Summary
Global repression of protein synthesis occurs in many stresses and has been attributed primarily to inhibition of translation initiation, although this mechanism may not always explain the full extent of repression. Here, using ribosome footprinting, we show that two hours of severe heat stress triggers global pausing of translation elongation at around codon 65 on most mRNAs in both mouse and human cells. The genome-wide nature of the phenomenon, its location and features of protein N-termini suggested the involvement of ribosome-associated chaperones. Following severe heat shock, the Hsp70’s interactions with translational machinery were markedly altered and its association with ribosomes reduced. Pre-treatment with mild heat stress or overexpression of Hsp70 protected cells from heat shock-induced pausing, while inhibition of Hsp70 activity triggered elongation pausing without heat stress. Our findings suggest that regulation of translation elongation in general, and by chaperones in particular, represents a major component of cellular stress responses.
Introduction
The cellular response to stress involves changes to many levels of gene regulation, including transcription, mRNA processing, and translation (Biamonti and Caceres, 2009; Gibson, 2008; Holcik and Sonenberg, 2005). Stress response pathways play important developmental and evolutionary roles, contributing to developmental robustness in varying environments (Akerfelt et al., 2010; Gibson, 2008; Jarosz and Lindquist, 2010; Lindquist, 2009). The heat shock response is one of the best characterized stress response pathways, during which heat shock proteins (HSPs), a class of molecular chaperones, are up-regulated in response to widespread protein mis-folding (Richter et al., 2010). Mis-folding stress and the heat shock response pathway in particular play specific developmental roles and are implicated in a variety of diseases. Up-regulation of chaperones is frequently observed in cancer, and chaperone inhibitors hold promise as antitumor agents (Calderwood et al., 2006; Whitesell and Lindquist, 2005). A number of studies monitoring incorporation of labeled amino acids have observed that protein synthesis is globally inhibited under various types of stresses, including heat shock (Bouche et al., 1979; Lindquist, 1980).
Both conserved and lineage-specific mechanisms are used to repress protein synthesis generally, while facilitating the expression of HSPs and other stress related proteins (Lindquist, 1980, 1981; Miller et al., 1979). Various mechanisms leading to global translational repression and selective up-regulation have been reported, generally involving regulation of translation initiation. Primary ways in which cells inhibit translation initiation globally are repression of cap recognition downstream of the mTOR pathway and repression of ternary complex recycling through phosphorylation of the initiation factor eIF2α (reviewed by (Sonenberg and Hinnebusch, 2009; Spriggs et al., 2010)). Both of these pathways are known to mediate inhibition of translation initiation in the response to heat stress (Duncan and Hershey, 1984; Vries et al., 1997). However, the extent of eIF2α phosphorylation, eIF4E dephosphorylation and sequestration by 4EBP that occurs under heat stress is relatively mild, and it has been suggested that the regulation of initiation cannot explain the degree of translational repression observed (Spriggs et al., 2010). Hence, there are still questions about how the full extent of translational repression is achieved during heat shock.
Here we investigated the genome-wide regulation of translation in response to heat shock using ribosome footprint profiling (Ingolia et al., 2009; Ingolia et al., 2011), which maps the locations of ribosomes on mRNAs at nucleotide resolution. Our analysis revealed widespread changes in translational regulation, including an unexpected regulatory response, in which translation elongation is globally paused after translation of ~ 65 amino acids. Exploration of the mechanism underlying this mode of translational regulation pointed to involvement of the Hsp70 family of chaperones.
Results
Ribosomes accumulate in the first 200 bases of mRNAs after severe heat shock
To better understand translational regulation genome-wide during chronic and acute heat stress in mammalian cells, we used ribosome footprint profiling to globally map the locations of individual ribosomes along mRNAs (Ingolia et al., 2009; Ingolia et al., 2011), in conjunction with RNA-Seq to assess mRNA abundance. Ribosome footprint and RNA-Seq libraries were prepared from NIH 3T3 mouse fibroblasts under normal growth conditions (Control, 37° C), and following 8 hours of mild (42° C, HS8M, chronic heat stress) or 2 hours of severe heat stress (44° C, HS2S, acute stress) (Fig. S1A-E). Neither of these conditions induced significant cell death, and cells appeared to fully recover 24 hours after withdrawal from heat stress (Fig. S1F). Changes in levels of total and ribosome-associated mRNA were observed for many genes following both mild and severe heat stress (Fig. S1A). As expected, heat shock proteins (HSPs) showed a significant upregulation in their translation levels under both conditions, as measured by footprint RPKM (Fig. S1G-I). Analytical polysome profiling revealed a substantial decrease in the proportion of heavy polysomes and the accumulation of monosomes in both mild and severe heat stress (Fig. 1A), consistent with previous observations and with a global reduction in translation initiation (McCormick and Penman, 1969). Notably, lighter polysome fractions representing 2-4 ribosomes actually increased somewhat in severe heat stress (Fig. 1A), discussed further below.
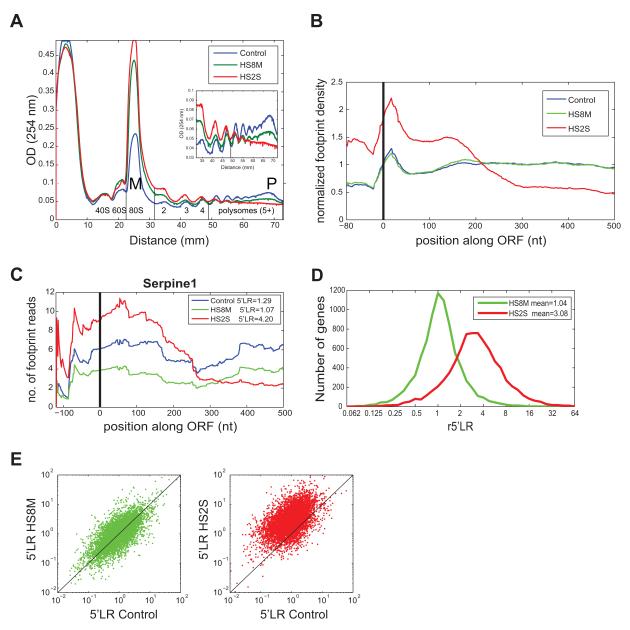
Figure 1. Heat shock induces a global increase in ribosome occupancy around 5′ ends of open reading frames.
(A) Polysome profiles of mouse fibroblasts (3T3 cells) in normal growth conditions (blue) or following mild heat shock (42° for 8 hours, green) or severe but not lethal heat shock (44° for 2 hours, red). The polysome region was defined as 5somes and higher. P:M (Polysome to Monosome) ratio is 0.79 in Control, 0.18 in HS8M and 0.08 in HS2S. Profile is representative of three replicate experiments, where relative P:M ratios in HS8M were 22.5% ± 0.9% of Control values and relative P:M ratios in HS2S were 7.2% ± 2.7% of Control. (B) Normalized footprint density along mRNAs (see Experimental procedures). (C) Raw footprint count per position (smoothed) across the Serpine1 transcript (ENSMUST00000041388, 5′ UTR and first 500 nt of the transcript are shown). (D) Distribution of r5′LR values in HS8M (green) and HS2S (red). Population means are indicated. (E) Changes in 5′LR values following heat shock. See Figure S1 and Table S1 for more details.
