bilayers Investigated by Solid-State NMR. Biochemistry. 2003;42(46):13725–13734. doi: 10.1021/bi035187w. [DOI] [PubMed] [Google Scholar]
- Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. Arginine-mediated RNA recognition: the arginine fork. Science. 1991;252(5010):1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Chan DI, Prenner EJ, Vogel HJ. Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim Biophys Acta. 2006;1758:1184–1202. doi: 10.1016/j.bbamem.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Chen J, Falla TJ, Liu H, Hurst MA, Fujii CA, Mosca DA, Embree JR, Loury DJ, Radel PA, Cheng CC, Gu L, Fiddes JC. Development of protegrins for the treatment and prevention of oral mucositis: Structure-activity relationships of synthetic protegrin analogues. Biopolymers. 2000;55:88–98. doi: 10.1002/1097-0282(2000)55:1<88::AID-BIP80>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Christiaens B, Symoens S, Verheyden S, Engelborghs Y, Joliot A, Prochiantz A, Vandekerckhove J, Rosseneu M, Vanloo B. Tryptophan fluorescence study of the interaction of penetratin peptides with model membranes. Eur J Biochem. 2002;269(12):2918–2926. doi: 10.1046/j.1432-1033.2002.02963.x. [DOI] [PubMed] [Google Scholar]
- Dathe M, Nikolenko H, Meyer J, Beyermann M, Bienert M. Optimization of the Antimicrobial Activity of Magainin Peptides by Modification of Charge. FEBS Lett. 2001;501(2–3):146–150. doi: 10.1016/s0014-5793(01)02648-5. [DOI] [PubMed] [Google Scholar]
- De Paëpe G. Dipolar Recoupling in Magic Angle Spinning Solid-State Nuclear Magnetic Resonance. Annu Rev Phys Chem. 2012;63:661–684. doi: 10.1146/annurev-physchem-032511-143726. [DOI] [PubMed] [Google Scholar]
- Derossi D, Chassaing G, Prochiantz A. Trojan peptides: the penetratin system for intracellular delivery. Trends Cell Biol. 1998;8(2):84–87. [PubMed] [Google Scholar]
- Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- Dietz GP, Stockhausen KV, Dietz B, Falkenburger BH, Valbuena P, Opazo F, Lingor P, Meuer K, Weishaupt JH, Schulz JB, Bähr M. Membrane-permeable Bcl-xL prevents MPTP-induced dopaminergic neuronal loss in the substantia nigra. J Neurochem. 2008;104(3):757–765. doi: 10.1111/j.1471-4159.2007.05028.x. [DOI] [PubMed] [Google Scholar]
- Doherty T, Su Y, Hong M. High-Resolution Orientation and Depth of Insertion of the Voltage-Sensing S4 Helix of a Potassium Channel in Lipid Bilayers. J. Mol. Bio. 2010;401(4):642–652. doi: 10.1016/j.jmb.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty T, Waring AJ, Hong M. Membrane-bound conformation and topology of the antimicrobial peptide tachyplesin-I by solid-state NMR. Biochemistry. 2006a;45:13323–13330. doi: 10.1021/bi061424u. [DOI] [PubMed] [Google Scholar]
- Doherty T, Waring AJ, Hong M. Peptide-lipid interactions of the beta-hairpin antimicrobial peptide tachyplesin and its linear derivatives from solid-state NMR. Biochim Biophys Acta. 2006b;1758:1285–1291. doi: 10.1016/j.bbamem.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Dorairaj S, Allen TW. On the thermodynamic stability of a charged arginine side chain in a transmembrane helix. Proceedings of the National Academy of Sciences. 2007;104(12):4943–4948. doi: 10.1073/pnas.0610470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochim Biophys Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- Fahrner RL, Dieckmann T, Harwig SS, Lehrer RI, Eisenberg D, Feigon J. Solution Structure of Protegrin-1, a Broad-spectrum Antimicrobial Peptide from Porcine Leukocytes. Chem Biol. 1996;3:543–550. doi: 10.1016/s1074-5521(96)90145-3. [DOI] [PubMed] [Google Scholar]
- Fernandez-Carneado J, Van Gool M, Martos V, Castel S, Prados P, De Mendoza J, Giralt E. Highly efficient, nonpeptidic oligo-guanidinium vectors that selectively internalize into mitochondria. J Am Chem Soc. 2005;127:869–874. doi: 10.1021/ja044006q. [DOI] [PubMed] [Google Scholar]
