Summary
Specification of the non-skeletogenic mesoderm (NSM) in sea urchin embryos depends on Delta signaling. Signal reception leads to expression of regulatory genes that later contribute to the aboral NSM regulatory state. In oral NSM, this is replaced by a distinct oral regulatory state in consequence of Nodal signaling. Through regulome wide analysis we identify the homeobox gene not as an immediate Nodal target. not expression in NSM causes extinction of the aboral regulatory state in the oral NSM, and expression of a new suite of regulatory genes. All NSM specific regulatory genes are henceforth expressed exclusively, in oral or aboral domains, presaging the mesodermal cell types that will emerge. We have analyzed the regulatory linkages within the aboral NSM gene regulatory network. A linchpin of this network is gataE which as we show is a direct Gcm target and part of a feedback loop locking down the aboral regulatory state.
Keywords: mesoderm, nodal signaling, oral/aboral axis, not, gcm, gataE, gene regulatory network
Introduction
At 24 h post fertilization (hpf) before the onset of gastrulation the non-skeletogenic mesoderm (NSM) of the early sea urchin embryo consists of a circular vegetal disc of about 60 cells (in Strongylocentrotus purpuratus) that is sharply divided into two domains. They display distinct regulatory states defined as the set of transcription factors present in each cell. About one-third of the NSM cells, located in a V-shaped sector directly beneath the oral ectoderm, give rise upon gastrulation to a particular mesenchymal cell type known as blastocoelar cells (Ruffins and Ettensohn, 1996). Following gastrulation, the blastocoelar cells differentiate into several kinds of functional immune cells of the embryo/larva (Rast et al., 2006; Hibino et al., 2006; Furukawa et al., 2009). However, the blastocoelar cell regulatory state originates prior to gastrulation when oral NSM cells commence to express exclusively specific transcription factors. The complementary aboral spatial regulatory state domain of the pre-gastrular NSM will give rise to pigment cells (Ruffins and Ettensohn, 1996). During gastrulation these cells embed themselves in the aboral ectoderm and execute a genetic program leading to pigment synthesis and a unique, dynamic, pseudopodial morphology. In this work we uncover a number of additional NSM transcription factor genes, and restudy the expression of others, with the conclusion that by gastrulation the regulatory states of the aboral and oral NSMs are entirely distinct and non-overlapping. Two other NSM derivatives, esophageal muscle cells, and coelomic pouch cells, are not specified until later, i.e., during gastrulation (Materna and Davidson, 2012; Ruffins and Ettensohn, 1996; Sweet et al., 2002). They do not figure in this study, which is confined to pre-gastrular specification mechanisms in the NSM.
Currently, knowledge regarding the origin, development, and ultimate pre-gastrular structure of the oral and aboral gene regulatory networks (GRNs) responsible for generating these regulatory states is very uneven. Much is known about the initiation of the aboral NSM GRN. Building on this in the present work, we have been able to provide a revised aboral NSM GRN that includes all known specifically expressed aboral NSM genes. The most upstream gene of the hierarchical aboral NSM GRN is gcm, which is activated as a direct target of Notch signaling (Ransick and Davidson, 2006), in response to presentation of the Delta ligand by the adjacent skeletogenic mesoderm (SM) (Materna and Davidson, 2012; Revilla-i-Domingo et al., 2007; Sweet et al., 2002). We showed recently by a synthetic approach that forced expression of gcm in cells of the skeletogenic lineage suffices to activate the whole of the downstream aboral NSM/pigment cell GRN in the complete absence of Notch signaling (Damle and Davidson, 2012). Regulatory interactions following gcm activation have been supported by direct cis-regulatory studies (Lee et al, 2007; Ransick and Davidson, 2012) including some on pigment cell differentiation genes (Calestani and Rogers, 2010). In contrast, almost nothing is yet known of the oral NSM GRN. Whole mount in situ hybridization studies in a number of laboratories have identified a handful of genes expressed specifically in oral NSM (among recent studies, Duboc et al., 2010; Poustka et al. 2007; Ransick and Davidson, 2006; Rizzo et al., 2006; Röttinger et al, 2006; Sharma and Ettensohn, 2011; Walton et al., 2009), but the functional interactions among these genes in general remain to be demonstrated. Almost the only causal relationship established is that specification of the oral NSM is dependent on the Nodal signal emanating from the oral ectoderm (Duboc et al., 2008; 2010). This accounts for the oral placement of this domain. However, it is probably impossible for any of the oral NSM genes to be considered direct targets of Nodal signaling because, as we show below, they are activated too many hours after all other known direct Nodal targets (e.g, Nam et al., 2007). The mechanistic link between Nodal signaling and the oral NSM regulatory state has remained completely obscure. We at least partially resolve this problem in the present work, in identifying a Nodal target, the homeodomain gene not, expression of which is in turn obligatory for proper oral NSM specification.
In S. purpuratus development the transcriptional activation of gcm (and of its immediate target genes) occurs in all cells of the endomesodermal lineage, while they yet consist of a single ring of 16 cells exposed to Delta ligand (Ransick and Davidson 2006). There follow two negative regulatory interactions that confine the aboral NSM GRN to mesodermal as opposed to endodermal descendants, and to aboral as opposed to oral NSM. When the endomesodermal ring divides radially to form two rings, the outer ring continues to express a Tcf–dependent endodermal GRN, while in the inner ring ongoing Notch signaling from the adjacent SM results in wholesale extinction of the endodermal GRN by interference with Tcf function, leaving only the mesodermal gcm network operating (Peter and Davidson, 2010; 2011). Institution of the oral NSM regulatory state later represses the aboral GRN in that segment. In this work we determine the polarity of these mechanisms. We find that not expression in the oral sector is the precondition for proper subdivision of the NSM. The observations imply a dominant spatial repression of key aboral genes by oral genes. But as we see in the following, the repression becomes mutual, in that expression of aboral genes also comes to exclude expression of oral genes.
Materials and Methods
Perturbation experiments
Morpholino antisense oligonucleotides (MASOs) were obtained from Gene-tools LLC and injected at 300 μM (100 μM for Nodal), in 0.12 M KCl. Injection volumes were ca. 5 pl. Sequences are as follows: Delta-MASO: CAAGAAGGCAGTGCGGCCGATCCGT; Ese-MASO: TTCCCTTCATGGCTGTAAAAACGAA, GataC-MASO: CATTAAAAGAAAATAACAAGTTCAC, GataE-MASO1: ACCACGCTTTGCTTCGTGTTTGGCC (translation block), GataE-MASO2: TCTCGTCTTGAGCCAGACTGCAATC (splice block), Nodal-MASO: TGCATGGTTAAAAGTCCTTAAAAAT, Not-MASO: GACATCAAGTTGGAACTCATCATAG, Prox1-MASO1: TGCATCCTCGACCTTAGACATTGGC (translation block), Prox1-MASO2: ACACCAAAAAGGACTTACCGTGAAC (splice block). Our GataE MASOs replace an earlier MASO (GACTTACACCGACCTGATGTGGCAT) that we find to have deleterious effects when injected at high concentrations. The γ-secretase inhibitor DAPT (Hughes et al., 2009) was dissolved in DMSO and added at 3 hpf to a final concentration of 8 μM (Materna and Davidson, 2012). Full length not transcript was amplified with tailed primers from cDNA and transcribed with T7 polymerase and polyadenylated in vitro. The sequence of not cDNA was independently confirmed by RNAseq (Tu et al., 2012) and has been submitted to genbank (JQ945921). It differs from a previously published Sp-Not sequence that appears to be a fusion of two unrelated genes (genbank, AF109903) (Peterson et al., 1999).
Embryo culture and RNA extraction
Sea urchin embryos were cultured at 15 °C and closely monitored for proper development. For lysis, sea water was removed before adding 350 μl RLT buffer from the Qiagen RNeasy Micro Kit (Qiagen). Embryo lysates were immediately stored at -70°C until use. RNA was extracted according to manufacturer's instructions.
RACE PCR and amplification of cDNA sequences
5’ ends of transcripts were identified using the Clontech SMART RACE PCR kit. PCR fragments were cloned and sequenced to identify the approximate transcription start site. The actual 5’ end of the six1/2 sequence differs from the gene prediction SPU_17397 (genbank accession numbers: prox1—JQ956375 , six1/2—JQ264781, z166—JQ945922).
Transcriptional profiling
Expression level was quantified with the Nanostring nCounter using our custom designed probe set for 183 genes. The samples were processed according to manufacturers’ instructions and data processed as described previously (Materna et al., 2010). This dataset was supplemented by QPCR for genes not included in the code set and fold changes were calculated using the poly-ubiquitin (ubq) and hmg1 genes as reference (Materna and Oliveri, 2008). Tables of all accession numbers, probe and primer sequences, as well as perturbation data are included in Supp. Table 1, 2 and 4.
Whole mount in situ hybridization
Whole mount in situ hybridization was conducted following standard procedures as described previously (Materna and Davidson, 2012; Ransick, 2004). For two color in situ staining, probes were labeled with digoxigenin or fluorescein. Following the first staining reaction, embryos were incubated in acidic glycine wash to strip the first antibody, followed by an additional blocking step and incubation with the second antibody. Fast-Red staining was performed first, followed by staining with NBT/BCIP. Primers for in situ templates are given in Supp. Table 3.
Nanotag analysis of reporter expression
A collection of 129 barcoded reporter constructs, containing Gcm Module E and GataE module 10, was injected together with Delta MASO or Gcm translation blocking MASO. RNA was extracted from embryos and quantified using a Nanostring code set for detection of barcodes. Data were analyzed as reported elsewhere (Nam and Davidson, 2012).
