Abstract
Calcium-binding photoproteins have been discovered in a variety of luminous marine organisms [1]. Recent interest in photoproteins from the phylum Ctenophora has stemmed from cloning and expression of several photoproteins from this group [2-5]. Additional characterization has revealed unique biochemical properties found only in ctenophore photoproteins, such as inactivation by light. Here we report the cloning, expression, and characterization of the photoprotein responsible for luminescence in the deep-sea ctenophore Bathocyroe fosteri. This animal was of particular interest due to the unique broad color spectrum observed in live specimens [6]. Full-length sequences were identified by BLAST searches of known photoprotein sequences against Bathocyroe transcripts obtained from 454 sequencing. Recombinantly expressed Bathocyroe photoprotein (BfosPP) displayed an optimal coelenterazine-loading pH of 8.5, and produced calcium-triggered luminescence with peak wavelengths closely matching the 493nm peak observed in the spectrum of live Bathocyroe fosteri specimens. Luminescence from recombinant BfosPP was inactivated most efficiently by UV and blue light. Primary structure alignment of BfosPP with other characterized photoproteins showed very strong sequence similarity to other ctenophore photoproteins and conservation of EF-hand motifs. Both alignment and structural prediction data provide more insight into the formation of the coelenterazine-binding domain and the probable mechanism of photoinactivation.
Keywords: calcium-activated photoproteins, ctenophore, bioluminescence
Introduction
Bioluminescence has been observed in many marine taxa [1]. In most organisms, light emission occurs via oxidation of a luciferin catalyzed by a protein intrinsic to the organism, not involving bacteria. When the protein coordinates with the luciferin first, and then requires the addition of a cofactor for light emission, the term “photoprotein” is used [7]. In cnidarians and ctenophores, divalent calcium ions bind to the photoprotein, inducing a conformational change that triggers oxidation of the bound luciferin coelenterazine, which subsequently leads to light emission. Several cnidarian genes encoding photoproteins have been cloned, expressed, optically characterized, and adapted for functional applications [8-13]. More recently, the first photoproteins from ctenophores have also been cloned and characterized [2-5]. These recent investigations have revealed that ctenophore photoproteins have previously unobserved biochemical characteristics, most notably their sensitivity to light.
This sensitivity to light, also described as photoinactivation, is characterized by the loss of luminescence activity upon exposure to light. Photoinactivation of photoproteins was first described in purified native photoprotein from the ctenophores Mnemiopsis (“mnemiopsin”) and Beroe (“berovin”) [14,15], and was also observed in recombinant Beroe photoprotein [4]. However, to date there has been no investigation focused on understanding the relation of this unique property to the unique elements in ctenophore photoprotein peptide sequences.
Here, we report the cloning of a photoprotein from the deep-sea lobate ctenophore Bathocyroe fosteri (BfosPP) using a novel bioinformatic approach. Characterization of recombinant BfosPP revealed that its optical characteristics match those of the native photoprotein in vivo, and shares the photoinactivation property found in ctenophore photoproteins. Computational prediction of BfosPP structure provides a more complete understanding of ctenophore photoproteins and insights into the nature of photoinactivation.
Materials and Methods
Preparation of cDNA , Sequencing, and Cloning
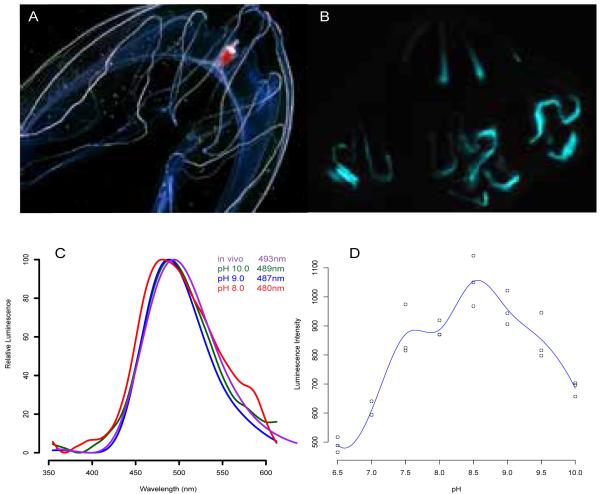
Live Bathocyroe fosteri (Figure 1A) were collected between 450 and 600 meters depth in Monterey Bay using remotely operated vehicles, and dark-adapted for bioluminescent images (Figure 1B) and spectral measurements (Figure 1C). RNA was extracted from frozen tissue using the RNeasy kit (Qiagen) and used to make cDNA libraries as previously described [16]. This material was used for 454 sequencing (Duke University). 454 shotgun reads were assembled into contiguous sequences using the de novo assembler NEWBLER (Roche) and uploaded onto a local BLAST database for homology-based searching. Berovin (GenBank Accession# 2HPK_A) and mnemopsin (GenBank Accession# ADD70248) amino acid sequences were used to search for photoprotein-like sequences. Primers with cloning restriction sites were designed to amplify photoproteins found in assembled contigs. Forward primer with XhoI restriction site 5′-CCT CCT CGA GAT GCC TAT TGA and reverse primer with NotI site 5′-CCT CGC GGC CGC TTA GTA TTT (IDT). First-strand cDNA synthesized from RNA served as the template for PCR and subsequent cloning into an expression vector described here [5].
