Abstract
Atomic force microscopy (AFM) was used to probe the mechanical and adherence properties of eight L. monocytogenes’ strains representative of the species’ two phylogenetic lineages I and II. From a functional perspective, lineage’ I strains were characterized by lower overall adhesion forces and higher specific and nonspecific forces compared to lineage’ II strains. From a structural perspective, lineage’ II strains were characterized by higher Young’s moduli and longer and stiffer biopolymers compared to lineage’ I strains. Both lineages’ I and II strains were similar in their grafting densities. Finally, our results indicated that epidemic and environmental strains of L. monocytogenes and irrespective of their lineage group were characterized by similar Young’s moduli of elasticties and adhesion forces at the cellular level. However, at the molecular level, epidemic strains were characterized by higher specific and nonspecific forces, shorter, denser and more flexible biopolymers compared to environmental strains.
Keywords: Listeria monocytogenes, Lineages’ I and II, epidemic, environmental, AFM, adhesion, mechanics and pathogenicity
Introduction
Listeria monocytogenes, a model organism for intracellular parasitism, are Gram-positive food-borne bacteria that are ubiquitous in nature [1]. L. monocytogenes can be isolated from a wide array of environmental sources including soil, sewage, food-processing plants and contaminated food [2]. Although literature studies indicate that all L. monocytogenes strains possess the single pathogenicity island (PAI) necessary for the in vivo internalization of the microbe within host cells, not all L. monocytogenes strains are pathogenic [3]. Pathogenic L. monocytogenes are known to cause severe human infections such as listeriosis and meningitis [4].
L. monocytogenes strains display both genetic and serotypic diversity [5]. Serotypic diversity arises from combinations of somatic (O) and flagellar (H) antigens, resulting in thirteen recognized serotypes within the species [6]. In comparison to serotyping, molecular subtyping methods such as pulse field gel electrophoresis allow for a more sensitive classification of L. monocytogenes population structures [7]. Based on subtyping, L. monocytogenes strains can be categorized into two major genetic groups named lineages’ I and II [8] and a less common, third lineage’ III [7, 9]. Lineage’ I strains are clonal and include strains of serotypes 1/2b, 3b, 3c and 4b [10]. Lineage’ II strains are largely diverse due to horizontal gene transfer and include strains of serotypes 1/2a, 1/2c and 3a [10]. Lineage’ III isolates are rare, overrepresented among isolates from food-production animals, underrepresented among human clinical cases and foods and largely represent strains of serotypes 4a and 4c [11]. Lineage’ I strains represent largely human host-adapted strains capable of causing disease due to their significantly higher pathogenic potential compared to strains of lineage II [12, 13]. In comparison, lineage’ II strains are environmentally-adapted and represent generic heterogeneous strains that are better suited to survive and multiply in the environment [14].
Despite significant differences in the genetic composition of lineages’ I and II strains, a clear distinction in the ability of the two lineages to cause diseases is still lacking [8]. For example, while strains of lineage II are known for their limited pathogenicity, some lineage II strains were shown to infect mammalian hosts [15]. Similarly, the infectivity of different serotypes appears to be randomly distributed among strains of lineages’ I and II. More than 90% of human listeriosis cases are attributed to only four serotypes (1/2a, 1/2b, 1/2c and 4b) with serotype 4b causing more than 50% of the listeriosis cases worldwide and accounting for nearly all of the outbreaks of human food-borne and perinatal listeriosis [5, 16]. Note that serotypes 1/2a and 1/2c belong to lineage’ II while 1/2b and 4b strains represent lineage’ I.
Traditionally, the ability of a bacterium to cause a disease has been largely determined by molecular epidemiology methods such as deoxyribonucleic acid (DNA) banding pattern-based subtyping methods [8]. However, it is clear that identifying the pathogenic potential of L. monocytogenes strains on the basis of serotyping and/or subtyping methods is not sufficiently distinctive. Therefore, the use of AFM can improve and complement our understanding of the relationship between the phylogenetic structural properties of L. monocytogenes’ strains and their variable functionalities such as survival in the environment, ability to colonize diverse surfaces and potential to infect multiple host species [17].
Here, we have used AFM to probe the adherence and mechanics of a set of L. monocytogenes strains representative of the species’ two major phylogenetic lineages. Some of the strains investigated were selected because they exhibited an unexpected epidemic functionality for their particular lineage. For example, environmental strains that belong to lineage I and epidemic strains that belong to lineage II were investigated. Due to their lack of representation among human clinical isolates [11], lineage’ III strains were not investigated.
Materials and Methods
Bacterial strains
Listeria monocytogenes strains investigated in this study were obtained from Prof. Douglas Call from the Allen School of Global Animal Health at Washington State University. Strains were either representative of lineage I or lineage II (Table 1). In selecting the strains above, caution was experienced to have environmental and epidemic strains representative of both lineages. The ability of L. monocytogenes strains used here to cause infections in mice was investigated by Dr. Call’s group using oral inoculation methods [18]. Their results revealed that mice inoculated with epidemic strains were significantly more invasive as a group and more likely to become infected than mice inoculated with environmental strains (P < 0.01) [18]. All epidemic strains investigated here were isolated from patients with L. monocytogenes infections and hence are pathogenic. In comparison, the environmental strains investigated here were isolated from contaminated food or soil. It has to be noted that both epidemic and environmental strains can be pathogenic because they have the pathogenicity genes, however only epidemic strains studied here were shown to really cause infections. A detailed description of how bacterial strains were cultured is given in the supplementary methods.
Table 1.