Results
Regulatory states of definitive oral and aboral NSM are distinet
The progression of regulatory states throughout NSM specification has not been studied systematically. But such knowledge is essential for determining the participants in the underlying GRNs, and for indicating order in the regulatory hierarchies (Materna and Oliveri, 2008). The following analyses include regulatory genes, i.e., transcription factors and signaling molecules, for which we and others had evidence of expression in the pre-gastrular NSM. Some genes previously not included in the provisional NSM GRNs emerged from a comprehensive Nanostring study of targets of Delta/Notch (D/N) signaling during the period of Delta presentation by the SM (Materna and Davidson, 2012). This work had demonstrated that expression of all NSM regulatory genes depends directly or indirectly on Notch signaling. For many genes included in the present study it was not clear whether they are expressed in oral or aboral NSM, which we resolved using double in situ hybridization.
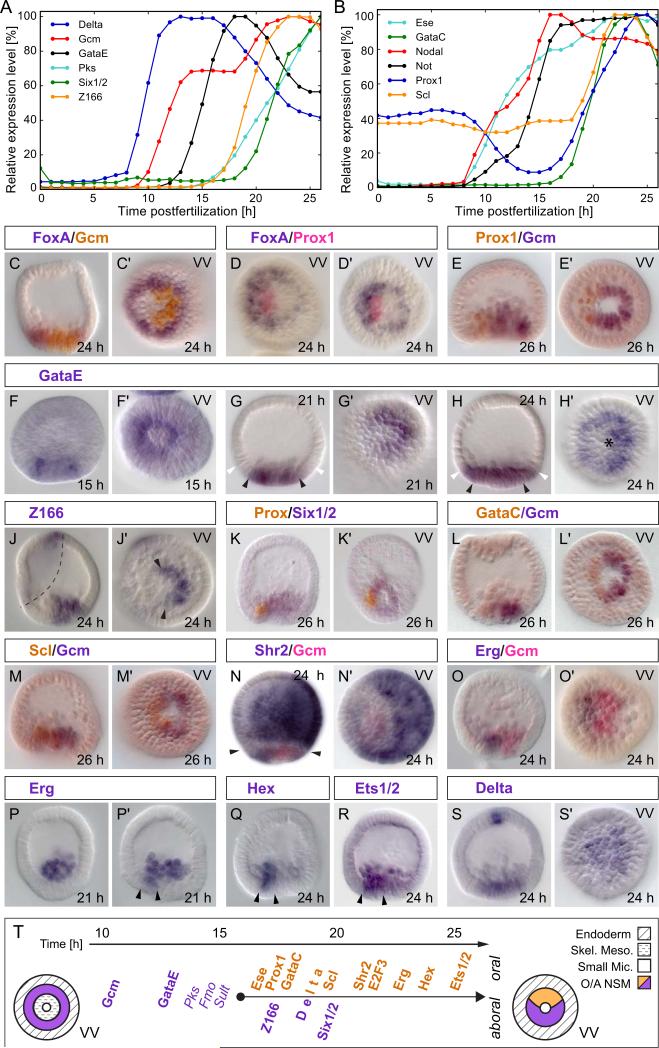
Figures 1A and B, excerpted from an earlier high resolution time course study of regulatory gene expression (Materna et al., 2010), show respectively that the regulatory states of both aboral and oral NSM are established sequentially and progressively over many hours, just as seen for the skeletogenic and endodermal domains (Oliveri et al, 2008; Peter and Davidson, 2010). Only some of the oral NSM genes that we treat in the following are included in Fig. 1B since for genes expressed simultaneously in other domains of the embryo (e.g. shr2, e2f3), whole embryo time courses are less informative. In the double in situ hybridizations shown in Fig. 1, gcm expression is used to define the aboral NSM (Ransick and Davidson, 2006), and foxA expression is used to define the endodermal domain (Oliveri et al., 2006; Peter and Davidson, 2010). Figs. 1C,C illustrate in a double in situ hybridization using these two probes how the asymmetrically located aboral NSM is immediately surrounded by the ring of endoderm cells at mesenchyme blastula stage.
Figure 1.
Expression profiles of NSM genes. A) Temporal expression profiles of NSM precursor/aboral NSM genes plus Delta ligand. B) Expression profiles for oral NSM genes and Nodal. Expression levels are given as a fraction of peak expression (Materna et al., 2010). C–S) Double in situ staining of NSM gene transcripts. C, D) foxA is expressed throughout the endoderm surrounding the NSM. G, H) gataE expression is restricted to the NSM until after 21 hpf (black arrows mark mesoderm boundary, white arrows mark endoderm boundary). Asterisk in H′ marks the vegetal pole. J) z166 expression in the NSM is restricted to the aboral segment (arrows in J’) but has a second expression compartment in the ciliated band (dashed line). N) shr2 is expressed in the oral NSM and in the aboral ectoderm separated by the endoderm (arrows in N). O–R) Once expressed in the NSM, erg, hex, and ets1/2 are restricted to the oral side marked by arrowheads in P’, Q, and R. Embryos presented in a lateral view with their oral side to the left. VV – vegetal view. T) Summary of gene expression profiles in NSM between 10 hpf and 25 hpf. Differentiation genes in italics.
We first consider new information included in Fig. 1 that concerns aboral NSM regulatory gene expression. The gataE gene had previously been studied for its role in endoderm formation (Lee and Davidson, 2007). It is activated in the endoderm by 24 hpf where it remains expressed throughout gastrulation (Lee and Davidson, 2004). But as Fig. 1A shows, gataE transcription ramps up much earlier in the NSM, only three hours after gcm transcription begins at 10 hpf, suggesting that gataE could be a direct target of Gcm (Bolouri and Davidson, 2003), which we show below is indeed almost certainly the case. As the in situ hybridizations in Fig. 1F–H illustrate, before its endodermal expression phase gataE is expressed identically with gcm (Ransick and Davidson, 2006), first in a ring (Fig. 1F), and then exclusively in the aboral NSM (Fig. 1G), and eventually in both aboral NSM and endoderm (Fig. 1H; compare to Fig. 1C). The zinc finger gene z166, which based on its domain structure is likely a transcription factor as well, is activated 3–4 hrs after gataE in the aboral NSM (Fig. 1A,J) (Materna et al., 2006). This is soon followed by the homeobox gene six1/2 (Fig. 1A,K; in Fig. 1K,K the opposing oral quadrant is marked by prox1 expression) (Poustka et al., 2007). The six1/2 cofactor eya is also expressed in the aboral NSM (not shown).
Figure 1B shows that of the oral NSM genes, ese is the earliest activated, but prior studies (Rizzo et al, 2006, Duboc et al., 2010) indicated that up to 15 hpf ese transcripts are ubiquitous in the embryo and have nothing specific to do with the oral NSM domain. However, following ingression of the skeletogenic cells, ese becomes specifically and exclusively expressed in oral NSM (Rizzo et al, 2006). Well before this, beginning by about 10 hpf, the homeodomain gene not is activated in the oral ectoderm, and as we describe in detail below, several hours prior to the activation of other oral NSM genes not transcription also occurs actively in the oral NSM. The not gene is the first regulatory gene to be expressed in the oral quadrant of the NSM. gataC, as seen in Fig. 1B, only begins to be transcribed significantly at about 17 hpf similar to prox1; both genes have almost identical activation kinetics between 18 and 24 hpf (Fig. 1B). Fig. 1D,E shows the expression of the prox1 gene (Poustka et al., 2007) in the fully established oral NSM at 24 hpf, inside the endodermal tier expressing foxA, and on the opposite side to gcm, which at this time is strictly aboral. Similar data are shown for gataC in Fig. 1L,L. The next oral NSM genes to turn on are scl (Fig. 1B,M), shr2 (Fig. 1N) and e2f3 (not shown). It significantly departs the baseline, which due to its exceptionally low transcript level (Materna et al., 2010) is higher than for other genes, at 19 hpf, at least two hours after prox1 and gataC. Both shr2 and e2f3 are expressed in the aboral ectoderm in addition to oral NSM, illustrated for shr2 in Fig. 1N. The next phase of maturation of the oral NSM regulatory state occurs at about the time SM ingression into the blastocoel is complete. At this time three genes that are first expressed in the SM, erg, hex, and ets1/2 are activated specifically in the oral NSM (Fig. 1O–R). The exact timing of erg activation varies even within a single batch of embryos. About a quarter of the embryos express erg in the oral NSM already at 21 hpf, but all embryos express it by 24 hpf. The hex gene is activated next, followed by ets1/2. Activation dynamics vary for ets1/2 in the NSM as they do for erg. Expression of the non DNA-binding co-factor lmo2 is also restricted to oral NSM (Duboc et al., 2010); and though initially expressed in the skeletogenic lineage the zinc finger gene z48 as well as the transcription factor foxN2/3, not further considered here, turn on in NSM prior to gastrulation (Materna et al., 2006; Tu et al., 2006). These genes follow the same pattern as erg, hex, and ets1/2 and so are likely to be specifically expressed in oral NSM after ingression of SM cells.
Following establishment of definitive and distinct oral and aboral NSM regulatory state domains, the only regulatory gene we are aware of that is expressed throughout the entire NSM, i.e., in the oral and aboral segment, is delta (Fig. 1S). As discussed elsewhere, delta is expressed wherever the repressor HesC is absent, as is the case in the NSM after about 19 hpf (Materna and Davidson, 2012; Revilla-i-Domingo et al., 2007; Smith and Davidson, 2008). The nemo-like kinase gene, nlk, is co-expressed with delta and their transcriptional regulation may be linked (Rottinger et al, 2006). However, by 24 hpf, expression of every one of the 13 genes encoding transcription factors examined here, expression within the NSM is confined to either the oral or aboral NSM domain. The non-overlapping oral and aboral regulatory states, and the sequence in which they are respectively assembled, are summarized in Fig. 1T.