Figure 1.
(A) Light image of Bathocyroe fosteri, (B) bioluminescence image taken without light. (C) Effect of pH on luminescence spectra: in vivo from live ctenophores (violet), and recombinant BfosPP regenerated at pH 10.0 (green), 9.0 (blue), and 8.0 (red); (D) Luminescence activity regenerated vs. pHs, optimum at 8.5.
Expression and Purification from E. coli cells
Sanger sequencing confirmed full length in-frame clones. Apophotoprotein (photoprotein lacking bound coelenterazine substrate) was expressed and purified as previously described [5]. Each step of purification was analyzed by polyacrylamide gel electrophoresis under denaturing conditions.
Luminescence Activity and Spectral Analysis
In vivo bioluminescent emission spectra were captured using an Ocean Optics QE65000 spectrometer with the attached fiber optic positioned exactly on the ctenophore upon mechanical stimulation. The Ocean Optics program SpectraSuite was used to collect the emission spectra.
The following conditions were used for luminescence assays: to load the protein with coelenterazine (“regenerate”), purified apophotoprotein was added to 50mM tris-HCl buffer pH 8.5 with 450mM NaCl, 5mM EDTA (Ambion), and 1mg/1ml coelenterazine in DMSO (Fisher) and incubated at 4°C for 16 hours in the dark. To determine the optimal pH for regeneration, this protocol was repeated for buffer adjusted in 0.5 increments using 10 N HCl over a range of 6.5 to 10. To measure activity, 200mM calcium acetate (Fisher) pH 6.8 was injected into aliquots of regenerated photoprotein and luminescent intensity was recorded with a custom-built photomultiplier-tube-based integrating sphere operated with a custom-built LabView user interface. Luminescence spectra were measured using a Roper Scientific back-illuminated CCD camera mounted to an Acton/Princeton Instruments SpectraPro monochromator. Emission spectra were collected using WinSpec software and exported to R where data were normalized and graphed using a spline curve-fit analysis. Protein absorbance was measured using 1mL UV-transparent plastic cuvettes in a Tecan Infinite 200 plate reader.
Photoinhibition Assays
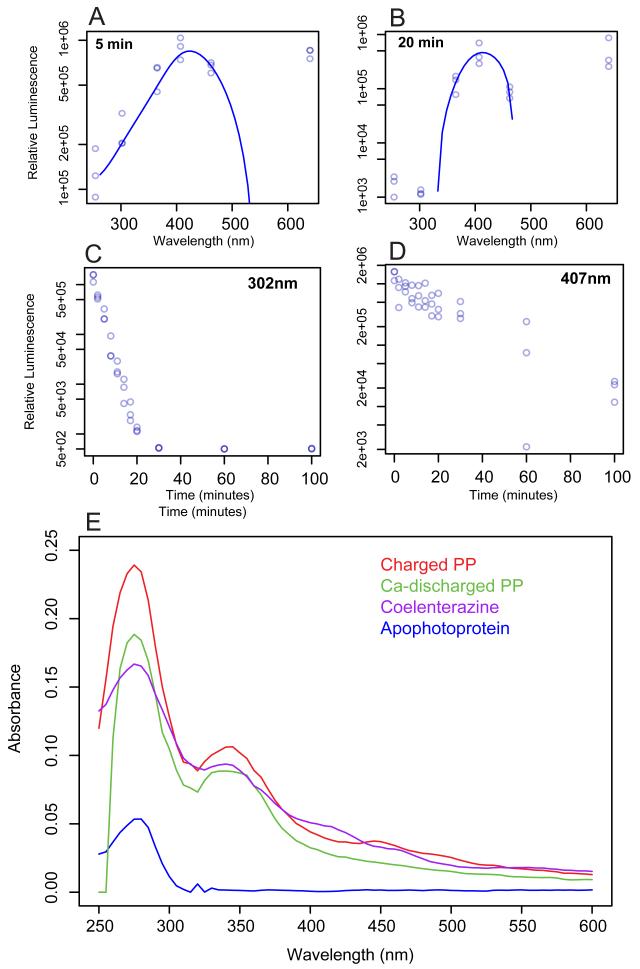
Regenerated photoprotein was aliquotted into a transparent 96-well plate on ice. Initial luminescence production was measured without prior exposure to light using a custom-built integrating sphere. The photoprotein was then exposed to light from LEDs emitting at maximal wavelengths of 254, 302, 365, 407, 462, and 640 nm with bandwidths between 2-10nm. Triplicate time increments were obtained for each wavelength. The integrated luminescence emission was used for analysis. Prior to each assay, the amount of energy per cm2 of each excitation source was measured using a digital power meter (Newport Instruments). The number of photons per second for each wavelength was calculated and reported in Figure 3.