The distribution of ribosome footprint sequence reads along individual mRNAs, or in a “metagene” analysis of reads mapping to all mRNAs, revealed a more complex picture (Fig. 1B, 1C). While changes in response to 8 hours of mild heat shock were generally modest, dramatic changes in relative ribosome occupancy occurred in response to two hours of severe heat shock in four distinct regions. These included increases in the 5′ UTR and the “initiation region” (reads indicating positioning of the ribosomal P site in the first 15 bases of the ORF), and a ~1.7-fold increase in the next ~180 nucleotides (nt) after the initiation region, which shifted to a ~1.7-fold decrease in the remainder of the ORF (Fig. 1B, Fig. S1J). The overall shape of this distribution remained unchanged whether gene-level normalization was used (as in Fig. 1B, Fig. S1J and K) or simply plotting raw read counts (Fig. S1L). Many individual genes which had sufficiently high read coverage displayed a distribution similar to the metagene distribution, as illustrated for Serpine1 (Fig. 1C, additional examples are shown in Fig. S1M). Similar biases in relative ribosome occupancy following severe heat shock were observed in a biological replicate experiment described below. These wholesale changes in ribosome occupancy along mRNAs imply major shifts in translational regulation in response to heat shock. The accumulation of ribosomes in the ~200 nt after the initiation site relative to the remainder of the ORF was particularly notable because it implied extensive post-initiation regulation of translation. Such a possibility has been noted (Ballinger and Pardue, 1983) but not specifically characterized previously in cellular responses to stress. We therefore chose to focus our efforts in this study on characterizing this phenomenon in detail and exploring its potential causes and consequences.
To objectively measure the extent of ribosome accumulation in the 5′ ends of ORFs, we defined the 5′ loading ratio (5′LR) of a gene as the ratio of the footprint read density between bases 16-195 (codons 6-65) of the ORF to the density along the remaining downstream positions in the ORF. This measure, which is analogous to measures used to quantify transcriptional pausing (Rahl et al., 2010), reflects the extent to which ORF-associated ribosomes are preferentially accumulated or depleted at the 5′ end of an ORF. To assess the effect of heat stress on translation of individual mRNAs, we calculated the relative loading ratio, r5′LR, defined as the ratio of the 5′LR under heat stress to that in control conditions. Following mild heat stress, no significant increase in 5′LR was observed in the metagene analysis or for most individual genes: r5′LR values were centered around 1.0 or slightly above (Fig. 1B, 1D, 1E). The distribution of 5′LR values increased dramatically following severe heat stress, with a mean r5′LR value above 3 (Fig. 1D). The increase in 5′LR in severe heat stress was apparent for most individual mRNAs examined (Fig. 1E and Table S1), indicating a global change in the translational machinery.
Translation elongation is transiently paused in severe heat stress
Multiple regulatory mechanisms might produce such a global increase in 5′LR, including: (i) transient (reversible) pausing of translation elongation around codon 65 on most of the gene’s mRNAs (resulting in accumulation of ribosomes 5′ of the pause); (ii) irreversible stalling of translation elongation around codon 65 on a subset of the gene’s mRNAs (also resulting in accumulation of ribosomes upstream); (iii) acceleration of translation elongation after codon 65; or (iv) premature termination of translation around codon 65 on a subset of a gene’s messages. To discriminate between these potential explanations for the increase in 5′LR after severe heat shock, we performed analytical polysome profiling following inhibition of translation initiation using the drug harringtonine, which blocks initiation after subunit joining by preventing elongation during the first rounds of peptide bond formation (Huang, 1975; Ingolia et al., 2011). Since treatment with harringtonine inhibits newly initiating monosomes while allowing previously engaged ribosomes to continue translating through termination, this drug enables the fates of elongating ribosomes to be followed under different conditions.
As expected, under control conditions, treatment with harringtonine resulted in a decrease in heavy polysomes and an increase in monosomes that was readily apparent after 1 minute and more complete after 3 minutes (Fig. 2A). Under severe heat shock conditions, one minute of harringtonine treatment had only a minimal effect on the polysome profile, but three minute treatment resulted in a distinct reduction in heavy polysomes, suggesting that most ribosomes are not irreversibly stalled (Fig. 2A, Fig. S2A). An acceleration of translation following codon 65 would predict more rapid rather than slower collapse of polysomes in heat shock conditions, and premature termination would also not be expected to slow the collapse of polysomes. The slower kinetics of this collapse are therefore most consistent with the model in which ribosomes are transiently paused during translation elongation in heat shock conditions.
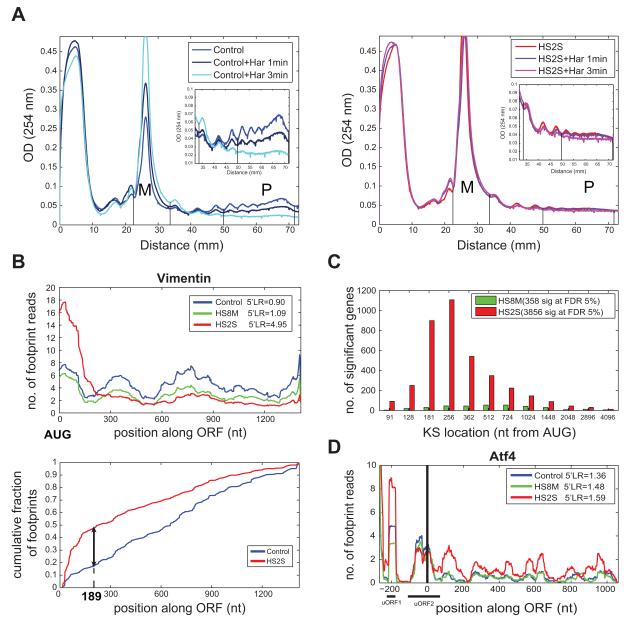
Figure 2. Ribosomes are paused after translating about 65 amino acids in severe heat shock.
(A) Polysome profiles of harringtonine treated Control (left graph, Control) and severe heat shock (right graph, HS2S) treated cells. Heat shocked or control cells were harvested after treatment with 0.1mM harringtonine for 1 minute (dark blue and purple lines for Control and HS2S respectively) or 3 minutes (cyan and pink lines for Control and HS2S respectively) or without any treatment (blue and red lines). P:M ratios are: 0.8 in Control, 0.33 in Control+Har 1min, 0.07 in Control+Har 3min, 0.10 in HS2S, 0.09 in HS2S+Har 1 min, and 0.05 in HS2S+Har 3min. A replicate experiment showed a similar polysome collapse (Figure S2A). (B) Illustration of the KS location and p-value calculation for Vimentin (ENSMUST00000028062). Cumulative plots of reads along the ORF (bottom panel) are shown for Control (blue) and HS2S (red) (see Experimental Procedures). (C) Distribution of the KS locations for all unique translated genes which were found to be significantly paused (at an FDR of 5%) in HS8M relative to Control (green) and in HS2S relative to Control (red). The number of significant genes is indicated, out of 5125 genes. (D) Raw footprint counts (smoothed) along the Atf4 transcript (ENSMUST00000109605) show an escape from the global pause. uORF positions are marked. See Figure S2 and Table S2 for more details.
To confirm that the reduced collapse rate of polysomes under heat stress after Harringtonine treatment was due to altered elongation rates, we used the translational inhibitor puromycin, which causes dissociation of ribosomes that are actively translating. Treatment with puromycin yielded a similar reduction in polysomes in heat stressed cells as under control conditions (Fig. S2B), indicating that under severe heat stress, as in control conditions, ribosomes are translating and are neither irreversibly stalled nor prematurely terminating. Together, these data support that the slowed kinetics of polysome collapse result from altered elongation rates.
Ribosome pausing was centered around codon 65 based on the metagene analysis, but such an averaged representation could hide variability between genes. To characterize this phenomenon in more depth, we calculated the inferred location and statistical significance of shifts in ribosome footprint density along individual genes using a modified Kolmogorov-Smirnov (KS) test. We reasoned that, if translation of an mRNA is paused in heat shocked cells, ribosomes should accumulate upstream of the pause, so the density of footprint reads should be significantly higher between the AUG and the location of the pause relative to the remainder of the ORF; this can be determined by the KS test (Supplemental Experimental Procedures). For example, this test identified a highly significant shift in footprint density at nucleotide 189 (codon 63) of the Vimentin ORF, a position we call the “KS location”, suggesting that ribosomes pause in this vicinity and accumulate upstream (Fig. 2B).