Requirement of Nodal signaling for establishment of the oral NSM regulatory state
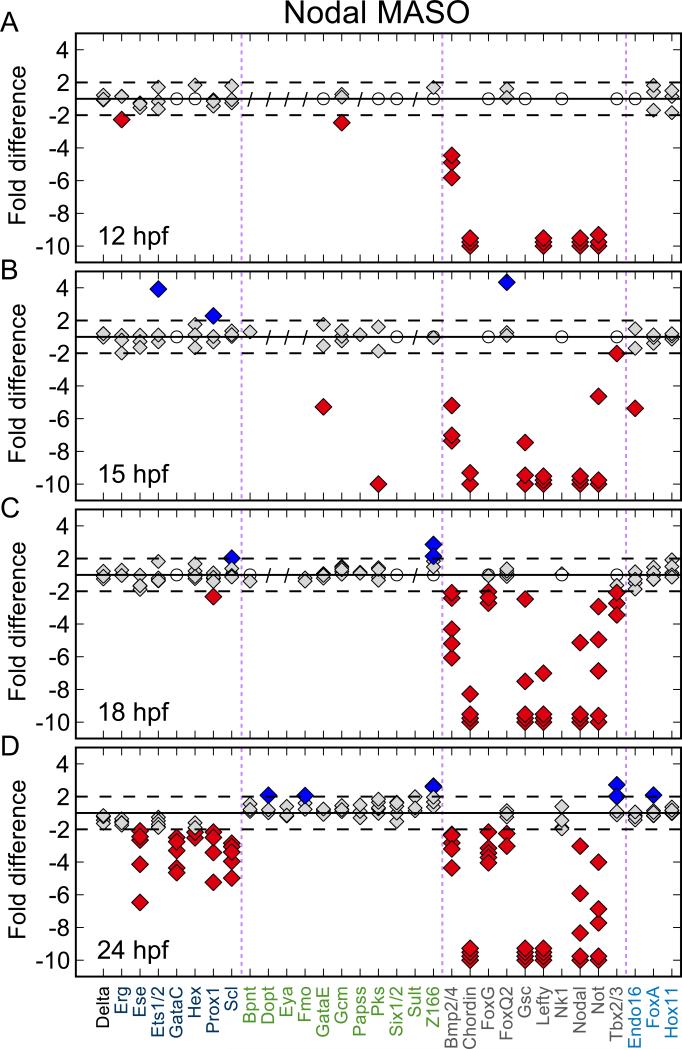
Earlier work has indicated that interference with nodal gene expression in the oral ectoderm disrupts oral gene expression across the embryo, including genes expressed in the NSM (Duboc et al., 2004; 2010; Su et al., 2009). To reveal the global effects of Nodal signaling on transcript levels of regulatory genes, we quantified transcript abundance in embryos bearing Nodal or Control MASO using Nanostring technology (Geiss et al., 2008). The analysis (Fig. 2) was carried out with a custom designed code set that contains gene specific probes for most of the regulatory genes expressed in a spatially restricted manner up to gastrulation in the S. purpuratus embryo (Materna et al., 2010). Additional measurements targeting pigment cell differentiation genes that were not included in the Nanostring code set were made by QPCR. All together, we examined the transcript levels of 206 genes. A table of all perturbation data is provided as Supp. Table 4.
Figure 2.
Effect of Nodal knockdown on transcript levels. Fold-difference of transcript abundance was calculated between experiment and control; 1-fold difference corresponds to no change. Negative values indicate lower transcript abundance in experiment compared to control. Each diamond represents a single experiment. Genes not significantly expressed (<50 transcripts/embryo) in treated and control embryos are marked with an open white circle. Genes not tested are marked with a slash. Dashed lines indicate threshold for significant changes. Fold differences of more than 10 fold are shown as 10 fold. Oral NSM genes—dark blue, aboral NSM genes—green, ectoderm genes—grey, endoderm genes—light blue. A, B) The first genes affected are the known Nodal targets bmp2/4, chordin, lefty, and nodal. D) At 24 hpf the transcript levels of all genes expressed specifically and exclusively in oral NSM are significantly reduced.
The first transcription factor genes specific to the oral NSM, prox1 and gataC, are transcriptionally activated in this region between 17 and 18 hpf (Fig. 1B,T), which is significantly later than the onset of Nodal transcription at about 8–9 hpf (Fig. 1B). As we see in Fig. 2A, direct targets of Nodal signaling are strongly affected by 12 hpf and include genes, such as bmp2/4, lefty, chordin, and nodal itself (Su et al., 2009; Nam et al., 2007; Range et al., 2007). Other transcriptional regulatory genes activated by Nodal signaling (Fig. 2B) are restricted to the ectoderm (e.g., gsc) as reported previously (Su et al., 2009). But the quantitative data of Fig. 2D also shows that Nodal MASO causes a significant reduction in transcript levels of all oral NSM transcription factor genes at the mesenchyme blastula stage (24 hpf), confirming that specification of oral NSM is defective in these conditions (Duboc et al., 2010). Of course this effect can only be detected after these genes have accumulated to significant levels in controls, which at 18 hpf is not yet the case. Nevertheless, the long interval between expression of nodal and activation of oral NSM specific genes is inconsistent with the kinetics of immediately successive gene interactions in this embryo (Bolouri and Davidson, 2003), and therefore it is inconsistent with the concept that the oral NSM regulatory genes are direct Nodal signaling targets (Duboc et al., 2010). The strong implication is that the Nodal input affects oral NSM regulatory genes indirectly, and that the response of these genes is at least one step removed from the immediate Nodal response gene(s) of the NSM.
Expression of the homeobox gene not
The homeobox gene not is very strongly affected by Nodal MASO even at the 12 hpf time point (Fig. 2A). In work to be presented elsewhere, we show not to be a direct transcriptional target of Nodal signaling (EL and EHD, unpublished), and not in turn plays a major role in initiating and patterning the oral ectoderm GRN (Li et al., 2012). In the following we show that the function of not, downstream of its activation by Nodal signaling, is essential for positioning the oral NSM and divergence ofNSM regulatory states following subdivision.
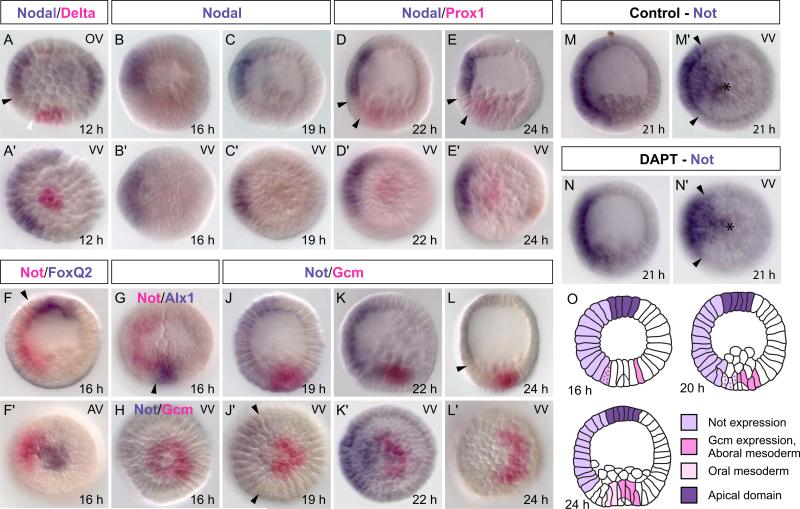
The detailed double whole mount in situ hybridization study of Fig. 3 reveals the spatial gene expression patterns of the nodal and not genes relative to other domains of the S. purpuratus embryo, between 12 hpf and 24 hpf. nodal transcription is confined to the oral ectoderm during this whole period; it is never expressed in any endomesodermal lineages (Fig. 3A–E); and though more broadly expressed laterally in the ectoderm at 12 hpf, thereafter nodal transcription is confined to a sector including only about one–fourth of the embryo circumference (Fig. 3A-E). In contrast, while expression of not overlaps that of nodal in the oral ectoderm, its expression domain borders the apical foxQ2 domain at the top of the embryo (Fig. 3F,F), and extends all the way down to the SM marked by alx at the vegetal pole (Fig. 3G). This means that NSM cells on the oral side transcribe not, and indeed, after 19 hpf expression of not extends right up to the domain of gcm expression that is strictly aboral at this time (Fig. 3J,J,K,K). This relation is transient, however, and by 24 hpf not message is confined to the ectoderm (Fig. 3L,L); i.e., it has by then ceased to be transcribed in the NSM.
Figure 3.
Spatial expression patterns of nodal and not. A) At 12 hpf nodal is expressed on the entire oral side of the ectoderm but is excluded from the endomesodermal tier in the vegetal half (black arrow in A marks the ectoderm/endomesoderm boundary; white arrowthe endomesoderm/SM boundary). D, E) prox1 and nodal are separated by the endoderm (between arrowheads). F–H) The not expression domain spans the entire oral side of the embryo and borders foxQ2 expressing cells apically (arrow in F) and alx1 expressing cells in the SM (arrow in G). J) gcm expression clears from not expressing, oral NSM cells. Arrowheads in J’ mark the boundaries of the not expression domain. L) At 24 hpf not expression has faded in the NSM and is restricted to ectoderm (arrowhead). M, N) Treatment of embryos with DAPT at 3 hpf interferes with Delta signaling from the SM and prevents activation of mesodermal genes. DAPT treatment does not affect expression of not in the NSM. Asterisks mark the vegetal pole. Unless otherwise noted embryos are presented in a lateral view with their oral side to the left. VV – vegetal view, AV – apical view, OV – oral view. O) Scheme summarizing the spatial expression of not and NSM genes.