Figure 3.
Photoinactivation of BfosPP at six wavelengths at (A) 5 minutes(B) 20 minutes, and over 100 minutes of exposure to light with maximum wavelengths of (C) 302nmand (D) 407nm. Calculated photons/sec/cm2 for each light source (×1014): 1.202 (254nm), 1.261 (302nm), 1.763 (365nm), 1.639 (407nm), 1.860 (462nm), 2.706 (640nm). (E) Absorption spectra for apophotoprotein without coelenterazine, charged with coeleneterazine, after calcium-discharge, and coelenterazine.
Bioinformatic Analysis
Sequences from previously cloned photoproteins were retrieved from NCBI using the following GenBank accession numbers: Clytin-I AB360787, Obelin U07128, Mitrocomin L31623, Aequorin (A.victoria) AY601106, Aequorin (A.coerulescens) AY236998, Bolinopsin CS447621, Berovin CS050690, Mnemiopsin-1 GQ231544, Mnemiopsin-2 GQ884175. Full-length coding sequences were translated and aligned using MAFFT with BLOSUM62 scoring matrix [17]. Conserved EF-hand motifs were identified in BfosPP by searching with the Pfam database (http://pfam.sanger.ac.uk/).
To estimate the best-fit model for the photoprotein gene phylogeny, we used the likelihood ratio test to determine the optimal substitution model. Likelihood scores were generated using PAUP* version 4.0a125 [18] and values for HKY85+⌈+I served as the null hypothesis. Scores were compared by hand. The best fit model was TN93+⌈, a sub model of GTR+⌈, where ⌈ = gamma distributed rates across sites. Trees were then constructed using PHYML plug-in [19] for Geneious Pro version 5.5.6 and 1000 bootstrap replicates were computed. In the final unrooted tree, only nodes showing support greater than 50% were labeled.
For protein structure analyses, the full-length amino acid sequence of BfosPP was uploaded to the I-TASSER server online platform (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) for both 3-dimensional structure and ligand-binding site predictions [20,21]. Models with highest confidence scores were downloaded as PDB files and analyzed in PyMol v.1.3 viewer.
Results
Identification of photoprotein sequences and expression
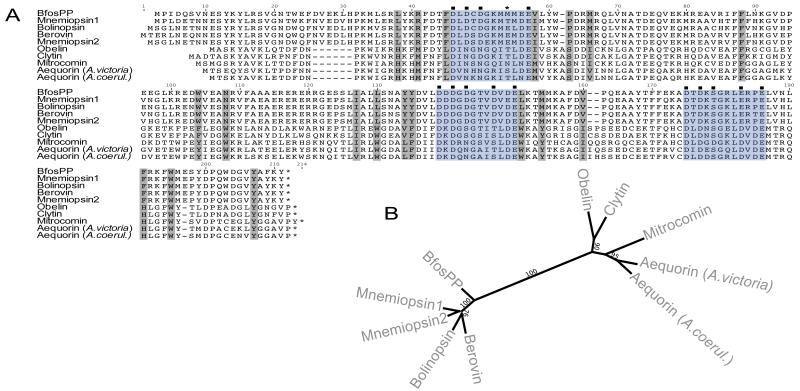
A total of 67894 reads of “Titanium” 454 sequencing were assembled into 8241 contigs with an average length of 300 base pairs after trimming. BLAST searches of assembled contigs using both berovin and mnemiopsin-1 as queries identified one contig with a full-length open reading frame of 206 amino acids. We have adopted the naming scheme presented in [5] to naming this protein BfosPP, where “Bfos” refers to the genus and species, and “PP” indicates a photoprotein. When its peptide sequence was aligned with those of other known cnidarian and ctenophore photoproteins (Figure 2), BfosPP showed highest amino acid sequence identity to mnemiopsin-1 at 78.6% and the lowest to aequorin at 43%. The amino acid sequence for Aequorea green fluorescent protein was also used to query Bathocyroe contigs, but no significant matches were found. Recombinant BfosPP was expressed in E. coli as soluble protein with an N-terminal 8-histidine and maltose binding protein (MBP), and subsequently cleaved using TEV protease to release untagged apoprotein with a single glycine residue preceding its N-terminal methionine.