Genome-wide KS analysis indicated that 75% of the translated mRNAs had a significant elongation pause under severe heat shock conditions at a false discovery rate (FDR) cutoff of 5%. The location detected by KS analysis beyond which footprint density began to plateau mostly occurred between 197 and 466 nt from the initiation codon (25th and 75th percentiles, Figs. 2C and S2C, Table S1), with a median estimated position of 263 nt. Thus, individual gene analysis indicated an elongation pause in the first few hundred bases for the majority of mouse mRNAs in response to severe heat shock, a phenomenon we call “5′ ribosomal pausing”. A ribosome paused in the first couple of hundred bases of an ORF might be expected to cause accumulation of a few ribosomes upstream, likely running as a light polysome, consistent with the observed increase in the light polysome fraction seen in severe heat shock (Fig. 1A).
While our analyses suggested that the vast majority of genes experience 5′ ribosome pausing in severe heat shock, the increase in 5′LR was not universal. One gene set of interest was the set of transcripts that were not significantly paused (based on KS test). Functional enrichment analysis of this set detected enrichment for transcriptional regulators (GO term “transcription factor activity”, p = 3.5 × 10−6, FDR cutoff of 1%), which might contribute to the transcriptional response to heat shock. We used a more stringent set of criteria to define “escapers” as mRNAs whose 5′ LR did not significantly change (or decreased) following severe heat shock (Supplemental Experimental Procedures). We did not observe HSPs to be among the escapers, suggesting that the up-regulation of translation of HSPs noted above (Fig. S1G-I) has other causes, e.g., increased mRNA levels and/or increased translation initiation. However, these criteria did identify 187 mRNAs that escaped translational pausing, including several transcription factors (Table S2). Interestingly, the Atf4 and Atf5 transcription factor genes were both among the escapers, with r5′LR values of 1.18 and 1.05, respectively, far below the mean value of around 3 (Fig. 2D, Fig. S2D). ATF5 is translationally induced following a variety of stresses, including heat shock (Zhou et al., 2008). The homologous ATF4 factor is a major regulator of oxidative, endoplasmic reticulum (ER) and amino acid starvation stress (Harding et al., 2000), but has not been previously connected to the heat shock response. Interestingly, both of these factors are known to be translationally up-regulated under stress conditions via a mechanism involving translation of upstream open reading frames (uORFs) (Harding et al., 2000; Zhou et al., 2008). Our observations suggest that escape from the global elongation and initiation blocks are used to regulate distinct subsets of genes.
Ribosome association of Hsp70 chaperones is altered during heat shock
We next sought to explore potential mechanisms underlying the general phenomenon of 5′ ribosomal pausing. The near-universal scope and relatively specific location of the pause suggested the involvement of a general aspect of translational elongation, which might be dependent more on the nascent peptide than gene-specific properties. Nascent peptides are bound by a variety of factors such as SRP (discussed below) and Hsp70 chaperones (Kramer et al., 2009), whose regulation is also the hallmark of the heat shock response. The cytosolic Hsp70 (70 kilodalton (kda) heat shock protein) chaperones HSC70 and HSP70 – the constitutive and inducible forms of the cytosolic Hsp70, respectively – are among the major chaperones in the cell that mediate protein folding. During translation, these proteins associate with ribosomes and interact with nascent chains as they emerge from the ribosome exit tunnel (Beckmann et al., 1990; Nelson et al., 1992). This function of hsp70 family proteins has been shown to be important for cell growth both in yeast and in mammals (Jaiswal et al., 2011; Nelson et al., 1992). Heat stress places a substantial burden on chaperones as a result of the unfolding of existing cytosolic proteins and, in response, cells up-regulate stress-specific homologs of these chaperones, including HSP70 (encoded by the Hspa1a and Hspa1b genes in mouse), which supplements the protein levels of the constitutive HSC70, encoded by Hspa8.
To assess the association of Hsp70 chaperones with the ribosome following heat shock, we fractionated polysome gradients from control or heat shocked cells, and measured the amount of HSC70/HSP70 that co-migrated with ribosomal proteins using a pan-HSP70 antibody. We detected a consistent 2- to 4-fold reduction in the association of HSC70/HSP70 with monosomes and polysomes in severe heat shock (Fig. 3A and 3B). Relatively little co-migration of HSC70/HSP70 was observed in the polysomal portion of the gradient following dissociation of ribosomes by EDTA treatment, supporting the specificity of the detected association (Fig. S3A). Significant depletion of HSC70/HSP70 was detected in several fractions ranging from monosomes to 5-somes (Fig. 3B). mRNAs with ribosomes paused at typical locations (around 200 nt) are expected to migrate mostly as lighter polysomes or monosomes, depending on whether additional ribosomes accumulate upstream of the pause. Reduced association of Hsp70 chaperones with translating ribosomes could affect the ribosome in various ways, including the exposure of nascent chains.
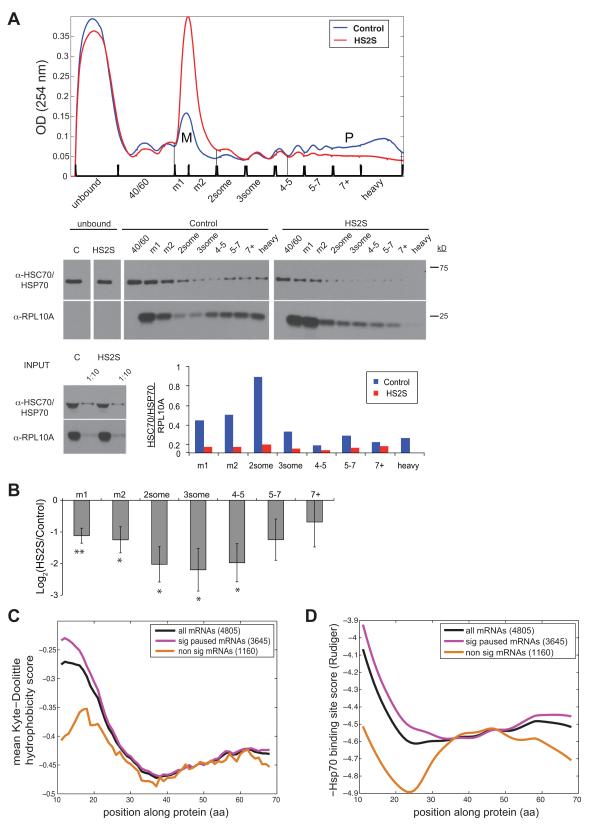
Figure 3. Association of HSC70 with polysomes is altered following heat shock.
(A) Western blot of HSC70/HSP70 (using pan-HSP70 antibody, see Experimental procedures for details) protein levels (middle panel) across a polysome gradient in Control and severe heat shock cells (HS2S, top panel). RPL10A was used to assess ribosome abundance in each fraction (middle panel). Levels of comigrating HSC70/HSP70 protein and RPL10A protein were measured using ImageJ software. Ribosomal association of HSC70/HSP70 was quantified by normalizing the amount of HSC70/HSP70 protein by the amount of RPL10A protein in each fraction (HSC70/HSP70 / RPL10A) for both control (blue bars) and heat shock (red bars) conditions. P:M ratio is 1.9 in Control and 0.18 in HS2S. Profile is representative of six experiments, where relative P:M ratios of HS2S were 8.5% ± 2.9% of Control. (B) Quantification and significance of the reduction in HSC70/HSP70 ribosome association following heat shock. Mean and standard error of the ratio (log2) of HSC70/HSP70 association (HSC70/HSP70 signal/RPL10A signal), in HS2S to control conditions, in each fraction, over 5 biological replicate experiments. Fractions in which HSC70/HSP70 and/or RPL10A signal was undetectable were excluded from analysis (see Methods for details). Significance of reduction in HSC70 ribosomal association is given by t-test. * indicates p < 0.05; ** indicates p < 0.01.