It is important to note that transcripts of not are present in the future oral NSM by 16 hpf (Figs. 1B, 3F,F), although they are likely to appear in these cells even earlier as a consequence of Nodal signaling (Fig. 2) (Li et al., 2012). For 2–3 hrs thereafter, gcm and gataE continue to be expressed in a circular pattern throughout the NSM (Fig. 1 F; Ransick and Davidson, 2006). Thus for a short time before the domains of oral and aboral NSM become mutually exclusive, gcm, gataE and not are co-expressed in the same cells, the future oral NSM.
Unlike the canonical genes that eventually constitute the oral NSM regulatory state (Materna and Davidson, 2012), not is unaffected by Delta/Notch (D/N) signaling. This is shown in Fig. 3M,M,N,N, where we see that transcription of not continues normally in the presence of DAPT (a γ-secretase inhibitor that prevents cleavage of the Notch intracellular domain and effectively blocks D/N signaling in sea urchin embryos; Hughes et al., 2009; Materna and Davidson, 2012). Thus the spatial location of not transcription in the NSM on the oral side of the embryo is solely due to Nodal signaling, which as Duboc et al. (2010) clearly show, extends down into this region.
Functional significance of not expression
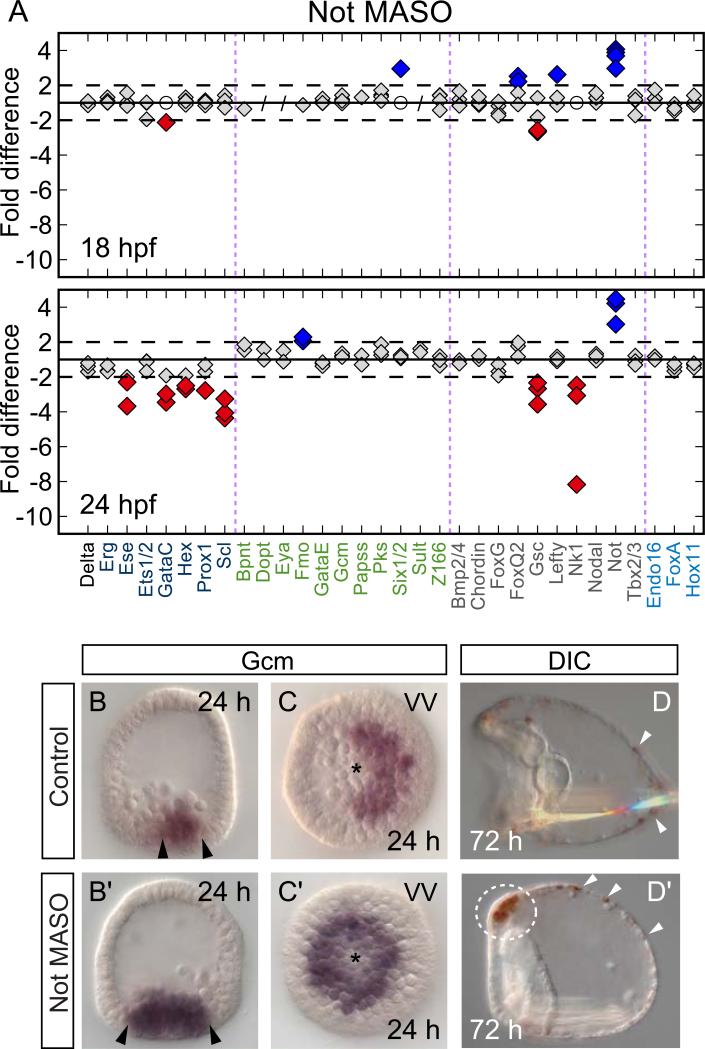
Global evaluation of the effects of blocking the translation of the not mRNA by MASO treatment revealed that only a small, specific set of genes is affected. These are all genes expressed either in the oral ectoderm or in the oral NSM (Fig. 4; Supp. Fig. 1) (Li et al., 2012). This MASO therefore has no general or off target effects and does not interfere with the overall health of the embryo. Gastrulation commences on time, but at pluteus stage gut development and spiculogenesis are somewhat delayed (Fig. 4D′). Results for the same selection of genes as shown for Nodal perturbations (Fig. 2) are shown in Fig. 4 A for Not MASO treated embryos. Here we see that at 24 hpf, transcript levels of all oral NSM regulatory genes which have by then been activated in controls are depressed several-fold by specific interference with not expression. These broad effects on the oral NSM regulatory state are comparable with those of Nodal MASO, but in contrast to the large number of oral ectoderm genes affected by Nodal MASO (directly and indirectly), the transcript levels of only a few oral ectoderm genes known to be Not targets, e.g., gsc and nk1 (Li et al., 2012), are depressed by Not MASO (see Supp. Fig. 1 and Supp. Table 4 for complete dataset). A main conclusion from the experiments in Fig. 4A is thus that not expression in the NSM is required for normal levels of expression of oral NSM genes at 24 hpf. This could be because not or a Not target gene serves as their activator, or because not expression abolishes a repressor of these genes.
Figure 4.
Not perturbation disrupts oral NSM specification A) Transcript levels of oral NSM genes are significantly lower in Not MASO injected embryos compared to controls by 24 hpf but only a few ectodermal genes are affected (e.g., gsc, nk1). Labels, symbols and cutoffs as in Fig. 2. B,C) gcm expression is restricted to aboral NSM at 24 hpf in controls. B’,C’) In Not MASO treated embryos gcm is consistently expressed throughout the entire ring of NSM cells. Asterisks mark the vegetal pole. D,D’) At 3 dpf a normal number of pigment cells have embedded themselves in the aboral ectoderm in controls and Not MASO injected embryos (white arrowheads). However, a dense group of pigment cells accumulates ectopically below the apical plate region (dashed circle in D’). Skeletogenesis and gut development are delayed postgastrulation. Embryos oriented with their oral side to the left. VV – vegetal view.
Not MASO experiments not only display the positive effect of not expression on oral NSM, but also the negative effects on aboral NSM genes. Thus in embryos bearing Not MASO the aboral NSM regulatory state domain increases in size at the expense of oral NSM (Fig. 4). Most strikingly, gcm expression persists throughout the entire circular NSM at 24 hpf in embryos treated with Not MASO while in controls, expression of gcm is extinguished in the oral segment by about 19 hpf. This effect can be seen clearly in Fig. 4B′,C′. The same is true for the gcm target gene gataE (not shown), while overexpression of not mRNA has the opposite effect, and decreases gataE transcript levels several fold (Supp. Table 4; this result further substantiates the specificity of the Not MASO). Since gcm expression alone is sufficient to confer pigment cell fate (Damle and Davidson, 2012) it is therefore not surprising that Not MASO causes an excess specification of pigment cells, that accumulate ectopically below the apical organ (Fig. 4D,D’). Whether this accumulation is due to intrinsic differences in these cells that likely derive from the oral NSM, or due to loss of Not function in the ectoderm is unknown. In conclusion, not gene expression is required for oral NSM clearance of gcm (and the downstream genes of the aboral NSM), as well as for normal activation of oral NSM genes.
An additional result from the Not MASO experiments was the observation that Not evidently represses itself, since the MASO caused a significant increase in the level of not transcripts (Fig. 4A) (Li et al., 2012). This conclusion is also consistent with the abrupt advent of the plateau of not transcript levels at 16 hpf following its prior steep rise that can be seen in the more extended timecourse (Materna et al., 2010); in the whole embryo not transcript levels decline monotonically after 25 hpf.