Figure 2.
(A) Amino-acid alignment of calcium-activated photoproteins from ctenonphores and cnidarians. EF-hand motifs I, II, IV shaded in blue, and hypothesized coelenterazine-binding sites in grey. Calcium-binding residues predicted by I-Tasser denoted by (■). Non-conservative substitution in BfosPP calcium-binding domain denoted by (*). (B) Phylogenetic tree of calcium-activated photoprotein genes using maximum likelihood with 1000 bootstap replicates.
Characterization
Determination of Luminescence Activity and Spectral Analysis
Once live B. fosteri were dark adapted, they produced blue-green light along their radial canals upon mechanical stimulation (Figure 1B). If live animals had been previously exposed to ambient light, they did not luminescence. The in vivo luminescence spectrum taken from a live dark-adapted individual had a peak wavelength 493nm (Figure 1C), which is at the high-wavelength end of the 30nm range previously reported [6]. Spectra for recombinant BfosPP regenerated and measured in buffer of pH 8, 9, and 10 varied only 9 nm from shortest to longest wavelength (480-489 nm). Relative intensity was normalized in order to compare only differences in color with regard to pH. These observations clearly show the luminescence signal to be green-blue; however, there was no “shoulder” observed in these spectra characteristic of fluorescent proteins.
BfosPP exhibited the highest luminescence intensity when the regeneration environment was pH 8.5 (Figure 1D) and when the buffer contained NaCl concentration similar to molarity of seawater (500 mM; results not shown). Between pH 7.5 and 9.5, the blue flash after calcium addition could be seen by eye. At pH 7.0 the intensity was too weak for the spectrometer to read.
We analyzed the absorbance spectra for three forms of BfosPP: purified photoprotein without coelenterazine (“apoPP”), photoprotein incubated with coelenterazine (“charged PP”), photoprotein after calcium has been added (“discharged PP), and that of coelenterazine (Figure 3E). The peak at 280nm was seen in each sample as is characteristic of aromatic amino acids found in proteins. A second major absorbance peak at 350nm, corresponding to coelenterazine, was observed in the spectrum of charged BfosPP, but not of apo-BfosPP, indicating that the recombinant protein had indeed been successfully loaded with the coelenterazine substrate.
Photoinactivation Assays
Following previous work suggesting that unlike those derived from cnidarians, ctenophore photoproteins are inactivated when exposed to light [14,15], we analyzed activity of coelenterazine-charged BfosPP after irradiation by individual wavelength bands across the UV and visual spectrum. BfosPP activity decreased over time across all wavelengths of light tested, except red light at 640 nm (Figure 3A,B). The rate of inactivation differed between wavelengths: Luminescence activity was almost gone (~1% remaining) after 20 minutes of exposure to the two shortest wavelengths 302 and 254nm compared to 407 nm where of 8% of original activity was observed after 60 minutes of exposure. Only the 407nm- and 640nm-exposed samples still produced light after 100 minutes, while all other samples were completely inactivated. Intensity of each LED was calculated to be the same order of magnitude (Figure 3).
Bioinformatic Analysis
Sequence Analysis
We aligned BfosPP to photoproteins that have currently been cloned from cnidarians and ctenophores (Figure 2). Pfam identified three EF-hand motifs which are highly conserved in photoproteins and responsible for the characteristic helix-loop-helix structure found in many other calcium-binding proteins [22]. Within the canonical EF-hand motif, there are twelve residues that form the calcium binding loop (highlighted in the alignment) and positions 1,3,5,7,9 and 12 are usually aspartic or glutamic acid, while glycine is the 6th residue. Analysis of BfosPP reveals one interesting substitution not conserved in other ctenophore or cnidarian photoproteins. In EF-hand I, methionine replaces a glutamic acid present in all other ctenophore photoproteins (except mnemiopsin-1, in which threonine replaces the acidic residue). The net charge for all three EF-hands remains negative and probably not entirely disruptive. However, this substitution could be responsible for a weakened affinity to the calcium ion.