(C) Hydrophobicity scores (see Supplemental Experimental procedures) of the first 70 amino acids encoded by all highly translated mRNAs (in black), of significantly paused mRNAs (in magenta) and of non-significantly paused mRNAs (in orange). The background representing the overall average Kyte-Doolittle hydrophobicity of all highly translated mRNAs (grey dashed line) is shown for reference. T-test p-value for the difference in the mean hydrophobicity values of the first 25 amino acids between all significantly paused mRNAs and non-significant mRNAs was 3.8 × 10−7. (D) Hsp70 binding site scores (Supplemental Experimental Procedures) were plotted as in (C). T-test p-value for the difference in the mean Hsp70 scores of the first 25 amino acids between all significantly paused mRNAs and non-significant mRNAs was 5.3 × 10−6.
See also Figure S3.
Significantly paused mRNAs have features associated with Hsp70 interaction
HSC70/HSP70 bind nascent chains generally, but have higher affinity for hydrophobic peptides, and hydrophobic peptides are expected to be more reliant on interaction with HSC70/HSP70 for proper folding (Hartl et al., 2011). The first 20-25 residues of proteins tend to be more hydrophobic than the rest of the protein, particularly for proteins containing a signal peptide (von Heijne, 1981), but also to a lesser extent for other proteins (Fig. 3C and S3C). We found that genes with significant 5′ ribosome pausing encoded more hydrophobic N-termini than did non-significantly paused genes (Fig. 3C, t-test p-value = 3.8 × 10−7). Hydrophobicity of the N-terminus also differed between mRNAs grouped by their KS location. Those with KS locations between codons 44-171 were associated with higher hydrophobicity of the N-terminus than the (smaller) sets of messages with more distally or more proximally located pause sites (Fig. S3E), suggesting that amino terminal hydrophobicity contributes to pausing in this region. Hsp70 is known to have a particularly high affinity for stretches of five hydrophobic residues flanked by up to four positively charged amino acids on each side (Rudiger et al., 1997). Analysis of the binding site content of protein N-termini using the scoring system proposed by Rudiger and colleagues revealed that significantly paused mRNAs had higher average Hsp70 interaction scores, reflecting greater propensity for Hsp70 binding (Fig. 3D), suggesting that these peptides would be more dependent on Hsp70 binding. Consistently, the N-termini of non-significantly paused mRNAs encoded significantly fewer strong Hsp70 binding sites (Fig. S3G). mRNAs that encoded signal peptides exhibited a similar degree of 5′ ribosome accumulation as mRNAs that did not (Fig. S3D), and differences in hydrophobicity between paused and non-paused mRNAs were observed for mRNAs with and without signal peptides (Fig. S3F), suggesting that this regulation occurs independently of the secretory pathway.
Both the location of this hydrophobic stretch at the N-terminus, and the higher hydrophobicity of significantly paused mRNAs are consistent with pausing being influenced by nascent peptides, around where they might be expected to first require interaction with chaperones.
Modulation of chaperone activity regulates translation elongation pausing
To test the potential role of Hsp70 chaperones in heat shock induced elongation pausing, we first analyzed thermotolerance. Chaperone expression is induced by mild heat shock (Kelley and Schlesinger, 1978; Lindquist, 1980, 1981) (Fig. S3B), explaining why mild heat shock treatment prior to severe shock has protective effects on cells in the phenomenon known as thermotolerance (Gerner and Schneider, 1975; Henle et al., 1978). Thermotolerance is dependent on protein synthesis and enables enhanced translation under severe heat shock conditions (Lindquist, 1980; Petersen and Mitchell, 1981). To assess how pre-induction of chaperones influences ribosome density patterns, we performed a thermotolerance experiment, pre-treating cells with 8 hours of mild heat shock prior to 2 hours of severe heat shock, along with a biological replicate of the control, mild and severe treatments. Pre-treatment of cells with mild heat shock resulted in partial rescue of the polysome collapse that occurred during severe heat shock (Fig. 4A), consistent with the expected relief from translational inhibition. Replicate ribosome footprint data yielded metagene profiles under control, mild and severe heat shock conditions (Fig. 4B) similar to those observed in Figure 1. Remarkably, pre-treatment with mild heat shock almost completely rescued the 5′ ribosome pausing observed after 2 hours of severe heat shock (Fig. 4B, Fig. S4A). These observations suggest that 5′ ribosomal pausing is regulatable and is reduced by factors induced by mild heat stress such as chaperones.
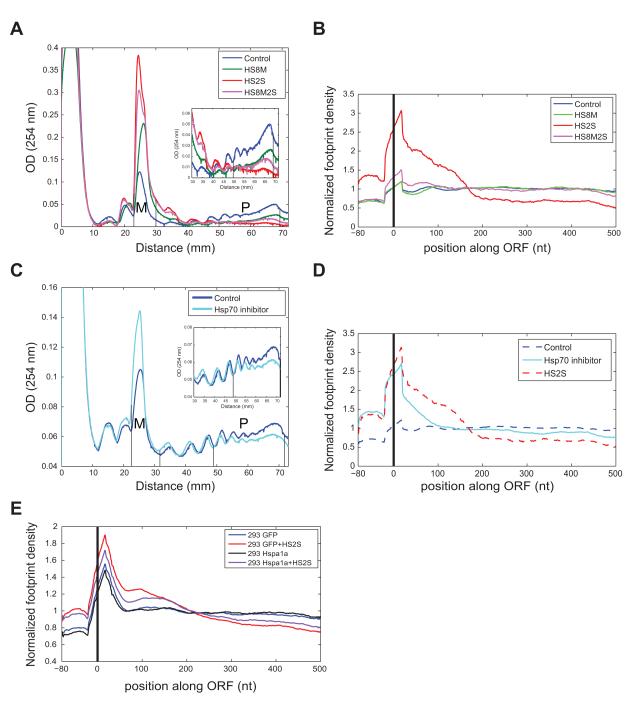
Figure 4. Modulation of chaperone activity regulates translation elongation pausing.
(A) Polysome profiles of a thermotolerance experiment. 3T3 cells were treated with mild (green), severe (red), or mild prior to severe (magenta) heat shock, or left untreated (blue). P:M ratios are 1.4 in Control, 0.33 in HS8M, 0.1 in HS2S, and 0.2 in HS8M2S. Profile is representative of two replicate experiments, where relative P:M ratios were 22.4% ± 1.3% of Control in HS8M, 5.8% ± 1.7% of Control in HS2S, and 12.2% ± 2.7% of Control in HS8M2S. (B) Normalized footprint density along mRNAs for a biological replicate experiment and for cells pre-treated with mild heat shock (8 h at 42° C) prior to severe heat shock (magenta). (C) Polysome profiles of cells treated with the Hsp70 inhibitor VER-155008 (Massey et al., 2010) at a 20 μM concentration (Hsp70 inhibitor, cyan line) or DMSO (Control, blue line) for 3hrs. P:M ratios are 1.52 in Control and 0.7 in Hsp70 inhibitor. Profile is representative of three replicate experiments, where relative P:M ratios for Hsp70 inhibitor were 46.5% ± 3.8% of Control. (D) Normalized footprint density along mRNAs for control cells after 3 hours of VER-155008 treatment (cyan). Control and severe heat shock plots (dotted lines) are identical to those in B. (E) Normalized footprint density along mRNAs for 293T cells overexpressing GFP or Hspa1a, with or without 2 hours of severe heat shock.