Aboral regulatory state effects on oral NSM genes
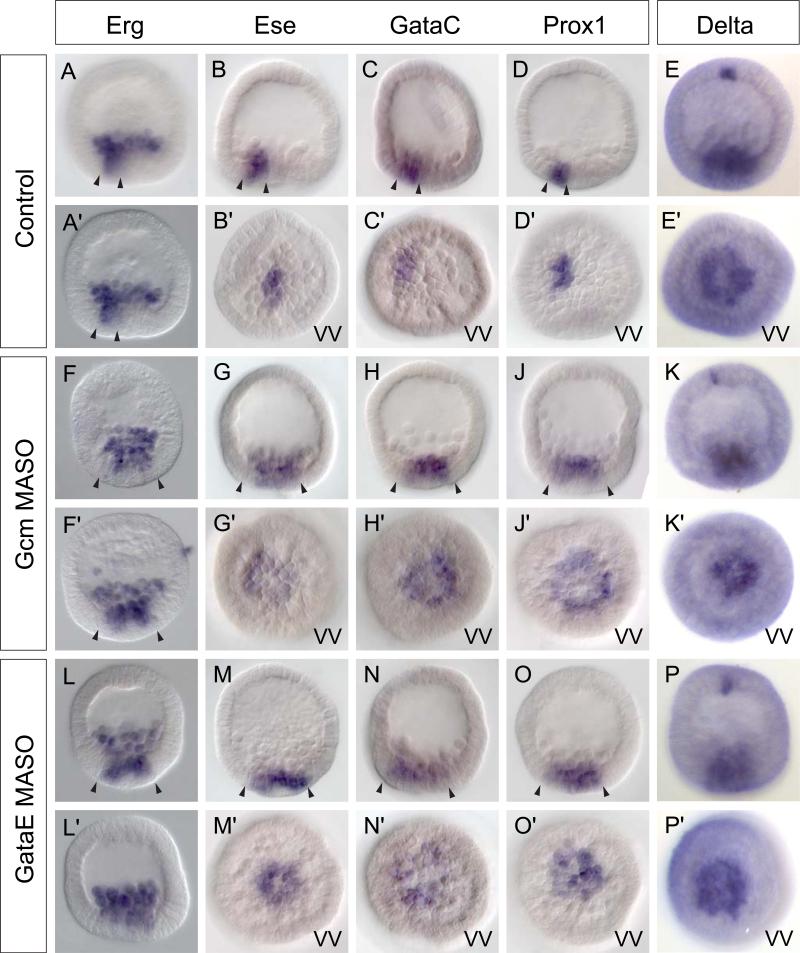
We have just seen that not gene expression is required to prevent the aboral NSM regulatory state from being maintained throughout the whole of the NSM. The experiments in Fig. 5 show an opposite effect exercised by aboral transcription factors on oral NSM genes: If gcm or gataE expression are blocked by treatment with MASOs we see an expansion of ese, prox1, gataC, and erg expression (the same is seen for shr2 expression; not shown). Instead of the 10 or 12 cells of the oral NSM segement normally expressing these genes at 24 hpf, they are now expressed ectopically within the NSM. In about one–third of embryos their transcripts are seen uniformly in all NSM cells (e.g., Fig. 5G -J, 5M -O; compare to controls in 5B – D ; additional examples are shown in Supp. Fig. 2). This result is similar to what has been observed following perturbation of Bmp2/4 (Duboc et al., 2010). However, interference with Bmp signaling leads to a dramatically expanded area of effective Nodal signaling, and thus an expansion of oral domains (Duboc et al., 2004), at least partially due to repression of aboral fates via not. Our perturbation of aboral NSM genes leaves the oral/aboral boundary outside the NSM unperturbed and is thus a more direct test of the interactions within the NSM. Gcm and GataE MASO both produce the same effect, and since Gcm is an upstream driver of gataE as we confirm below, the actual spatial repressor is either GataE or a gene downstream of it. Yet the data in Fig. 6 showing the quantitative effects of Gcm MASO and GataE MASO (see also Supp. Fig. 3) on transcript levels of these oral NSM genes indicates very little change. Since more cells are expressing these genes in the presence of these MASOs, this means that their levels of expression are lower per cell. Thus, paradoxically, GataE or one of its downstream targets is both required for the normal level of expression of oral NSM genes within the oral NSM segment, and also for constraining expression of oral NSM genes outside the oral NSM. The first of these interactions must be executed early while gataE expression overlaps with oral NSM gene expression; the second after the oral and aboral NSM domains have become exclusive. An additional conclusion is that the oral NSM GRN responds to some activator that is present throughout the NSM, and thus its normal level of expression is the output ofboth specific oral NSM and pan-NSM drivers.
Figure 5.
Spatial effects of interference with aboral NSM specification on expression of oral NSM genes. A–D) In wild type embryos the oral NSM is made up of about a dozen cells. F–J, L–O) Treatment with Gcm and GataE MASO causes ectopic expression of oral genes within the NSM. Arrowheads indicate the width of the expression compartment in lateral views. G’–J’, L’–O’) In about one-third of embryos, oral NSM genes are expressed uniformly throughout the entire ring of NSM cells reminiscent of delta expression. E, K, P) Expression of delta is not affected by GataE and Gcm MASO injections. All embryos are 24 hpf. Oral side oriented to the left. VV – vegetal view.
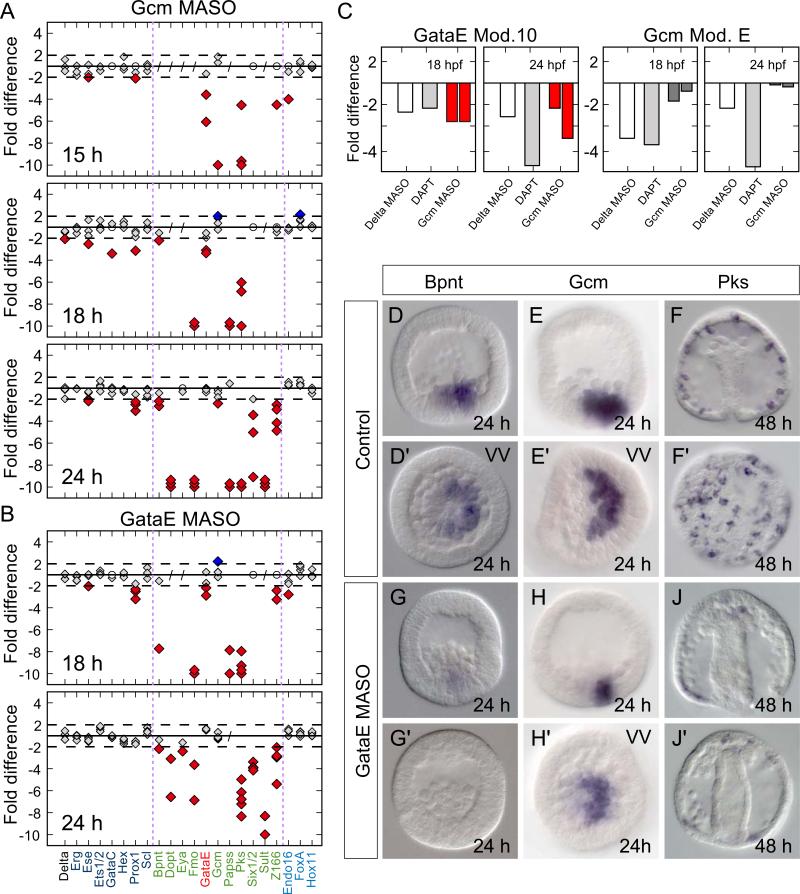
Figure 6.
gataE is a central node in the pre-gastrular aboral NSM GRN. A) Quantitative evaluation of transcript levels following Gcm MASO injection. B) GataE MASO injection causes reduced abundance of aboral NSM genes similar to Gcm knockdown. Labels, symbols and cutoffs in as in Fig. 2. C) gataE Module 10 transcriptional activity is strongly reduced following Delta MASO injection or DAPT treatment. Gcm MASO injection causes a significant reduction in transcriptional output of Module 10. Activity of Gcm module E serves as a control. It receives an input from Notch but not Gcm. D–J) In situ staining of aboral NSM gene transcripts confirms the quantitative effects and shows that markers of pigment cell differentiation are strongly reduced following GataE MASO injection. F, J) Perturbed embryos develop few, if any, pigment cells as confirmed by the almost complete absence of pks expressing cells at late gastrulation. Oral side oriented to the left. VV – vegetal view.
Additional linkages in the GRN of the definitive aboral NSM
The gcm cis-regulatory system has been established as the earliest direct target of D/N signaling (Ransick and Davidson, 2006). Gcm provides a direct input into pigment cell differentiation genes, which begin to be expressed even before gastrulation (Calestani et al., 2003; Calestani and Rogers, 2010). Specification and differentiation of pigment cells is thus mediated by a relatively shallow regulatory hierarchy. Nonetheless, the aboral NSM GRN retained several gaps that we have now been able to fill.
A Nanostring analysis of the consequences of interference with gcm expression was carried out to provide a comprehensive indication of the downstream gene set, supplemented by QPCR analysis of an enhanced set of pigment cell differentiation genes (Fig. 6A). As expected, Gcm MASO causes severe decreases in transcript levels of the differentiation genes bpnt, dopt, fmo, papss, pks, and sult. The bpnt and papss genes are transcribed maternally and these RNAs are initially ubiquitous, but transcripts become localized to the NSM at blastula stage (Röttinger et al., 2006). In situ hybridization confirms localization of bpnt transcripts in aboral NSM at 24 hpf (Fig. 6D,D). Fig. 6A also shows that at 24 hpf Gcm MASO significantly depresses six1/2, and the same is true of another recently discovered aboral NSM gene, z166.
An important result is that embryos in which the Gcm factor is depleted display a significant reduction in the level of gataE transcripts at the 15 and 18 hour time points (Fig. 6A). As noted above, gataE is expressed congruently with gcm in all NSM cells during this period. The gataE gene had previously been identified as a direct D/N target (Lee et al., 2007; Lee, 2007; Materna and Davidson, 2012). But a conundrum remained from a cis-regulatory study of the gataE gene (Lee, 2007). This was the implication of an additional positive input into the gataE gene, an input also downstream of D/N signaling, because stronger depression of activity of the relevant gataE cis-regulatory module (“Module 10”) is observed when all Delta expression is blocked using Delta MASO than when the Su(H) target sites in this module are mutated (Lee, 2007). Our results suggest that the additional input in the early NSM could be from Gcm itself (interference with neither gcm nor delta expression affects later gataE expression in the endoderm; Materna and Davidson, 2012).
This inference was strengthened by experiments utilizing the Module 10 gataE cis-regulatory expression construct, which had been previously shown to recapitulate accurate expression at this developmental stage (Lee et al., 2007). In tests using either Delta MASO or DAPT, Module 10 construct output decreased 2–4 fold (Fig. 6C). However, a very similar effect on Module 10 construct output was obtained by blocking gcm expression, in the absence of any interference with D/N signaling (Fig. 6C). Within the 600 bp Module 10 we identified two short sequences (ACCCGTAG; ACCCGTGT) that bear resemblance to the Gcm consensus site (IUPAC sequence: RCCCGYAT). But whether these putative binding sites are functional remains to be confirmed. Nevertheless, our findings indicate that Gcm is likely to provide the missing Notch-dependent input into the gataE gene during its early NSM phase of expression.