Amino acid residues involved in coelenterazine binding have been confirmed for aequorin [23] and obelin [24] but not yet for a ctenophore photoprotein. Binding residues from cnidarian photoproteins align to hypothesized residues for ctenophores (Figure 2) showing conserved hydrophobic amino acids that may form the hydrophobic binding pocket. Between ctenophore and cnidarian photoproteins, four positions in the putative coelenterazine-binding pocket display non-conservative substitutions. Two of these substitute hydrophobic amino acids for positively charged histidines found in cnidarian photoproteins, while the other two replace methionine and tryptophan (found in cnidarian photoproteins) with arginine and asparagine, respectively, leading to conservation of both net hydrophobicity and charge in the binding pocket.
Photoprotein Phylogeny
The photoprotein phylogeny was constructed using the full-length alignment of ten known photoprotein sequences (Figure 2B). In this unrooted tree, genes from each phylum group together with strong branch support (100%) and branch lengths suggest these groups are highly divergent. Within the ctenophore clade, there is moderate bootstrap support for bolinopsin clustering with berovin (76% support) instead of with the Mnemiopsis genes (64%). Interestingly, BfosPP branches outside the other ctenophore genes with very strong support (100%). The cnidarian photoproteins are also not monophyletic but exhibit much stronger branch support for each clade compared to the ctenophores (Figure 2B).
Structure and Binding-site Predictions
The top five structural models returned from the I-TASSER server had the best C-score of −1.12 and the lowest of −2.77. This score (typically from −5 to 2) represents the quality of each model where a higher score means higher confidence in the predicted model. The estimated accuracy of the top model was 0.57±0.14 (TM-score) and 7.8±4.4 Å (RMSD). Both are standards for measuring similarity between the predicted model and a native structure. TM scores above 0.5 signify model has correct topology whereas values less than 0.17 could mean random similarity. The top five best threading templates were obelin (PDB:1jf0), berovin (2hpKA), calmodulin-like protein (3kheA) and calexcitin (2ccmA).
Amino acid residues predicted to bind to the ligand C2-hydroperoxy-coelenterazine were reported by COFACTOR, the function-predicting algorithm implemented in I-TASSER. Aequorin (PDB: 1ej3) served as the template protein with a C-score of 0.24 and in this case, scores range from 0 to 1. Many of the predicted binding residues overlap with those originally hypothesized based on primary sequence alignment.
Discussion
Characterization of Recombinant Bathocyroe Photoprotein
Currently, only five calcium-activated photoproteins from four species of ctenophores have been cloned and expressed, including the first photoprotein from Bathocyroe fosteri reported here. With this recent addition, it is clear that photoproteins derived from ctenophores are biochemically distinct from the cnidarian photoproteins. We determined the optimal environment for coelenterazine binding and calcium-activated luminescence in BfosPP to be pH 8.5, with salt concentration similar to ocean molarity (500mM). These conditions are similar to the natural environment of Bathocyroe and to those published for berovin [4] and mnemiopsin-1,2 [2,3]. They differ markedly from those of cnidarian photoproteins, whose activity is highest at pH ~7.0 and without salt [7].
A previous study collected emission spectra from 18 Bathocyroe fosteri and found substantial variation (459-492nm) [6]. Our spectra were measured from only two animals, but in both cases the maximum emission wavelength was 493nm, the high end of the wavelength range previously recorded. This indicates that unlike cnidarians such as Aequorea victoria, wavelength variation in the luminescent emission of the ctenophore Bathocyroe fosteri is not caused by energy transfer from its photoprotein to an associated fluorescent protein. Instead, this wavelength variation could be partially explained by changes in the pH of the microenvironment within the photocyctes (light-emitting cells), since we also observed the emission spectra of recombinant BfosPP to be slightly blue-shifted at lower pH. In this study, we were not able to measure luminescence spectra below pH 8.0 due to the lower luminescence intensity of the photoprotein in that pH range. However we speculate that within the photocytes, the native photoprotein may be stable at lower pHs due to components in the cellular environment not replicated under our in vitro experimental conditions, enabling the emission spectrum to shift toward shorter wavelengths while maintaining high luminescence activity.