- Fischer R, Fotin-Mleczek M, Hufnagel H, Brock R. Break on through to the other side-biophysics and cell biology shed light on cell-penetrating peptides. Chem Bio Chem. 2005;6:2126–2142. doi: 10.1002/cbic.200500044. [DOI] [PubMed] [Google Scholar]
- Frankel AD, Pabo CO. Cellular Uptake of the Tat Protein from Human lmmunodeficiency Virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- Freites JA, Tobias DJ, von Heijne G, White SH. Interface connections of a transmembrane voltage sensor. Proc Natl Acad Sci U S A. 2005;102(42):15059–15064. doi: 10.1073/pnas.0507618102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. Arginine-rich Peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J Biol Chem. 2001;276(8):5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- Glukhov E, Stark M, Burrows LL, Deber CM. Basis for selectivity of cationic antimicrobial peptides for bacterial versus mammalian membranes. J Biol Chem. 2005;280(40):33960–33967. doi: 10.1074/jbc.M507042200. [DOI] [PubMed] [Google Scholar]
- Gratton J-P, Yu J, Griffith JW, Babbitt RW, Scotland RS, Hickey R, Giordano FJ, Sessa WC. Cell-permeable peptides improve cellular uptake and therapeutic gene delivery of replication-deficient viruses in cells and in vivo. Nat Med. 2003;9:357–362. doi: 10.1038/nm835. [DOI] [PubMed] [Google Scholar]
- Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55(6):1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- Gullion T, Schaefer J. Rotational-echo double resonance NMR. J Magn Reson. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Guo Q, Zhao G, Hao F, Guan Y. Effects of the TAT peptide orientation and relative location on the protein transduction efficiency. Chem Biol Drug Des. 2012;79(5):683–690. doi: 10.1111/j.1747-0285.2011.01315.x. [DOI] [PubMed] [Google Scholar]
- Haack T, Peczuh MW, Salvatella X, Sanchez-Quesada J, de Mendoza J, Hamilton AD, Giralt E. Surface recognition and helix stabilization of a tetraaspartate peptide by shape and electrostatic complementarity of an artificial receptor. J Am Chem Soc. 1999;121:11813–11820. [Google Scholar]
- Herce HD, Garcia AE. Molecular dynamics simulations suggest a mechanism for translocation of the HIV-1 TAT peptide across lipid membranes. Proc Natl Acad Sci U S A. 2007;104(52):20805–20810. doi: 10.1073/pnas.0706574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M. Resonance assignment of 13C/15N labeled proteins by two- and three-dimensional magic-angle-spinning NMR. J Biomol NMR. 1999;15:1–14. doi: 10.1023/a:1008334204412. [DOI] [PubMed] [Google Scholar]
- Hong M. Oligomeric structure, dynamics, and orientation, of membrane proteins from solid-state NMR", Structure. Structure. 2006;(14):1731–1740. doi: 10.1016/j.str.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Hong M, Fritzsching KJ, Williams JK. Hydrogen-Bonding Partner of the Proton-Conducting Histidine in the Influenza M2 Proton Channel Revealed From 1H Chemical Shifts. J Am. Chem. Soc. 2012a doi: 10.1021/ja307453v. ASAP online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Su Y. Structure and dynamics of cationic membrane peptides and proteins: insights from solid-state NMR. Protein Sci. 2011;20(4):641–655. doi: 10.1002/pro.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Zhang Y, Hu F. Membrane protein structure and dynamics from NMR spectroscopy. Annu Rev Phys Chem. 2012b;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu KN, Yau WM, Tycko R. Detection of a transient intermediate in a rapid protein folding process by solid-state nuclear magnetic resonance. J Am Chem Soc. 2010;132(1):24–25. doi: 10.1021/ja908471n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huster D, Yao XL, Hong M. Membrane protein topology probed by H-1 spin diffusion from lipids using solid-state NMR spectroscopy. J Am. Chem. Soc. 2002;124(5):874–883. doi: 10.1021/ja017001r. [DOI] [PubMed] [Google Scholar]
- Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Frequency selective heteronuclear dipolar recoupling in rotating solids: Accurate 13C-15N distance measurements in uniformly 13C,15N labeled peptides. J Am Chem Soc. 2001;123(3507–3519) doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- Jing X, Yang M, Kasimova MR, Malmsten M, Franzyk H, Jorgensen LFC, Nielsen HM. Membrane adsorption and binding, cellular uptake and cytotoxicity of cell-penetrating peptidomimetics with α-peptide/β-peptoid backbone: Effects of hydrogen bonding and α-chirality in the β-peptoid residues. Biochim Biophys Acta. 2012;1818(11):2660–2668. doi: 10.1016/j.bbamem.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Khafagya E-S, Morishitaa M, Isowab K, Imaib J, Takayamaa K. Effect of cell-penetrating peptides on the nasal absorption of insulin. J Contolled Release. 2009;133(2):103–108. doi: 10.1016/j.jconrel.2008.09.076. [DOI] [PubMed] [Google Scholar]
- Letoha T, Gaál S, Somlai C, Czajlik A, Perczel A, Penke B. Membrane translocation of penetratin and its derivatives in different cell lines. J Mol Recognit. 2003;16:272–279. doi: 10.1002/jmr.637. [DOI] [PubMed] [Google Scholar]
- Li L, Vorobyov I, Allen TW. Potential of Mean Force and pKa Profile Calculation for a Lipid Membrane-Exposed Arginine Side Chain. J Phys Chem B. 2008;112(32):9574–9587. doi: 10.1021/jp7114912. [DOI] [PubMed] [Google Scholar]
- Li S, Hong M. Protonation, Tautomerization, and Rotameric Structure of Histidine: A Comprehensive Study by Magic-Angle-Spinning Solid-State NMR. J Am. Chem. Soc. 2011;133(5):1534–1544. doi: 10.1021/ja108943n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Su Y, Luo W, Hong M. Water-Protein Interactions of an Arginine-Rich Membrane Peptide in Lipid Bilayers Investigated by Solid-State Nuclear Magnetic Resonance Spectroscopy. J Phys Chem B. 2010;114(11):4063–4069. doi: 10.1021/jp912283r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW. Membrane Pores Induced by Magainin. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- Luo W, Hong M. Conformational Changes of an Ion Channel Detected Through Water–Protein Interactions Using Solid-State NMR Spectroscopy. J Am. Chem. Soc. 2010;132(7):2378–2384. doi: 10.1021/ja9096219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani F, Lindberg S, Langel U, Futaki S, Gräslund A. Mechanisms of cellular uptake of cell-penetrating peptides. 2011 doi: 10.1155/2011/414729. 414729 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangoni ML, Rinaldi AC, Di Giulio A, Mignogna D, Bozzi A, Barra D, Simmaco M. Structure-function relationships of temporins, small antimicrobial peptides from amphibian skin. Eur J Biochem. 2000;267:1447–1454. doi: 10.1046/j.1432-1327.2000.01143.x. [DOI] [PubMed] [Google Scholar]
- Mani R, Cady SD, Tang M, Waring AJ, Lehrer RI, Hong M. Membrane-dependent oligomeric structure and pore formation of a b-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc Natl Acad Sci USA. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim Biophys Acta. 1999;1462(1–2):1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K, Murase O, Tokuda H, Funakoshi S, N NF, Miyajima K. Orientational and Aggregational States of Magainin 2 in Phospholipid Bilayers. Biochemistry. 1994;33:3342–3349. doi: 10.1021/bi00177a027. [DOI] [PubMed] [Google Scholar]