GataE in turn plays a major role in the aboral NSM GRN. Two different GataE MASOs were utilized (see Supp. Fig. 3). Fig. 6A,B shows a striking similarity in the effects of GataE MASO and the effects of Gcm MASO. Since Gcm is a driver of gataE, the implication is that gataE provides direct inputs into the affected genes. As Calestani and Rogers (2010) showed, differentiation genes such as pks receive both inputs directly, in a feed forward relation. Figs. 6G,G show that in the absence of gataE expression embryos fail to express bpnt at 24 hpf. At 48 hpf GataE MASO embryos lack pigment cells altogether, as shown by the absence of pks expression (Fig. 6J,J). Transcript levels of six1/2 and z166 are also greatly reduced in embryos bearing GataE MASO, about equally to embryos bearing Gcm MASO, so gataE is likely to provide direct inputs into these genes of the aboral GRN; the delayed activation of these genes compared to that of gcm can be explained satisfactorily in this way. However, in contrast to gcm which continues to be expressed in differentiated pigment cells (Ransick and Davidson, 2006; 2012) there is no detectable expression of gataE in dispersed pigment cells (Lee and Davidson, 2004). The key roles of GataE are thus confined to multiple participations in the pregastrular aboral NSM specification GRN.
Discussion
Two major clarifications devolve from the detailed measurements presented in this work. First, taken together with prior results, these studies now provide a more comprehensive architectural image of the aboral NSM gene control network as it is formulated through pre-gastrular development, from its initiation, to the activation of differentiation genes. Second, we have identified not as the gene that transforms the Nodal signal into a transcriptional regulatory input initiating the spatially confined gene interactions which in turn generate the oral NSM regulatory state. And third, we have shown that mutual repressive interactions between oral and aboral NSM GRNs are required to maintain their separate domains. Knowledge of the internal linkages of the oral NSM remains sporadic however, but as a result of the current study at least some of the inputs, outputs and functions of this GRN are now in focus.
The aboral NSM GRN
Understanding of the GRN that controls specification of the aboral NSM has grown incrementally since the initial studies, which demonstrated that the aboral NSM gives rise to pigment cells, and that the specification of the aboral NSM depends on Notch signaling in response to presentation of the Delta ligand by the adjacent SM (Sherwood and McClay, 1999; Sweet et al., 2002). A major step was the cis-regulatory demonstration of the direct interaction of activated Su(H) with the early control module of the primary Notch target, the gcm gene (Ransick and Davidson, 2006). A recent study illuminated the use of a different (i.e., “late”) cis-regulatory module of gcm, which controls its expression after the initiating Delta signal has ceased (Ransick and Davidson, 2012). This work demonstrated the institution of a feedback relation with six1/2, which serves to stabilize the aboral NSM regulatory state in addition to positive gcm auto-regulation. Here we establish the crucial link between Notch signaling, gcm, gataE and six1/2.
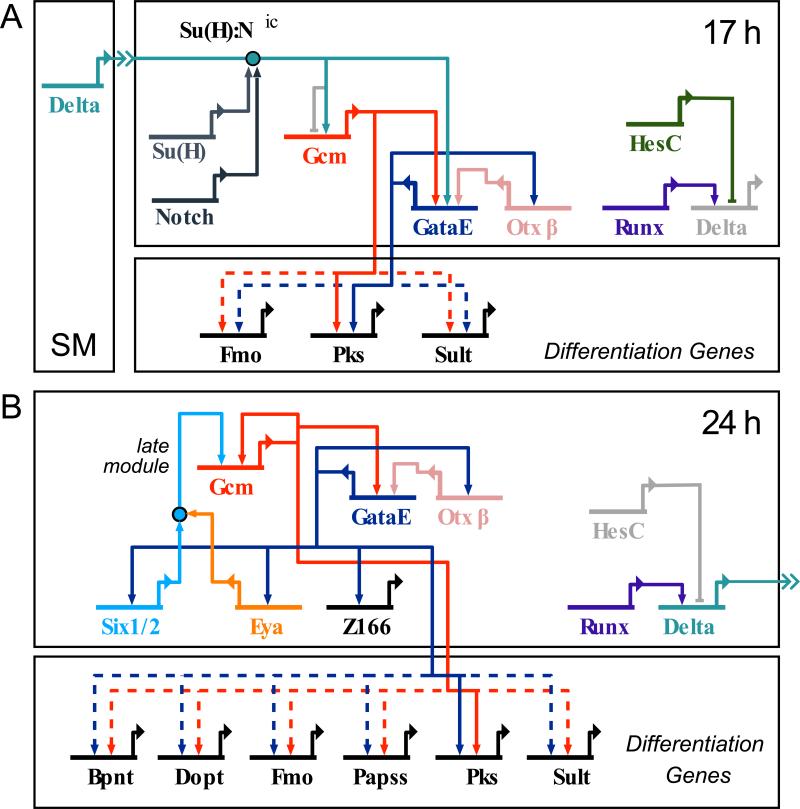
Genetic regulatory circuitry is interesting and important in different ways but of most general interest, the GRN explains the developmental phenomenology. For instance the GRNs in Fig. 7 explain how and why reception of the Delta signal produces the pigment cell regulatory state and hence pigment cell specification. It explains the roles of gcm, and illustrates their multiplicity in the hierarchical process. First the gcm gene acts as the primary, novel, regulatory state pioneer activated by the D/N signal (Fig. 7A). Next it sets in train an immediately downstream circuit including activation of a player of major significance, gataE. Next it locks in the pigment cell regulatory state by engaging in a three gene feedback loop (Fig. 7B). Then it serves as a driver of differentiaiton genes, i.e., pigment synthesis genes. We can now understand another phenomenon, the sufficiency of forced gcm expression in the total absence of Notch signaling for conversion of another mesodermal cell type to pigment cell fate (Damle and Davidson, 2012). An interesting sidelight afforded by knowledge of the circuitry is that not all circuit elements are equally important: in this forced expression experiment the feed forward Notch signal input directly into gataE is absent, so we can regard that as a robust aspect of design rather than an essential aspect.
Figure 7.
GRN for establishment and maintenance of the aboral NSM regulatory state. A) Prior to NSM subdivision Delta signaling emanating from the skeletogenic mesoderm (SM) activates gcm. Both Delta and Gcm activate gataE transcription as part of a coherent feed forward loop. Delta signaling also serves to clear endoderm genes by interference with TCF/β-catenin, the main driver of endoderm genes. Presence of HesC prevents delta expression. B) Following separation of NSM segments, the aboral NSM regulatory state is locked down through positive feedback between gataE, six1/2–eya and gcm. This subcircuit is thus independent of the initial Delta input. Repression of delta is relieved after clearance of HesC.
There remain unresolved questions of course. One outstanding issue is the identity of the aboral NSM gene which represses oral NSM genes resulting in the expanded pattern of oral gene expression when either Gcm MASO or GataE MASO is applied. There are currently no known target genes for z166 and its function remains unresolved. And among the differentiation genes only for pks is there direct evidence of the driver linkages into the GRN (Calestani and Rogers, 2010). Finally, though this refers to all the NSM, more recent results show that the control of hesC expression needs further examination. hesC clearance from the NSM is what allows delta to be expressed there after about 19 hpf (Sweet et al, 2002; Revilla-i-Domingo et al., 2007; Smith and Davidson, 2008). As we see in Figs. 1 and 5, this is the only regulatory gene within the NSM that does not respect the oral/aboral regulatory state segregation.
Initiation and maintenance of the oral NSM regulatory state
As stated above, the GRN operating in the oral NSM has yet to be elucidated. However, a complicating problem is that two of the key early oral NSM genes, prox1 and ese, are expressed zygotically in a global fashion before becoming exclusively transcribed in the oral NSM. The prox1 gene is transcribed from early in development (as well as maternally), and interruption of early prox1 transcript processing with MASOs produces cleavage stage lethality. GataC MASO treatment does not affect any regulatory genes pre-gastrulation (Supp. Fig. 4; Solek and Rast, personal communication); and Ese MASO treatment only produces a mild reduction of prox1 transcript (Supp. Fig. 4). Thus current analyses of regulatory transactions in the specification of the oral NSM begin with the expression patterns of genes that for the most part are functionally unconnected.
In addition to clarifying the total exclusiveness of oral vs. aboral NSM regulatory states following subdivision, this study has revealed the identity of the gene which initiates the proper establishment of the oral NSM regulatory state. This is the Nodal target gene not. The evidence, shown in Figs. 2, 3, and 4, and Supp. Fig. 1, can be summarized briefly as follows: (i) nodal transcription precedes transcription of the not gene, and transcription of not precedes subdivision of the mesoderm (Fig. 1), as required for its proposed role in the NSM. (ii) Downregulation of all oral NSM genes caused by Nodal MASO is exactly paralleled by the effects of Not MASO (Figs. 2 and 4, 24 hpf time points), also as required if not is the effector gene which explains the role of Nodal in the oral NSM (Duboc et al., 2004; Duboc et al., 2010; Su et al., 2009). (iii) Interference with expression of not prevents proper establishment of the oral NSM. In its place gcm, gataE and the aboral regulatory state continue to be expressed in the whole NSM rather than clearing from the oral segment (Fig. 4); therefore expression of the not gene is functionally causal for normal oral NSM specification and disappearance of the aboral NSM regulatory state. Expression of not in the oral segment of the NSM begins prior to 16 hpf (Fig. 3G,H) while clearance of gcm and gataE transcripts from the oral NSM occurs after 18 hpf (Fig. 3J,J′). Thus the early expressed genes of the aboral GRN, in particular gcm and gataE, are co-expressed on the oral side with the not gene for several hours. Therefore, Not could be acting as a direct repressor of gcm (or of both of these genes) though less direct explanations cannotbe excluded. (iv) expression of not is impervious to interference with D/N signaling (Fig. 3), but every oral NSM gene requires the SM Delta signal for its subsequent expression (Materna et al., 2012); therefore, though not expression is required for oral NSM specification, an additional Notch-sensitive input is also required. (v) Although gcm and gataE eventually cease to be expressed in the oral NSM, their transcription overlaps with that of oral NSM genes for a significant amount of time. They could therefore provide an initial input into the transcriptional regulators of the oral NSM, consistent with the depression of prox1 expression seen in both Gcm and GataE MASO (Fig. 6A,B).