Photoinactivation
Rapid inactivation of coelenterazine-charged BfosPP by UV-and 462nm light matches results for native mnemiopsin reported nearly four decades ago [15]. In that study, inactivation was attributed to the absorption of coelenterazine, while our results suggest that it is mostly caused by UV-absorption by the protein. The absorbance spectra for all three forms of BfosPP have the same UV-absorption peak at 280nm corresponding to the protein, as well as a 345 nm peak corresponding to coelenterazine for all but the apophotoprotein. We have also observed that when apo-BfosPP is exposed to light, activity is not diminished if it is subsequently regenerated with coelenterazine in the dark. These results suggest that the apoprotein itself is not changed by light exposure, but rather that light exposure alters the conformation of coelenterazine in the binding pocket such that the protein is unable to emit light. Alternatively, excited state energy from UV photons absorbed by aromatic amino acids proximal to the bound coelenterazine may participate Förster resonance energy transfer to the coelenterazine, leading to photo-oxidation. Consistent with both of these hypotheses, the photoinactivation reaction was found to be reversible when recombinant BfosPP and native purified photoprotein were re-incubated with coelenterazine.
Anctil and Shimomura [25] discuss the mechanism of photoinactivation in Mnemiopsis and suggest, based on the narrow pH range of activity, that binding of coelenterazine is dependent on a highly ionizable group. They found that oxygen was needed for coelenterazine to bind and regenerate photoinactivated native purified mnemiopsin [25]. They propose that light splits both coelenterazine and oxygen from the photoprotein, inactivating coelenterazine in the process. Our results are consistent with this hypothesis since we did not observe a coelenterazine absorbance peak in the spectrum of photoinactivated BfosPP (results not shown).
Structure and Sequence Analysis
I-TASSER uses multiple solved structures as templates to generate predicted structures of input peptide sequences based on primary sequence alignment and secondary structure prediction. Since only one crystal structure is known for the group of ctenophore photoproteins, I-TASSER used other known photoproteins and calcium-binding proteins. The structural model predicted for BfosPP is similar to other photoproteins despite the differences in primary sequence. Ctenophore photoproteins are divergent from cnidarians, with the substitution rate along the dividing branch close to saturation. However, both groups have evolved the same overall structure and oxidize the same substrate. Interestingly, BfosPP does not group with the other lobate ctenophore Bolinopsis infundibulum, which is found at similar depths in the ocean. In fact, BfosPP is most similar to mnemiopsin-1, from another lobate ctenophore found in warmer shallow water. A single-gene tree of only 5 sequences is not enough to clarify these differences, but more sequence information from a variety of ctenophores may provide better insight into the evolution of these proteins.
Interestingly, previous mutagenesis experiments targeting the obelin (cnidarian) active site have identified key residues involved in coelenterazine binding that are substituted in ctenophores [26,27]. Some of the residues involved in stabilizing coelenterazine in obelin (Tyr138, His175, Trp179, Tyr190) and in generating or stabilizing the excited state (His22, Trp92) [26] are non-conservatively replaced in BfosPP. Most notably His175 is replaced by the non-hydrogen-bonding phenylalanine (Table 1). When previous mutagenesis experiments in aequorin replaced the same histidine residue with alanine, phenylalnine or tryptophan, all luminescence activity was lost [28]. Thus replacement of this histidine in ctenophore photoproteins might be partly responsible for photoinactivation, since there is no residue available for hydrogen bonding with Tyr202 (numbered according to BfosPP) which stabilizes the 2-hydroperoxy group of bound coelenterazine, where decarboxylation occurs [26]. Even though previous experiments with aequorin reported loss of activity upon substitution of histidine, it is possible these experiments were not performed in the dark, and therefore loss of activity may have been due to subsequent photoinactivation. In addition to this single mutation, the overall hydrophobicity of the active site in ctenophores was calculated to be significantly less than cnidarians [9]. This increase in residues available for hydrogen bonding may also destabilize the binding of coelenterazine. However, we note that residues highlighted as forming the coelenterazine binding cavity (Figure 3A) are based on only cnidarian photoprotein experiments. Further mutagenesis experiments must be completed on ctenophore photoproteins to better understand the mechanism of photoinactivation.
Table 1.