See also Figure S4.
To more specifically look at the role of HSC7/HSP70 chaperones in the phenomenon, we turned to explore whether inhibition of Hsp70 activity under control conditions would inhibit translation elongation. To test this possibility, cells were subject to a short treatment of a small molecule drug, VER-155008, that inhibits the ATPase activity of both constitutive and inducible Hsp70 proteins, an activity that is required for their chaperone function (Massey et al., 2010). Treatment with the Hsp70 inhibitor for a short period led to a reproducible reduction in heavy polysomes and an accumulation of monosomes (Fig. 4C), supporting the direct involvement of hsp70 proteins in the control of translation in mammalian systems. Footprint profiling revealed a pronounced accumulation of ribosomes near the 5′ ends of mRNAs in cells treated with the drug for 3 hours in the absence of stress (Fig. 4D).
Analysis of individual mRNAs revealed that ~2500 mRNAs were significantly paused following treatment with Hsp70 inhibitor, the majority of which were also significantly paused under severe heat shock (overlap of 2072, p = 10−34) with a similar but somewhat broader distribution of inferred KS locations (Fig. S4C). Similar to what we observed under severe heat shock (Fig. 3C), mRNAs that were significantly paused following hsp70 inhibitor treatment encoded significantly more hydrophobic N-termini than non-paused mRNAs (p = 5 × 10−7, Fig. S4F). The inferred KS locations were in the vicinity of those seen under severe heat shock for 60% of the mRNAs, comparing the same mRNAs in both experiments (Fig. S4D, S4E), while the metagene analysis showed accumulation somewhat 5′ to that observed in heat shock. Differences between the effects of inhibitor treatment and severe heat shock on footprint profiles may reflect incomplete inhibition of hsp70 activity, or functions of hsp70s that do not require ATP hydrolysis. Similar to the trends observed in severe heat stress, more proximal KS locations correlated with greater N-terminal hydrophobicity, while more distal KS locations were associated with reduced N-terminal hydrophobicity (Fig. S4G).
To verify that Hsp70 chaperones directly effect ribosome accumulation and to explore this phenomenon in another species, we overexpressed the inducible HSP70 (Hspa1a) in human 293T cells. We first subjected 293T cells to 2 hours of severe heat stress. Following severe heat shock, 293T cells showed a similar 5′ ribosome accumulation to that observed in mouse 3T3 cells, with a similar pause location at around 200 nt into the ORF based on metagene analysis (Fig. 4E), of somewhat smaller magnitude than that seen in 3T3 cells. This observation extends the phenomenon of heat stress-induced 5′ ribosomal accumulation to a second mammalian species.
Ectopic expression of Hsp70s is generally inhibitory to cell growth (Feder et al., 1992), but we were able to achieve moderate levels of overexpression of HSP70, about 1.5-fold in excess over HSC70/HSP70 levels observed in normal growth conditions (Fig. S4H). We note that, as is commonly observed in transformed lines, 293T cells express both HSC70 and HSP70 under control conditions (Fig. S4H). HSP70 overexpressing cells were then subjected to 2 hours of severe heat shock. Strikingly, this moderate ectopic expression of Hsp70 resulted in a partial rescue of the elongation pausing following severe heat shock, as seen in Figure 4E (See also Fig. S4I and S4J). The observations that Hsp70 inhibitor treatment leads to 5′ ribosome pausing in the absence of stress, and that moderate overexpression of Hsp70 partially relieves heat shock induced elongation pausing, directly implicate Hsp70 chaperone in the regulation of translation elongation during severe heat shock.
The Hsp70-translational machinery interactome: characterization and stress-associated changes
Hsp70 chaperones are known to bind numerous proteins (Calloni et al., 2012; Kampinga and Craig, 2010). Given our results, it was of particular interest to characterize interactions between Hsp70s and the translation machinery. Consistent with a recent observation (Jaiswal et al., 2011) that the ribosomal cochaperone DNAJC2 (MPP11) does not influence the ribosomal association of HSC70 (the predominant Hsp70 protein in 3T3 cells in both control conditions and after 2 hours of severe heat shock), knockdown of this factor in 3T3 cells did not appreciably affect the distribution of ribosomes under normal growth conditions (data not shown).
To explore potential ways in which Hsp70 chaperones may influence elongation pausing, we next sought to characterize direct interactions between Hsp70s and the translation machinery using the high-throughput LUMIER with BACON assay (LUMIER with Bait control, Experimental Procedures) (Taipale et al., 2012). Using this assay, we examined the interaction of 504 clones representing a broad panel of 377 translation-related genes (Table S3) in a 293T cell line stably expressing Renilla-tagged HSC70. We found that HSC70 interacts with about half of these factors to varying extents (213 genes, above the 75th percentile of the estimated background luminescence, Fig. 5A), and with 109 of them strongly (at the 99.9th percentile of background, Figs. S5A and S5B). The known Hsp70 client proteins HSF-1 (Abravaya et al., 1992) and P53 (Zylicz et al., 2001) were among the top interactors (ranked 4 and 33 in their luminescence score), as were a handful of co-chaperones which were included as positive controls. We next asked whether these interactions changed in response to stress. The LUMIER assay is based on Renilla luciferase tagging, and Renilla luciferase activity is markedly reduced following heat shock (data not shown). Thus, for application of this method we instead treated cells with another proteotoxic stress, the proteasome inhibitor Bortezomib. Of course, some aspects of the cellular responses to Bortezomib and heat stress may differ. But the resulting interaction data showed a global reduction in the interaction of HSC70 with ribosomal proteins (Fig. S5C), paralleling the reduced ribosomal association of Hsp70 obtained with severe heat shock.
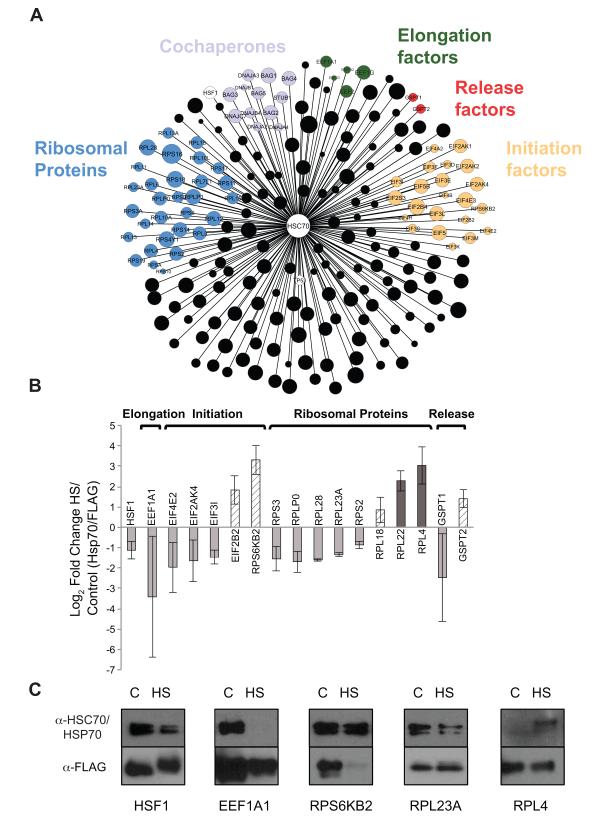
Figure 5. The Hsp70 chaperone – translation machinery interactome.