In sum, we have learned that not transcription is the obligatory initiator of the regulatory train of events culminating in the definitive oral NSM GRN. The not gene is not to be considered an intrinsic part of this network; as is so often the case for genes that perform pioneer functions in embryonic specification, not expression does not continue in the NSM, as this gene represses itself (Fig. 4) (Li et al., 2012) resulting in transient expression there. Instead the specific role of the not gene is likely to deal with the developmentally pre-existent aboral NSM GRN so as to permit installation of the oral NSM GRN. This places the NSM role of not expression in an interesting evolutionary light.
For it is the oral NSM, not the aboral NSM, that is pleisiomorphic for echinoderms. The pigment cell lineage is likely an invention of the echinoid branch of the eleutherozoan echinoderms. Current phylogeny (Pisani et al., 2011) shows the eleutherozoan outgroup to the echinoids, ophiuroids and holothuroids, to be the asteroids or sea stars, the divergence dating back to the origins of the eleutherozoans, probably in the lower Ordovician if not before. Indeed sea stars have no pigment cell lineage and expression of gcm and gataE is conspicuously absent from embryonic sea star mesoderm (Hinman and Davidson, 2007; McCauley et al., 2010). Nor is mesoderm formation Notch-dependent in sea stars as it is in sea urchins (Hinman and Davidson, 2007). On the other hand, the sea star embryonic mesoderm generates blastocoelar cell types similar to those descendent from oral NSM of sea urchins. In both species these cells, as noted earlier, have immune functions in the larva (Hibino et al., 2006; Furukawa et al., 2009). Furthermore, the sea star embryonic mesoderm regulatory state resembles that of the oral NSM of sea urchins, including transcripts of gataC, etsl/2, hex, and erg. In vertebrates the same cohort of genes expressed in the sea urchin oral NSM is utilized widely in hematopoiesis and lymphangiogenesis. Thus for example gataC, scl, and lmo2 form a complex that regulates hematopoietic genes (Lecuyer and Hoang, 2004; Matthews and Visvader, 2003); erg is an ortholog of the vertebrate fli1 gene and together with scl and gataC orthologues is important in hematopoiesis (Pimanda et al, 2007); and orthologs of scl, ese and ets1/2 are involved in specification and function of endothelial cells as well as of many immune cell types (De Val and Black, 2009; Porcher et al., 1996). The prox1 gene is also expressed in endothelial cells where it establishes the lymphatic lineage (Johnson et al., 2008). Thus the regulatory state expressed in the oral NSM is anciently associated with deuterostome immune and hematopoietic system diversification. From this vantage point we can see that the function of the not device and its downstream wiring is to carve out from the evolutionarily superimposed pigment cell regulatory state a mesodermal domain in which the pleisiomorphic echinoderm immune GRN is enabled to develop.
Supplementary Material
Supplemental Figure 1
Not MASO injection causes specific defects.RNA was extracted from MASO injected embryos and transcript levels of 183 genes were quantified using the NanoString nCounter. The counts obtained for each gene in perturbed embryos are plotted against those of control embryos. The MASOaffects only a few genes in the codeset significantly, thus confirming the crude phenotypic assessment that besides thesespecific defects development proceeds unperturbed. By 24 hpf only oral NSM genes and a few oral ectoderm genes are affected in repeat experiments. The dotted lines indicate a threshold of 2-fold change. Genes present with ca. 25 transcripts or less per embryo are marked with an open, gray circle.Transcription levels were estimated from previous quantification data (Materna et al., 2010);
Supplemental Figure 2
Perturbation of aboral NSM specification causes ectopic expression ofproxl within the NSM. Additional examples after Gcm MASO and GataE MASO injection. All embryos show a significantly increased number of proxl expressing cells even though the spatial arrangement does not always resemble a perfect ring(e.g., D, E, H). Embryos are 24 hpf and presented in a vegetal view unless otherwise noted. LV—lateral view.
Supplemental Figure 3
GataE splice blocking MASO affects transcript abundance of a gene set comparable to GataE translation blocking MASO and causes a significant reduction in transcript levels of aboral NSM/pigment cell differentiation genes. Transcript abundance in injected and control embryos was determined with the Nanostring nCounter and supplemented with QPCR data for pigment cell differentiation genes. Fold-difference of transcript abundance was calculated for each gene; 1-fold difference corresponds to no change, negative values indicate lower transcript abundance ininjected compared to control embryos. Each diamond represent a single experiment. Dashed lines indicate the threshold for significant changes. Oral NSM genes are labeled dark blue, aboral NSM genes green, ectoderm genes grey, endoderm genes light blue.
Supplemental Figure 4
Perturbation effects of GataC and Ese knockdown on transcript levels.A) GataC MASO injection has no significant effect on transcript levels of the genes included in the codeset. B) Ese MASO injection affects only few genes, but, among others, the oral NSM gene proxl. Transcript abundance in injected and control embryos was determined with the Nanostring nCounter and supplemented with QPCR data for pigment cell differentiation genes. Fold-difference of transcript abundance was calculated for each gene; 1-fold difference corresponds to no change, negative values indicate lower transcript abundance in injected compared to control embryos. Each diamond represent a single experiment. Genes not significantly expressed in experiment and control are marked with an open white circle. Dashed lines indicate the threshold for significant changes. Oral NSM genes are labeled dark blue, aboral NSM genes green, ectoderm genes grey, endoderm genes light blue.
Highlights.
Early mesoderm segments display distinct and non-overlapping regulatory states.
Regulome wide analysis identified the pan-oral not gene as a target of Nodal signaling.
not disrupts maintenance ofthe aboral regulatory state and institutes oral mesoderm.
A three-way feedback loop in the aboral mesoderm locks down the aboral regulatory state.
Acknowledgements
We would like to thank Jongmin Nam for his help with Nanotag experiments and Gilson Sanchez for help with in situ stainings. Many thanks to Cynthia Solek and Jonathan Rast for sharing unpublished data. We are greatful for many discussions with Joel Smith and Celina Juliano. Research was supported by NIH grant HD037105 and by the Lucille P. Markey Charitable Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calestani C, Rast JP, Davidson EH. Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development. 2003;130:4587–4596. doi: 10.1242/dev.00647. [DOI] [PubMed] [Google Scholar]
- Calestani C, Rogers DJ. Cis-regulatory analysis of the sea urchin pigment cell gene polyketide synthase. Dev. Biol. 2010;340:249–255. doi: 10.1016/j.ydbio.2010.01.026. [DOI] [PubMed] [Google Scholar]
- Damle SS, Davidson EH. Synthetic in vivo validation of gene network circuitry. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1548–1553. doi: 10.1073/pnas.1119905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, Black BL. Transcriptional control of endothelial cell development. Dev. Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Lapraz F, Besnardeau L, Lepage T. Lefty acts as an essential modulator of Nodal activity during sea urchin oral-aboral axis formation. Dev. Biol. 2008;320:49–59. doi: 10.1016/j.ydbio.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Duboc V, Lapraz F, Saudemont A, Bessodes N, Mekpoh F, Haillot E, Quirin M, Lepage T. Nodal and BMP2/4 pattern the mesoderm and endoderm during development of the sea urchin embryo. Development. 2010;137:223–235. doi: 10.1242/dev.042531. [DOI] [PubMed] [Google Scholar]
- Duboc V, Röttinger E, Besnardeau L, Lepage T. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev. Cell. 2004;6:397–410. doi: 10.1016/s1534-5807(04)00056-5. [DOI] [PubMed] [Google Scholar]
- Geiss G, Bumgarner R, Birditt B, Dahl T, Dowidar N, Dunaway D, Fell H, Ferree S, George R, Grogan T, James J, Maysuria M, Mitton J, Oliveri P, Osborn J, Peng T, Ratcliffe A, Webster P, Davidson E, Hood L. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Furukawa R, Takahashi Y, Nakajima Y, Dan-Sohkawa, Kaneko H. Defense system by mesenchyme cells in bipinnaria larvae of the starfish, Asterina pectinifera. Dev. Comp. Immunol. 2009;33:205–215. doi: 10.1016/j.dci.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, Buckley KM, Brockton V, Nair SV, Berney K, Fugmann SD, Anderson MK, Pancer Z, Cameron RA, Smith LC, Rast JR. The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JN, Dodge N, Rathjen PD, Rathjen J. A novel role for gamma-secretase in the formation of primitive streak-like intermediates from ES cells in culture. Stem Cells. 2009;27:2941–2951. doi: 10.1002/stem.218. [DOI] [PubMed] [Google Scholar]
- Johnson NC, Dillard ME, Baluk P, McDonald DM, Harvey NL, Frase SL, Oliver G. Lymphatic endothelial cell identity is reversible and its maintenance requires Proxl activity. Genes Dev. 2008;22:3282–3291. doi: 10.1101/gad.1727208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Hoang T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp. Hematol. 2004;32:11–24. doi: 10.1016/j.exphem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Lee PY. PhD thesis. Caltech; 2007. Function and regulation of the Strongylocentrotus GataE gene. [Google Scholar]