- McDermott AE. Structure and dynamics of membrane proteins by magic angle spinning solid-state NMR. Annu Rev Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- Mitchell DJ, Steinman L, Kim DT, Fathman CG, Rothbard JB. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Pept. Res. 2000;56(5):318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- Persson D, Thorén PE, Nordén B. Penetratin-induced aggregation and subsequent dissociation of negatively charged phospholipid vesicles. FEBS letters. 2001;505(2):307. doi: 10.1016/s0014-5793(01)02843-5. [DOI] [PubMed] [Google Scholar]
- Renault M, Cukkemane A, Baldus M. Solid-state NMR spectroscopy of complex molecules. Angew Chem Int Ed. 2011;49:8346–8357. doi: 10.1002/anie.201002823. [DOI] [PubMed] [Google Scholar]
- Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- Rienstra CM, Hohwy M, Hong M. 2D and 3D 15N-13C-13C NMR chemical shift correlation spectroscopy of solids: assignment of MAS spectra of peptides. J Am Chem Soc. 2000;122(10979–10990) [Google Scholar]
- Rothbard JB, Jessop TC, Wender PA. Adaptive translocation: the role of hydrogen bonding and membrane potential in the uptake of guanidinium-rich transporters into cells. Adv Drug Deliv Rev. 2005;57(4):495. doi: 10.1016/j.addr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Roumestand C, Louis V, Aumelas A, Grassy G, Calas B, Chavanieu A. Oligomerization of Protegrin-1 in the Presence of DPC Micelles. A Proton High-resolution NMR Study. FEBS Lett. 1998;421:263–267. doi: 10.1016/s0014-5793(97)01579-2. [DOI] [PubMed] [Google Scholar]
- Rouser G, Nelson GJ, Fleischer S, Simon G. Biological Membranes. 2nd ed. Vol. 1. London, New York: Academic Press; 1968. pp. 5–69. [Google Scholar]
- Sanchez-Quesada J, Seel C, Prados P, de Mendoza J, Dalcol I, Giralt E. Anion helicates: double strand helical self-assembly of chiral bicyclic guanidinium dimers and tetramers around sulfate templates. J Am Chem Soc. 1996;118(1):277–278. [Google Scholar]
- Schmidt N, Mishra A, Lai G, Wong G. Arginine-rich cell-penetrating peptides. FEBS Letters. 2009 doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- Schmidt NW, Tai KP, Kamdar K, Mishra A, Lai GH, Zhao K, Ouellette AJ, Wong GC. Arginine in α-defensins: differential effects on bactericidal activity correspond to geometry of membrane curvature generation and peptide-lipid phase behavior. J Biol Chem. 2012;287(26):21866–21872. doi: 10.1074/jbc.M112.358721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug KA, Lindner W. Noncovalent binding between guanidinium and anionic groups: focus on biological- and synthetic-based arginine/guanidinium interactions with phosph[on]ate and sulf[on]ate residues. Chem Rev. 2005;105:67–114. doi: 10.1021/cr040603j. [DOI] [PubMed] [Google Scholar]
- Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In Vivo Protein Transduction: Delivery of a Biologically Active Protein into the Mouse. Science. 1999;285(5433):1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- Shai Y, Oren Z. From "Carpet" Mechanism to de-novo Designed Diastereomeric Cell-selective Antimicrobial Peptides. Peptides. 2001;22(10):1629–1641. doi: 10.1016/s0196-9781(01)00498-3. [DOI] [PubMed] [Google Scholar]
- Sperandeo P, Dehò G, Polissi A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim Biophys Acta. 2009;1791:594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Steiner H, Hultmark D, Engstrom Å, Bennich H, Boman HG. Sequence and specificity of two antimicrobial proteins involved in insect immunity. Nature. 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- Su Y, DeGrado WF, Hong M. Orientation, dynamics, and lipid interaction of an antimicrobial arylamide investigated by 19F and 31P solid-state NMR spectroscopy. J Am Chem Soc. 2010a;132(26):9197–9205. doi: 10.1021/ja103658h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Doherty T, Waring AJ, Ruchala P, Hong M. Roles of Arginine and Lysine Residues in the Translocation of a Cell-Penetrating Peptide from 13C, 31P, and 19F Solid-State NMR. Biochemistry. 