(A) HSC70-translational machinery interactome in a network representation, was drown using Cytoscape. Proteins are represented by circles, and the size of the circle corresponds to the HSC70 interaction score (log2([HSC70, luminescence]/[FLAG ELISA]). Different groups of interest are indicated. (B) Hsp70 interactome changes upon 2 hours of severe heat shock was tested using co-IP. Blots were quantified using ImageJ and shown are log2 of Hsp70 (interaction) normalized to FLAG (expression), of proteins that changed more than 1.5 fold. Striped bars represent proteins with increased normalized interaction and significant destabilization, defined as a significant reduction in FLAG levels – at least 3 fold in one replicate and at least 2 fold in the other. Dark grey bars represent increased interactions with Hsp70 with stable protein levels. Light grey bars represent decreased interactions without significant destabilization. Mean and standard deviation of two experiments is shown. (C) co-IP WB data for several proteins. See Supplemental Experimental procedures, Figure S5, Table S3 and Table S4 for more details.
To explore potential changes in the interactions of the endogenous Hsp70 chaperones with the translational machinery following heat stress, we used a lower-throughput co-immunoprecipitation assay. This assay was applied following 2 hours of severe heat shock to each of a set of 48 translation-related proteins, including primarily translation factors and ribosomal proteins (and several positive controls) representing a spectrum of basal HSC70 interaction scores (Figs. 5B, 5C, S5D and Table S4). In this assay, transfected FLAG tagged proteins were pulled down and protein expression quantified by anti-FLAG antibody, while interaction with the endogenous Hsp70s was measured using a pan-Hsp70 antibody. Because the FLAG tags were located at the C terminus, this assay measures interaction with mature rather than nascent proteins. Interaction scores were then calculated as the ratio of [Hsp70]/[FLAG], controlling for effects on expression. HSF-1, the heat shock transcription factor, had reduced interaction following heat stress (Fig. 5B and 5C), consistent with its phosphorylation, trimerization, and concomitant activation of its transcriptional activity (Bjork and Sistonen, 2010).
Heat shock is known to cause mis-folding of pre-existing proteins, which are then detected by Hsp70 and other chaperones for refolding. If the chaperones are unable to refold those proteins, they are targeted for degradation, thus preventing accumulation and aggregation of misfolded proteins in the cell (Hartl et al., 2011). We classified each of the tested proteins according to the change in its Hsp70 interaction score (increased or decreased), and protein levels (stable or destabilized, as measured by FLAG), following heat shock. Several proteins showed a large reduction in their overall protein levels while their interaction with Hsp70 chaperones remained unchanged or was elevated, resulting in an increase in their interaction scores (Fig. 5B). This group is expected to consist of proteins that become misfolded and degraded as a result of severe heat shock, and hence show an increase in Hsp70 interaction. Among those are the release factor GSPT2 (eRF3 subunit), and the initiation regulator RPS6KB2.
Only two proteins showed a consistent up-regulation in their interaction with Hsp70 without a respective decrease in expression, RPL22 and RPL4 (Fig. 5B). Interestingly, both these proteins protrude into the interior of the ribosome exit tunnel, and have been shown to play a role in gene specific elongation stalling in bacteria and eukaryotes (Wilson and Doudna Cate, 2012).
The largest class of changes included proteins that were relatively stable, and showed reduced Hsp70 interactions (Fig. 5B). We expect this group to reflect regulatory interactions. Hsp70 was found to interact with five elongation factors under normal growth conditions (Fig. 5A), out of which EEF1A1 was found to have reduced Hsp70 interactions during heat stress. Several ribosomal proteins also showed this type of reduced interaction with Hsp70 following heat shock, further supporting the reduced association of Hsp70 with ribosomes during heat stress. In particular, RPL23A, which is located on the outside of the exit tunnel, was downregulated in its interaction with Hsp70 following severe heat shock (Fig. 5B and 5C). This supports the notion that Hsp70 chaperones are present in proximity to the exit tunnel under normal growth conditions, and their association is reduced under severe heat shock.
Discussion
In this study we identify a novel aspect of the cellular response to heat stress, in which ribosomes accumulate at the 5′ ends of open reading frames of most mRNAs, apparently as a result of temporary pausing of translation elongation (Fig. 6). Under heat stress, cellular priorities shift from growth to cytoprotective functions, including prevention of protein unfolding and aggregation. Reduced HSC70/HSP70 association with ribosomes and altered interactions with the translational machinery were observed following 2 hours of severe heat stress. These changes may serve several functions, including reversible inhibition of translation elongation that, together with inhibition of translation initiation, may help to reduce cellular protein production and the associated burden on the cellular chaperone machinery. Reversibility of the elongation pause by induction of chaperones would allow cells to rapidly accelerate protein synthesis and growth once they have effectively adapted to the stress associated with protein mis-folding, or following a return to non-stress conditions.
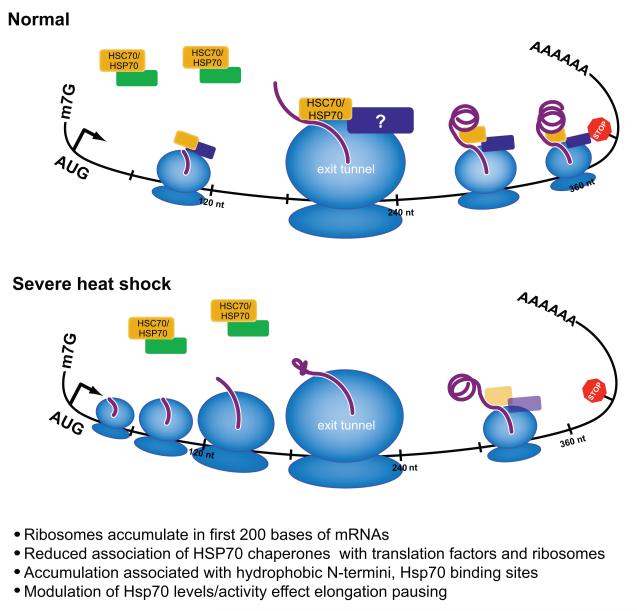
Figure 6. Heat shock induced translation elongation pausing is modulated by Hsp70 chaperones.
During severe heat shock, ribosomes often pause at 5′ ends of mRNAs after translating about 65 amino acids (Figs. 1, 2), resulting in accumulation of ribosomes upstream and depletion of ribosomes downstream of this point; Following severe heat shock, HSC70/HSP70 association with the ribosome is reduced (Fig. 3A, 3B). As Hsp70 proteins interact with nascent peptide chains as they emerge from the exit tunnel (Beckmann et al., 1990; Nelson et al., 1992), nascent chains might be exposed. More significantly paused mRNAs tend to encode N-termini with stronger Hsp70 requiring characteristic (Fig. 3C and 3D). Modulation of Hsp70 levels, by thermotolerance or overexpression, rescue heat shock induced elongation pausing, while inhibition of Hsp70 activity leads to pausing in the absence of stress (Fig. 4). Hsp70 chaperones interact with various translation factors and ribosomal proteins, and this interaction is regulated following heat shock (Fig. 5). HSC70/HSP70 may affect ribosome dynamics via its interaction elongation factors, regulation of ribosomal proteins in the exit tunnel, or through reduced interaction with nascent chains.
We observed elongation pausing in heat stress in both mouse and human cells. These findings, together with a much earlier report pointing to changes in elongation rates with heat shock in Drosophila cells (Ballinger and Pardue, 1983), suggest that the mechanism may be a deeply conserved feature of the response to proteotoxic stress. Our data show that the levels and activity of Hsp70s impact elongation pausing. A variety of possible mechanisms for these effects can be imagined, including altered interactions of Hsp70 with elongation factors, Hsp70-dependent regulation of ribosomal proteins in the exit tunnel, or aberrant exposure of nascent peptides in heat stress. Multiple chaperones are known to interact with ribosomes and with nascent peptides in both prokaryotes and eukaryotes (Kramer et al., 2009), and it is possible that additional ribosome-associated chaperones play a role in translation elongation under normal or stress conditions.