- Lee PY, Davidson EH. Expression of Spgatae, the Strongylocentrotus purpuratus ortholog of vertebrate GATA4/5/6 factors. Gene Expr. Patterns. 2004;5:161–165. doi: 10.1016/j.modgep.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Lee PY, Nam J, Davidson EH. Exclusive developmental functions of gatae cis-regulatory modules in the Strongylocentrorus purpuratus embryo. Dev. Biol. 2007;307:434–445. doi: 10.1016/j.ydbio.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Materna SC, Davidson EH. Direct and indirect control of oral ectoderm regulatory gene expression by Nodal signaling in the sea urchin embryo. Dev. Biol. 2012 doi: 10.1016/j.ydbio.2012.06.022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Davidson EH. A comprehensive analysis of Delta signaling in pre-gastrular sea urchin embryos. Dev. Biol. 2012;364:77–87. doi: 10.1016/j.ydbio.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Howard-Ashby M, Gray RF, Davidson EH. The C2H2 zinc finger genes of Strongylocentrotus purpuratus and their expression in embryonic development. Dev. Biol. 2006;300:108–120. doi: 10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Materna SC, Nam J, Davidson EH. High accuracy, high-resolution prevalence measurement for the majority of locally expressed regulatory genes in early sea urchin development. Gene Expr. Patterns. 2010;10:177–184. doi: 10.1016/j.gep.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Oliveri P. A protocol for unraveling gene regulatory networks. Nat. Protoc. 2008;3:1876–1887. doi: 10.1038/nprot.2008.187. [DOI] [PubMed] [Google Scholar]
- Matthews JM, Visvader JE. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 2003;4:1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccauley BS, Weideman EP, Hinman VF. A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev. Biol. 2010;340:200–208. doi: 10.1016/j.ydbio.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Nam J, Davidson EH. Barcoded DNA-tag reporters for multiplex cis-regulatory analysis. Plos One. 2012;7:e35934. doi: 10.1371/journal.pone.0035934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J, Su YH, Lee PY, Robertson AJ, Coffman JA, Davidson EH. Cis-regulatory control of the nodal gene, initiator of the sea urchin oral ectoderm gene network. Dev. Biol. 2007;306:860–869. doi: 10.1016/j.ydbio.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development. 2006;133:4173–4181. doi: 10.1242/dev.02577. [DOI] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev. Biol. 2010;340:188–199. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. A gene regulatory network controlling the embryonic specification of endoderm. Nature. 2011;474:635–639. doi: 10.1038/nature10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KJ, Harada Y, Cameron RA, Davidson EH. Expression pattern of Brachyury and Not in the sea urchin: comparative implications for the origins of mesoderm in the basal deuterostomes. Dev. Biol. 1999;207:419–431. doi: 10.1006/dbio.1998.9177. [DOI] [PubMed] [Google Scholar]
- Pimanda JE, Ottersbach K, Knezevic K, Kinston S, Chan WYI, Wilson NK, Landry J, Wood AD, Kolb-Kokocinski A, Green AR, Tannahill D, Lacaud G, Kouskoff V, Göttgens B. Gata2, Flil, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17692–17697. doi: 10.1073/pnas.0707045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani D, Feuda R, Peterson KJ, Smith AB. Resolving phylogenetic signal from noise when divergence is rapid: a new look at the old problem of echinoderm class relationships. Mol. Phylogenet. Evol. 2012;62:27–34. doi: 10.1016/j.ympev.2011.08.028. [DOI] [PubMed] [Google Scholar]
- Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- Poustka AJ, Kiihn A, Groth D, Weise V, Yaguchi S, Burke RD, Herwig R, Lehrach H, Panopoulou G. A global view of gene expression in lithium and zinc treated sea urchin embryos: new components of gene regulatory networks. Genome Biol. 2007;8:R85. doi: 10.1186/gb-2007-8-5-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Range R, Lapraz F, Quirin M, Marro S, Besnardeau L, Lepage T. Cis-regulatory analysis of nodal and maternal control of dorsal-ventral axis formation by Univin, a TGF-beta related to Vg1. Development. 2007;134:3649–3664. doi: 10.1242/dev.007799. [DOI] [PubMed] [Google Scholar]
- Ransick A. Detection of mRNA by in situ hybridization and RT-PCR. Methods Cell. Biol. 2004;74:601–620. doi: 10.1016/s0091-679x(04)74024-8. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev. Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. Cis-regulatory logic driving glial cells missing: Self-sustaining circuitry in later embryogenesis. Dev. Biol. 2012;364:259–267. doi: 10.1016/j.ydbio.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast JP, Smith LC, Loza-Coll M, Hibino T, Litman G. Genomic insights into the immune system of the sea urchin. Science. 2006;314:952–956. doi: 10.1126/science.1134301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus). Dev. Biol. 2006;300:35–48. doi: 10.1016/j.ydbio.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Rottinger E, Croce J, Lhomond G, Besnardeau L, Gache C, Lepage T. Nemo-like kinase (NLK) acts downstream of Notch/Delta signalling to downregulate TCF during mesoderm induction in the sea urchin embryo. Development. 2006;133:4341–4353. doi: 10.1242/dev.02603. [DOI] [PubMed] [Google Scholar]
- Ruffins SW, Ettensohn CA. A fate map of the vegetal plate of the sea urchin (Lytechinus variegatus) mesenchyme blastula. Development. 1996;122:253–263. doi: 10.1242/dev.122.1.253. [DOI] [PubMed] [Google Scholar]
- Sharma T, Ettensohn CA. Regulative deployment of the skeletogenic gene regulatory network during sea urchin development. Development. 2011;138:2581–2590. doi: 10.1242/dev.065193. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, McClay DR. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development. 1999;126:1703–1713. doi: 10.1242/dev.126.8.1703. [DOI] [PubMed] [Google Scholar]
- Smith J, Davidson EH. Gene regulatory network subcircuit controlling a dynamic spatial pattern of signaling in the sea urchin embryo. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20089–20094. doi: 10.1073/pnas.0806442105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Li E, Geiss GK, Longabaugh WJ, Kramer A, Davidson EH. A perturbation model of the gene regulatory network for oral and aboral ectoderm specification in the sea urchin embryo. Dev. Biol. 2009;329:410–421. doi: 10.1016/j.ydbio.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet HC, Gehring M, Ettensohn CA. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development. 2002;129:1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- Tu Q, Cameron RA, Worley KC, Gibbs RA, Davidson EH. Gene structure in the sea urchin Strongylocentrotus purpuratus based on transcriptome analysis. Genome Res. 2012 doi: 10.1101/gr.139170.112. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: phylogeny and embryonic expression. Dev. Biol. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Walton KD, Warner J, Hertzler PH, McClay DR. Hedgehog signaling patterns mesoderm in the sea urchin. Dev. Biol. 2009;331:26–37. doi: 10.1016/j.ydbio.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1
Not MASO injection causes specific defects.RNA was extracted from MASO injected embryos and transcript levels of 183 genes were quantified using the NanoString nCounter. The counts obtained for each gene in perturbed embryos are plotted against those of control embryos. The MASOaffects only a few genes in the codeset significantly, thus confirming the crude phenotypic assessment that besides thesespecific defects development proceeds unperturbed. By 24 hpf only oral NSM genes and a few oral ectoderm genes are affected in repeat experiments. The dotted lines indicate a threshold of 2-fold change. Genes present with ca. 25 transcripts or less per embryo are marked with an open, gray circle.Transcription levels were estimated from previous quantification data (Materna et al., 2010);
Supplemental Figure 2
Perturbation of aboral NSM specification causes ectopic expression ofproxl within the NSM. Additional examples after Gcm MASO and GataE MASO injection. All embryos show a significantly increased number of proxl expressing cells even though the spatial arrangement does not always resemble a perfect ring(e.g., D, E, H). Embryos are 24 hpf and presented in a vegetal view unless otherwise noted. LV—lateral view.
Supplemental Figure 3
GataE splice blocking MASO affects transcript abundance of a gene set comparable to GataE translation blocking MASO and causes a significant reduction in transcript levels of aboral NSM/pigment cell differentiation genes. Transcript abundance in injected and control embryos was determined with the Nanostring nCounter and supplemented with QPCR data for pigment cell differentiation genes. Fold-difference of transcript abundance was calculated for each gene; 1-fold difference corresponds to no change, negative values indicate lower transcript abundance ininjected compared to control embryos. Each diamond represent a single experiment. Dashed lines indicate the threshold for significant changes. Oral NSM genes are labeled dark blue, aboral NSM genes green, ectoderm genes grey, endoderm genes light blue.
Supplemental Figure 4
Perturbation effects of GataC and Ese knockdown on transcript levels.A) GataC MASO injection has no significant effect on transcript levels of the genes included in the codeset. B) Ese MASO injection affects only few genes, but, among others, the oral NSM gene proxl. Transcript abundance in injected and control embryos was determined with the Nanostring nCounter and supplemented with QPCR data for pigment cell differentiation genes. Fold-difference of transcript abundance was calculated for each gene; 1-fold difference corresponds to no change, negative values indicate lower transcript abundance in injected compared to control embryos. Each diamond represent a single experiment. Genes not significantly expressed in experiment and control are marked with an open white circle. Dashed lines indicate the threshold for significant changes. Oral NSM genes are labeled dark blue, aboral NSM genes green, ectoderm genes grey, endoderm genes light blue.