2009;48(21):4587–4595. doi: 10.1021/bi900080d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Hong M. Conformational disorder of membrane peptides investigated from solid-state NMR line widths and line shapes. J Phys Chem B. 2011;115(36):10758–10767. doi: 10.1021/jp205002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Mani R, Doherty T, Waring AJ, Hong M. Reversible Sheet-Turn Conformational Change of a Cell-Penetrating Peptide in Lipid Bilayers Studied by Solid-State NMR. J Mol Biol. 2008a;381(5):1133–1144. doi: 10.1016/j.jmb.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Mani R, Hong M. Asymmetric Insertion of Membrane Proteins in Lipid Bilayers by Solid-State NMR Paramagnetic Relaxation Enhancement: A Cell-Penetrating Peptide Example. J Am Chem Soc. 2008b;130(27):8856–8864. doi: 10.1021/ja802383t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Waring AJ, Ruchala P, Hong M. Membrane-bound dynamic structure of an Arginine-rich cell-penetrating peptide, the protein transduction domain of HIV TAT, from solid-state NMR. Biochemistry. 2010b;49(29):6009–6020. doi: 10.1021/bi100642n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Waring AJ, Ruchala P, Hong M. Structures of β-Hairpin Antimicrobial Protegrin Peptides in Lipopolysaccharide Membranes: Mechanism of Gram Selectivity Obtained from Solid-State Nuclear Magnetic Resonance. Biochemistry. 2011;50(12):2072–2083. doi: 10.1021/bi101975v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundlass NK, Raines RT. Arginine residues are more effective than lysine residues in eliciting the cellular uptake of onconase. Biochemistry. 2011;50(47):10293–10299. doi: 10.1021/bi200979k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A, Wu Z, Tucker K, Yang D, Lu W, Lubkowski J. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci. 2006;15:2749–2760. doi: 10.1110/ps.062336606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takechi Y, Tanaka H, Kitayama H, Yoshii H, Tanaka M, Saito H. Comparative study on the interaction of cell-penetrating polycationic polymers with lipid membranes. Chem Phys Lipids. 2011;165(1):51–58. doi: 10.1016/j.chemphyslip.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Tang M, Hong M. Structure and mechanism of b-hairpin antimcrobial pepetides in lipid bilayers from solid-state NMR spectroscopy. Molecular BioSystems. 2009;(5):317–322. doi: 10.1039/b820398a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Waring AJ, Hong M. Phosphate-Mediated Arginine Insertion Into Lipid Membranes and Pore Formation by a Cationic Membrane Peptide from Solid-State NMR. J Am Chem Soc. 2007;129(37):11438–11446. doi: 10.1021/ja072511s. [DOI] [PubMed] [Google Scholar]
- Tang M, Waring AJ, Hong M. Arginine dynamics in a membrane-bound cationic beta-hairpin peptide from solid-state NMR. Chembiochem. 2008a;9(9):1487–1492. doi: 10.1002/cbic.200800005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Waring AJ, Lehrer RI, Hong M. Effects of guanidinium-phosphate hydrogen bonding on the membrane-bound structure and activity of an arginine-rich membrane peptide from solid-state NMR spectroscopy. Angew, Chem Int Ed Engl. 2008b;47(17):3202–3205. doi: 10.1002/anie.200705993. [DOI] [PubMed] [Google Scholar]
- Thundimadathil J, Roeske RW, Guo L. Effect of Membrane Mimicking Environment on the Conformation of a Pore-Forming (xSxG)6 Peptide. Biopolymers. 2006;84:317–328. doi: 10.1002/bip.20470. [DOI] [PubMed] [Google Scholar]