Analyzing the Hsp70-translational machinery interactome, we found that Hsp70 interacted with five elongation factors under normal growth conditions, and that its interaction with EEF1A1 was reduced following severe heat stress. This raises the possibility that Hsp70’s altered interaction with EEF1A1 in heat stress might influence elongation rates, though whether and how this could contribute to elongation pausing is unclear. A handful of initiation factors also had decreased interactions with Hsp70 under heat stress, suggesting a possible connection between Hsp70 and regulation of translation initiation under heat shock beyond its previously reported role. However, any link to initiation factors is likely distinct from Hsp70’s role in elongation pausing.
Our analysis identified RPL4 and RPL22, the only two ribosomal proteins that extend into the wall of the ribosomal exit tunnel forming a “constriction” (Wilson and Doudna Cate, 2012), as the only two stable proteins in the Hsp70-translational machinery interactome that showed a consistent increase in their overall extent of Hsp70 interaction. The exit tunnel itself was long considered a passive region in the ribosome. More recently, evidence has emerged that structural changes involving RPL22 and RPL4 can occur inside the exit tunnel, leading to regulatory effects on elongation (Berisio et al., 2003; Wilson and Doudna Cate, 2012). For example, the bacterial L22 protein interacts with the nascent peptide of SecM (Nakatogawa and Ito, 2002) and the eukaryotic L4 protein interacts with the nascent peptide of a uORF of the CMV protein gp48 (Bhushan et al., 2010), both leading to greater constriction of the exit tunnel and stalling of elongation. One possibility is that during heat shock Hsp70 chaperones regulate the conformation of the exit tunnel via interactions with RPL22 and/or RPL4.
The overall association of HSC70/HSP70 with ribosomes was down-regulated following 2 hours of severe heat stress (Figs. 3A, 3B), and consistent reduction in the interaction with several ribosomal proteins was shown by co-IP (Fig. 5B and 5C). A number of ribosomal proteins have previously been shown to efficiently incorporate into ribosomes when C-terminally tagged, e.g., RPL22 (Sanz et al., 2009), RPL23a (Inada et al., 2002; Ross et al., 2007), RPL18 and RPL16 (Halbeisen et al., 2009), suggesting that our assay often detected interaction with proteins incorporated into ribosomes. The extent of pausing correlated with hydrophobicity of the N-termini and with presence of Hsp70 binding motifs. Hsp70 proteins have evolved to recognize exposed hydrophobic patches, particularly when flanked by basic residues, as a sign of a misfolded or aggregated proteins. In the absence of ribosome-associated Hsp70, nascent peptides with stronger Hsp70 binding motifs might be more dependent on Hsp70 and thus have a greater tendency to misfold or aggregate, potentially impacting the efficiency of translation. More distant KS locations were associated with reduced N-terminal hydrophobicity (Figs. 3C, 3D and S3E), consistent with ribosomes being able to translate further downstream before pausing. These observations suggest that exposure of nascent peptides might impact elongation. Finally, RPL23A (Rpl25 in yeast), which serves as the docking site for nascent chain binding accessory factors (Kramer et al., 2009), interacted strongly with Hsp70 under normal growth conditions, and showed reduced interactions under heat stress (Figs. 5B, 5C). This observation suggests that Hsp70 is present in the vicinity of the exit tunnel under normal growth conditions, but much less so during heat stress. Indeed Hsp70 has been proposed in the past to play a role in aiding in the passage of nascent peptides through the exit tunnel (Nelson et al., 1992). Together, the above observations suggest that nascent peptides emerging from the ribosome might be involved in the process of elongation pausing (Fig. 6).
The prevalence of elongation pausing in the heat stress responses of both mouse and human cells make it an intriguing phenomenon to explore in other stresses and organisms. Elongation pausing could facilitate translational repression under certain conditions, such as proteotoxic stresses, while induction of chaperones might counteract stress-induced elongation pausing. For example, elevated levels of chaperones are often observed in cancer (Mosser and Morimoto, 2004; Whitesell and Lindquist, 2005). This phenomenon may therefore be relevant to a broad spectrum of stresses and diseases.
Experimental procedures
Heat shock and drug treatment conditions
Mouse fibroblast 3T3 cells were plated at low density and then treated with mild (8 hours at 42° C) or severe heat shock (2 hours at 44° C) to induce chronic or acute heat stress responses, respectively (Supplemental Experimental Procedures). Harringtonine treatments were performed at 0.1 mM harringtonine for 3 minutes, or 1 minute of 0.1 mM harringtonine followed by 2 minutes of 0.1 mg/ml cycloheximide (CHX), which was added to the media. Puromycin treatment was done at 1 mg/ml for 3 minutes. Treatment with the Hsp70 inhibitor VER-155008 (Massey et al.) (Tocris Bioscience) was performed at 20 μM for 3 hours.
Ribosome footprint profiling
Ribosome footprinting was performed as described by (Ingolia et al., 2009) with modifications described in Supplemental Experimental Procedures.
Transfections
Transfection conditions are detailed in the Supplemental Experimental Procedures.
Mapping of footprint reads
Ribosome footprint reads were trimmed from the 3′ end from 36 bases to 32 bases, and multiple trailing adenosine (A) bases were removed. Next, footprint reads of size 22-32 bases were mapped to the mouse genome (mm9) or the human genome (hg18) using Bowtie (Langmead et al., 2009). Reads mapping uniquely to exonic positions were subsequently mapped to Ensembl gene and transcript annotation files (downloaded from UCSC) using custom python and Perl scripts (Supplemental Experimental Procedures).
Polysome profiling
3T3 cells were treated with 0.1 mg/ml cycloheximide (CHX) for 5 minutes, then lysed (Supplemental Experimental Procedures). Profiles were normalized by loading equal amounts of RNA OD units of all samples in a single experiment. To compare between experiments we used relative P:M ratio, calculated as the P:M ratio in the given treatment divided by that in the corresponding Control condition and expressed as a percentage.
Protein association with polysome fractions
After ultracentrifugation, 24 equal volume fractions were collected and pooled as described in Figures 3A top panel and precipitated by addition ice cold acetone and overnight incubation at −20° C. Significance of the reduction in ribosomal association of HSC70/HSP70 was calculated by t-test over 5 replicates using MATLAB. Further details provided in Supplemental Experimental Procedures.
Antibodies
Antibodies are detailed in the Supplemental Experimental Procedures.
Footprint density plots and tests of pause significance and location
All uniquely mapping exonic footprint reads were mapped to Ensembl mRNA transcripts using their genomic annotations in the ensGene table from UCSC. For each transcript, a profile was generated as a vector containing the number of reads for which the 5′ end mapped 12 nt 5′ of each position of the transcript, and then normalized by dividing by the mean number of reads per position along the first 450 nucleotides of the CDS, similarly to (Ingolia et al., 2009). Presented in the figures are the averaged profiles for the filtered unique set in each condition (see Supplemental Experimental Procedures for further details on this and on KS analysis). An elevation of ribosome density at the 5′ ends of yeast genes, which was gradually decreasing up to ~600 bases of the ORF, has been observed in yeast under normal growth conditions in the first footprinting study (Ingolia et al., 2009). This elevation has been attributed to effects of tRNA abundance on translational efficiency (Qian et al., 2012; Tuller et al., 2010).
Hydrophobicity profiles and Hsp70 binding site scoring
Hydrophobicity profiles and Hsp70 binding site scoring are described in the Supplemental Experimental Procedures.