Amino acid residues involved in coelenterazine binding. Numbering of ctenophore residues according to alignment in Figure 2. Shaded residues have been identified in the active site of obelin and are non-conservative substitutions in ctenophores.
| I-TASSER predicted |
Ctenophores | Cnidarians |
|---|---|---|
| Y | Leu38 | His |
| Y | Arg41 | Met |
| Y | Phe45 | Leu |
| Y | Val/Ile58 | Ile/Met |
| Y | Phe63 | Ser |
| N | Met66 | Ile/Val |
| Y | Phe88 | Phe |
| Y | Trp104 | Phe/Tyr |
| Y | Asn108 | Trp |
| N | Ile127 | Ile/Val |
| Y | Leu130 | Trp |
| Y | Tyr134 | Val/Leu |
| Y | Tyr135 | Phe |
| Y | Leu151 | Trp |
| Y | Met154 | Tyr |
| N | Val160 | Ile |
| N | Phe191 | His |
| Y | Trp195 | Trp |
| N | Tyr207 | Tyr |
Supplementary Material
Figure S1. Stereoview for 3-dimensional model of BfosPP predicted by I-Tasser server with bound peroxy-coelenterazine.
Highlights.
-
-
We have expressed and characterized the first photoprotein from Bathocyroe fosteri
-
-
Emission spectra of BfosPP was at the upper limit of previous in vivo observations
-
-
This photoprotein is inactivated when exposed to certain wavelengths of light
-
-
There are non-conservative substitutions in the putative binding-pocket of ctenophores
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Refrences
- [1].Haddock SHD, Moline MA, Case JF. Bioluminescence in the Sea. Annu. Rev. Marine. Sci. 2010;2:443–93. doi: 10.1146/annurev-marine-120308-081028. [DOI] [PubMed] [Google Scholar]
- [2].Jafarian V, Sariri R, Hosseinkhani S, Aghamaali M-R, Sajedi RH, Taghdir M, Hassannia S. A unique EF-hand motif in mnemiopsin photoprotein from Mnemiopsis leidyi: Implication for its low calcium sensitivity. Biochemical and Biophysical Research Communications. 2011;413:164–70. doi: 10.1016/j.bbrc.2011.08.022. [DOI] [PubMed] [Google Scholar]
- [3].Aghamaali M-R, Jafarian V, Sariri R, Molakarimi M, Rasti B, Taghdir M, Sajedi RH, Hosseinkhani S. Cloning, Sequencing, Expression and Structural Investigation of Mnemiopsin from Mnemiopsis leidyi: An Attempt Toward Understanding Ca2+-Regulated Photoproteins. Protein J. 2011;30:566–74. doi: 10.1007/s10930-011-9363-8. [DOI] [PubMed] [Google Scholar]
- [4].Markova SV, Burakova LP, Golz S, Malikova NP, Frank LA, Vysotski ES. Light-sensitive photoprotein berovin from the bioluminescent ctenophore Beroe abyssicola: A novel type of Ca2+-regulated photoprotein. FEBS Journal. 2012;279:856–70. doi: 10.1111/j.1742-4658.2012.08476.x. [DOI] [PubMed] [Google Scholar]
- [5].Schnitzler CE, Pang K, Powers MP, Reitzel AM, Ryan JF, Simmons D, Tada T, Park M, Gupta J, Brooks SY, Blakesley RW, Yokoyama S, Haddock SHD, Martidale MQ, Baxevanis AD. Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes. BMC Biology. 2012 doi: 10.1186/1741-7007-10-107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Haddock SHD, Case JF. Bioluminescence spectra of shallow and deep-sea gelatinous zooplankton: ctenophores, medusae and siphonophores. Marine Biology. 1999;133:571–82. [Google Scholar]
- [7].Shimomura O. Bioluminescence: Chemical Principles And Methods. World Scientific; 2006. [Google Scholar]
- [8].Inouye S, Noguchi M, Sakaki Y, Takagi Y, Miyata T, Iwanaga S, Miyata T, Tsuji FI. Cloning and sequence analysis of cDNA for the luminescent protein aequorin. Proceedings of the National Academy of Sciences. 1985;82:3154–8. doi: 10.1073/pnas.82.10.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Inouye S, Tsuji FI. Cloning and sequence analysis of cDNA for the Ca2+-activated photoprotein, clytin. FEBS Letters. 1993;315:343–6. doi: 10.1016/0014-5793(93)81191-2. [DOI] [PubMed] [Google Scholar]
- [10].Fagan TF, Ohmiya Y, Blinks JR, Inouye S, Tsuji FI. Cloning, expression and sequence analysis of cDNA for the Ca2+-binding photoprotein, mitrocomin. FEBS Letters. 1993;333:301–5. doi: 10.1016/0014-5793(93)80675-k. [DOI] [PubMed] [Google Scholar]