- Torchilin VP, Rammohan R, Weissig V, Levchenko TS. TAT peptide on the surface of liposomes affords their efficient intracellular delivery even at low temperature and in the presence of metabolic inhibitors. Proc Natl Acad Sci U S A. 2001;98:8786–8791. doi: 10.1073/pnas.151247498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu Rev Biophys Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmschneider MB, Ulmschneider JP. Membrane adsorption, folding, insertion and translocation of synthetic trans-membrane peptides. J. Membr. Biol. 2008;25(3):245–257. doi: 10.1080/09687680802020313. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- Vorobyov I, Li L, Allen TW. Assessing atomistic and coarse-grained force fields for protein-lipid interactions: the formidable challenge of an ionizable side chain in a membrane. J Phys Chem B. 2008;112(32):9588–9602. doi: 10.1021/jp711492h. [DOI] [PubMed] [Google Scholar]
- Weissig V, D'Souz GG. Organelle-Specific Pharmaceutical Nanotechnolog. Chapter 22. Hoboken, New Jersey: John Wieley & Sons, Inc; 2012. p. 403. [Google Scholar]
- Wender PA, Galliher WC, Goun EA, Jones LR, Pillow TH. The design of guanidinium-rich transporters and their internalization mechanisms. Advanced Drug Delivery Reviews. 2008;60(4–5):452. doi: 10.1016/j.addr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: Peptoid molecular transporters. Proc Natl Acad Sci USA. 2000;97(24):13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annual Reviews of Biophysics and Biomolecular Structure. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Hong T, Waring A, Lehrer RI, Hong M. Solid-state NMR Investigations of Peptide-lipid Interaction and Orientation of a Beta-sheet Antimicrobial Peptide, Protegrin. Biochemistry. 2002;41(31):9852–9862. doi: 10.1021/bi0257991. [DOI] [PubMed] [Google Scholar]
- Yao XL, Schmidt-Rohr K, Hong M. Medium- and Long-Distance 1H–13C Heteronuclear Correlation NMR in Solids. J Magn Reson. 2001;149(1):139–143. [Google Scholar]
- Ye J, Fox SA, Cudic MR EM, Lauer JL, Fields GB, Terentis AC. Determination of penetratin secondary structure in live cells with Raman microscopy. J Am Chem Soc. 2010;132(3):980. doi: 10.1021/ja9043196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman MR, Yount NY. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol Rev. 2003;55(1):27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- Yoo J, Cui Q. Does arginine remain protonated in the lipid membrane? Insights from microscopic pKa calculations. Biophys J. 2008;94(8):L61. doi: 10.1529/biophysj.107.122945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci USA. 1987;54:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech SG, Wand AJ, McDermott AE. Protein structure determination by high-resolution solid-state NMR spectroscopy: application to microcrystalline ubiquitin. J Am Chem Soc. 2005;127:8618–8626. doi: 10.1021/ja0503128. [DOI] [PubMed] [Google Scholar]
- Zhang W, Smith SO. Mechanism of penetration of Antp(43–58) into membrane bilayers. Biochemistry. 2005;44:10110–10118. doi: 10.1021/bi050341v. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Doherty T, Li J, Lu W, Barinka C, Lubkowski J, Hong M. Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR. J Mol Biol. 2010a;397(2):408–422. doi: 10.1016/j.jmb.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu W, Hong M. The membrane-bound structure and topology of a human α-defensin indicate a dimer pore mechanism for membrane disruption. Biochemistry. 2010b;49(45):9770–9782. doi: 10.1021/bi101512j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Blatter XL, Seelig A, Seelig J. Protein Transduction Domains of HIV-1 and SIV TAT Interact with Charged Lipid Vesicles. Binding Mechanism and Thermodynamic Analysis. Biochemistry. 2003;42(30):9185–9194. doi: 10.1021/bi0346805. [DOI] [PubMed] [Google Scholar]