Hsp70 – translation machinery interactome
LUMIER-BACON assay was done as in Taipale et al. (Taipale et al., 2012), using a 293T cell line stably expressing Renilla tagged HSC70 (Hspa8). Co-IP with endogenous Hsp70 chaperones was performed as in (Taipale et al., 2012). Further details provided in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Global translation elongation pause at 5′ end of ORFs in severe heat shock
Modulation of Hsp70 levels/activity affects elongation pausing
Pausing is associated with hydrophobic N-termini
Hsp70 shows reduced ribosome association and altered interactions in heat stress
Acknowledgments
High throughput sequencing data have been submitted to GEO (accession number GSE32060). We thank Wendy Gilbert and members of her lab for helpful advice and suggestions, use of equipment throughout this study, and for critical reading of the manuscript. We thank Eric Wang for help with mapping of footprint data and for useful discussions, Sandro Santigata and Luke Whitesell for helpful advice, and Daniel Treacy for help with footprint library preparation. R. S. is an Awardee of the Weizmann Institute of Science – National Postdoctoral Award Program for Advancing Women in Science. This work was supported by an EMBO long-term fellowship and the Machiah foundation (R. S.), NIGMS fellowship number F32GM095060 (J. A. H.), and by grants from the NIH (C. B. B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abravaya K, Myers MP, Murphy SP, Morimoto RI. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger DG, Pardue ML. The control of protein synthesis during heat shock in Drosophila cells involves altered polypeptide elongation rates. Cell. 1983;33:103–113. doi: 10.1016/0092-8674(83)90339-2. [DOI] [PubMed] [Google Scholar]
- Beckmann RP, Mizzen LE, Welch WJ. Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- Berisio R, Schluenzen F, Harms J, Bashan A, Auerbach T, Baram D, Yonath A. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat Struct Biol. 2003;10:366–370. doi: 10.1038/nsb915. [DOI] [PubMed] [Google Scholar]
- Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, Beckmann R. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol Cell. 2010;40:138–146. doi: 10.1016/j.molcel.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Biamonti G, Caceres JF. Cellular stress and RNA splicing. Trends Biochem Sci. 2009;34:146–153. doi: 10.1016/j.tibs.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Bjork JK, Sistonen L. Regulation of the members of the mammalian heat shock factor family. FEBS J. 2010;277:4126–4139. doi: 10.1111/j.1742-4658.2010.07828.x. [DOI] [PubMed] [Google Scholar]
- Bouche G, Amalric F, Caizergues-Ferrer M, Zalta JP. Effects of heat shock on gene expression and subcellular protein distribution in Chinese hamster ovary cells. Nucleic Acids Res. 1979;7:1739–1747. doi: 10.1093/nar/7.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Calloni G, Chen T, Schermann SM, Chang HC, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl FU. DnaK Functions as a Central Hub in the E. coli Chaperone Network. Cell Rep. 2012;1:251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Duncan R, Hershey JW. Heat shock-induced translational alterations in HeLa cells. Initiation factor modifications and the inhibition of translation. J Biol Chem. 1984;259:11882–11889. [PubMed] [Google Scholar]
- Feder JH, Rossi JM, Solomon J, Solomon N, Lindquist S. The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes Dev. 1992;6:1402–1413. doi: 10.1101/gad.6.8.1402. [DOI] [PubMed] [Google Scholar]
- Gerner EW, Schneider MJ. Induced thermal resistance in HeLa cells. Nature. 1975;256:500–502. doi: 10.1038/256500a0. [DOI] [PubMed] [Google Scholar]
- Gibson G. The environmental contribution to gene expression profiles. Nat Rev Genet. 2008;9:575–581. doi: 10.1038/nrg2383. [DOI] [PubMed] [Google Scholar]
- Halbeisen RE, Scherrer T, Gerber AP. Affinity purification of ribosomes to access the translatome. Methods. 2009;48:306–310. doi: 10.1016/j.ymeth.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Henle KJ, Karamuz JE, Leeper DB. Induction of thermotolerance in Chinese hamster ovary cells by high (45 degrees) or low (40 degrees) hyperthermia. Cancer Res. 1978;38:570–574. [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Huang MT. Harringtonine, an inhibitor of initiation of protein biosynthesis. Mol Pharmacol. 1975;11:511–519. [PubMed] [Google Scholar]
- Inada T, Winstall E, Tarun SZ, Jr., Yates JR, 3rd, Schieltz D, Sachs AB. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA. 2002;8:948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell. 2011 doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal H, Conz C, Otto H, Wolfle T, Fitzke E, Mayer MP, Rospert S. The chaperone network connected to human ribosome-associated complex. Mol Cell Biol. 2011;31:1160–1173. doi: 10.1128/MCB.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330:1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley PM, Schlesinger MJ. The effect of amino acid analogues and heat shock on gene expression in chicken embryo fibroblasts. Cell. 1978;15:1277–1286. doi: 10.1016/0092-8674(78)90053-3. [DOI] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. Varying patterns of protein synthesis in Drosophila during heat shock: implications for regulation. Dev Biol. 1980;77:463–479. doi: 10.1016/0012-1606(80)90488-1. [DOI] [PubMed] [Google Scholar]
- Lindquist S. Regulation of protein synthesis during heat shock. Nature. 1981;293:311–314. doi: 10.1038/293311a0. [DOI] [PubMed] [Google Scholar]
- Lindquist S. Protein folding sculpting evolutionary change. Cold Spring Harb Symp Quant Biol. 2009;74:103–108. doi: 10.1101/sqb.2009.74.043. [DOI] [PubMed] [Google Scholar]
- Massey AJ, Williamson DS, Browne H, Murray JB, Dokurno P, Shaw T, Macias AT, Daniels Z, Geoffroy S, Dopson M, et al. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother Pharmacol. 2010;66:535–545. doi: 10.1007/s00280-009-1194-3. [DOI] [PubMed] [Google Scholar]
- McCormick W, Penman S. Regulation of protein synthesis in HeLa cells: translation at elevated temperatures. J Mol Biol. 1969;39:315–333. doi: 10.1016/0022-2836(69)90320-9. [DOI] [PubMed] [Google Scholar]
- Miller MJ, Xuong NH, Geiduschek EP. A response of protein synthesis to temperature shift in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979;76:5222–5225. doi: 10.1073/pnas.76.10.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DD, Morimoto RI. Molecular chaperones and the stress of oncogenesis. Oncogene. 2004;23:2907–2918. doi: 10.1038/sj.onc.1207529. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- Petersen NS, Mitchell HK. Recovery of protein synthesis after heat shock: prior heat treatment affects the ability of cells to translate mRNA. Proc Natl Acad Sci U S A. 1981;78:1708–1711. doi: 10.1073/pnas.78.3.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8:e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Ross CL, Patel RR, Mendelson TC, Ware VC. Functional conservation between structurally diverse ribosomal proteins from Drosophila melanogaster and Saccharomyces cerevisiae: fly L23a can substitute for yeast L25 in ribosome assembly and function. Nucleic Acids Res. 2007;35:4503–4514. doi: 10.1093/nar/gkm428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. Embo J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover K, Karras G, Lindquist S. Quantitative Analysis of Hsp90-Client Interactions Reveals Principles of Substrate Recognition. Cell. 2012 doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- von Heijne G. On the hydrophobic nature of signal sequences. Eur J Biochem. 1981;116:419–422. doi: 10.1111/j.1432-1033.1981.tb05351.x. [DOI] [PubMed] [Google Scholar]
- Vries RG, Flynn A, Patel JC, Wang X, Denton RM, Proud CG. Heat shock increases the association of binding protein-1 with initiation factor 4E. J Biol Chem. 1997;272:32779–32784. doi: 10.1074/jbc.272.52.32779. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Wilson DN, Doudna Cate JH. The structure and function of the eukaryotic ribosome. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- Zylicz M, King FW, Wawrzynow A. Hsp70 interactions with the p53 tumour suppressor protein. Embo J. 2001;20:4634–4638. doi: 10.1093/emboj/20.17.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.