- [11].Inouye S. Cloning, expression, purification and characterization of an isotype of clytin, a calcium-binding photoprotein from the luminous hydromedusa Clytia gregarium. J Biochem. 2008;143:711–7. doi: 10.1093/jb/mvn024. [DOI] [PubMed] [Google Scholar]
- [12].Inouye S, Sahara Y. Expression, purification and characterization of a photoprotein, clytin, from Clytia gregarium. Protein Expression and Purification. 2007;53:384–9. doi: 10.1016/j.pep.2006.12.014. [DOI] [PubMed] [Google Scholar]
- [13].Illarionov BA, Bondar VS, Illarionova VA, Vysotski ES. Sequence of the cDNA encoding the Ca 2+-activated photoprotein obelin from the hydroid polyp Obelia longissima. Gene. 1995;153:273–4. doi: 10.1016/0378-1119(94)00797-v. [DOI] [PubMed] [Google Scholar]
- [14].Ward WW, Seliger H. Properties of mnemiopsin and berovin, calcium-activated photoproteins from the ctenophores Mnemiopsis species and Beroe ovata. Biochemistry. 1974;13:1500–10. doi: 10.1021/bi00704a028. [DOI] [PubMed] [Google Scholar]
- [15].Ward WW, Seliger H. Action spectrum and quantum yield for the photoinactivation of mnemiopsin, a bioluminescent photoprotein from the ctenophore Mnemiopsis sp. Photochemistry and Photobiology. 1976;23:351–63. doi: 10.1111/j.1751-1097.1976.tb07260.x. [DOI] [PubMed] [Google Scholar]
- [16].Ewen-Campen B, Shaner N, Panfilio KA, Suzuki Y, Roth S, Extavour CG. The maternal and early embryonic transcriptome of the milkweed bug Oncopeltus fasciatus. BMC Genomics. 2011;12 doi: 10.1186/1471-2164-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Swofford DL. PAUP. Phylogenetic Analysis Using Parismony (* and Other Methods). Version 4. Sinauer Associates; Sunderland, Massachusetts: 2002. [Google Scholar]
- [19].Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- [20].Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:1–8. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nakayama S, Kretsinger RH. Evolution of the EF-hand family of proteins. Annual Review of Biophysics and Biomolecular Structure. 1994;23:473–507. doi: 10.1146/annurev.bb.23.060194.002353. [DOI] [PubMed] [Google Scholar]
- [23].Head J, Inouye S, Teranishi K, Shimomura O. The crystal structure of the photoprotein aequorin at 2.3 angstrom resolution. Nature. 2000;405:372–6. doi: 10.1038/35012659. [DOI] [PubMed] [Google Scholar]
- [24].Deng L, Markova SV, Vysotski ES, Liu Z-J, Lee J, Rose J, Wang B-C. Crystal structure of a Ca2+-discharged photoprotein: implications for mechanisms of the calcium trigger and bioluminescence. J Biochem. 2004;279:33647–52. doi: 10.1074/jbc.M402427200. [DOI] [PubMed] [Google Scholar]
- [25].Anctil M, Shimomura O. Mechanism of photoinactivation and re-activation in the bioluminescence system of the ctenophore Mnemiopsis. Biochemical Journal. 1984;221:269–272. doi: 10.1042/bj2210269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu Z-J, Stepanyuk GA, Vysotski ES, Lee J, Markova SV, Malikova NP, Wang B-C. Crystal structure of obelin after Ca2+-triggered bioluminescence suggests neutral coelenteramide as the primary excited state. Proc Natl Acad Sci USA. 2006;103:2570–5. doi: 10.1073/pnas.0511142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vysotski ES, Lee J. Ca2+-regulated photoproteins: structural insight into the bioluminescence mechanism. Acc. Chem. Res. 2004;37:405–15. doi: 10.1021/ar0400037. [DOI] [PubMed] [Google Scholar]
- [28].Ohmiya Y, Tsuji FI. Bioluminescence of the Ca2+-binding photoprotein, aequorin, after histidine modification. FEBS Letters. 1993;320:267–70. doi: 10.1016/0014-5793(93)80600-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Stereoview for 3-dimensional model of BfosPP predicted by I-Tasser server with bound peroxy-coelenterazine.