A summary of the cellular overall adhesion forces quantified using AFM between eight L. monocytogenes strains that represent both lineages' I and II and a model surface of Si3N4 in water. In addition, the specific and nonspecific force components decoupled by using Poisson statistical analysis and the estimated nanomechanical properties using WLC modeling, Hertz modeling and steric modeling of the data are also included. Finally, the total protein content measured for the various strains is included. Errors reported here represent the standard errors of the mean.
| Strain(LMB) | 469a* | 496a * | 503b * | 508a, * | 429a§ | 446a§ | 437b§ | 622b§ |
|---|---|---|---|---|---|---|---|---|
| Serotype | 4b | 4b | 1/2a | 1/2b | 4b | 4b | 1/2a | 1/2c |
| Analysis of adhesion forces measured by AFM and estimated using Poisson statistical method | ||||||||
| AFM average adhesion force (Fad, nN) | 0.126 ± 0.002 | 0.224 ± 0.007 | 0.175 ± 0.004 | 0.121 ± 0.002 | 0.175 ± 0.12 | 0.141 ± 0.003 | 0.207 ± 0.003 | 0.107 ± 0.003 |
| Specific force (Fi, nN) | 0.091 | 0.088 | 0.089 | 0.077 | 0.075 | 0.046 | 0.084 | 0.054 |
| Nonspecific force (Fo, nN) | 0.096 | 0.081 | 0.092 | 0.077 | 0.093 | 0.061 | 0.119 | 0.059 |
| Number of peaks | 963 | 367 | 568 | 652 | 647 | 635 | 630 | 297 |
| # of hydrogen bonds | 0.33 | 1.63 | 0.93 | 0.57 | 1.10 | 1.73 | 1.05 | 0.89 |
| Hertz modeling of the AFM force-indentation data | ||||||||
| E(kPa) | 156.7 ± 2.0 | 137.2 ± 2.3 | 153.5 ± 1.8 | 144.4 ± 2.4 | 150.8 ± 1.5 | 142.9 ± 2.5 | 154.7 ± 1.5 | 145.4 ± 2.2 |
| r2 | 0.86 | 0.91 | 0.87 | 0.89 | 0.92 | 0.92 | 0.89 | 0.88 |
| n | 153 | 158 | 157 | 215 | 158 | 101 | 108 | 164 |
| Steric modeling of the AFM approach force-distance data | ||||||||
| L (nm) | 87.7 ± 8.4 | 68.9 ± 1.6 | 63.3 ± 1.9 | 62.6 ± 2.9 | 104.8 ± 2.9 | 90.6 ± 1.3 | 143.7 ± 11.6 | 229.1 ± 11.9 |
| Γ × 10−16(m−2) | 3.22 ± 0.17 | 1.65 ± 0.07 | 1.70 ± 0.07 | 3.44 ± 0.17 | 0.89 ± 0.04 | 1.56 ± 0.04 | 3.62 ± 0.20 | 1.68 ± 0.11 |
| r2 | 0.95 | 0.97 | 0.86 | 0.92 | 0.93 | 0.90 | 0.97 | 0.93 |
| n | 189 | 210 | 167 | 201 | 196 | 168 | 190 | 168 |
| WLC modeling of the AFM retraction force-distance data | ||||||||
| Lp (nm) | 0.31 ± 0.02 | 0.32 ± 0.04 | 0.34 ± 0.02 | 0.28 ± 0.02 | 0.41 ± 0.04 | 0.35 ± 0.03 | 0.36 ± 0.04 | 0.39 ± 0.03 |
| Lc (nm) | 1000.4 ± 72.0 | 605.8 ± 50.9 | 595.8 ± 48.1 | 637.5 ± 45.8 | 517.0 ± 25.6 | 1299.2 ± 62.9 | 916.2 ± 82.0 | 993.0 ± 61.1 |
| r2 | 0.84 | 0.87 | 0.84 | 0.84 | 0.78 | 0.78 | 0.80 | 0.83 |
| n | 48 | 26 | 50 | 50 | 37 | 39 | 28 | 53 |
| Colorimetric measurements of the total proteins' concentration in cells | ||||||||
| Total protein concentration (mg/mL) | 0.078 | 0.060 | 0.042 | 0.091 | 0.017 | 0.082 | 0.090 | 0.045 |
Strains that represent lineage I,
Strains that represent lineage II,
Strains that represent epidemic strains and
Strains that represent environmental strains.
AFM experiments
Bacterial cells grown until late exponential phase of growth were attached to gelatin-coated mica disks according to previously published protocols [19]. A detailed description of the conducted AFM experiments is provided in the supplementary methods.
Quantification of AFM adhesion forces
AFM retraction curves were individually analyzed to quantify the adhesion forces between the bacterial surface biopolymers and silicon nitride (Si3N4) in water. Each pull-off peak in the retraction curve was counted as an adhesion peak with similar magnitude and opposite sign [20]. Adhesion forces in this manuscript are thus positive numbers. The adhesion forces measured by AFM represent the sum of all specific and nonspecific forces measured between the AFM Si3N4 tip and the bacterial surface biopolymers. The nonspecific forces for our system (bacteria-Si3N4-water) were considered as the sum of Lifshitz-van der Waals and electrostatic interactions while specific forces were represented by hydrogen bonds.
Poisson statistical analysis of AFM adhesion forces
The overall cellular adhesion force measured using AFM can be decoupled into specific and nonspecific forces using a Poisson statistical analysis method developed originally by Beebe et al (see supplementary methods for details) [65]. Briefly, specific and nonspecific forces can be estimated by:
| (1) |
A linear regression curve of σF2 versus µF can be used to determine the specific individual bond rapture force (Fi) as the slope of the line and the nonspecific forces (Fo) can be calculated then from the intercept (-Fi Fo). For our measurements, σF2 and µF were taken as the variance and the average force for all adhesion events measured on each cell.
Elasticity of bacterial cells
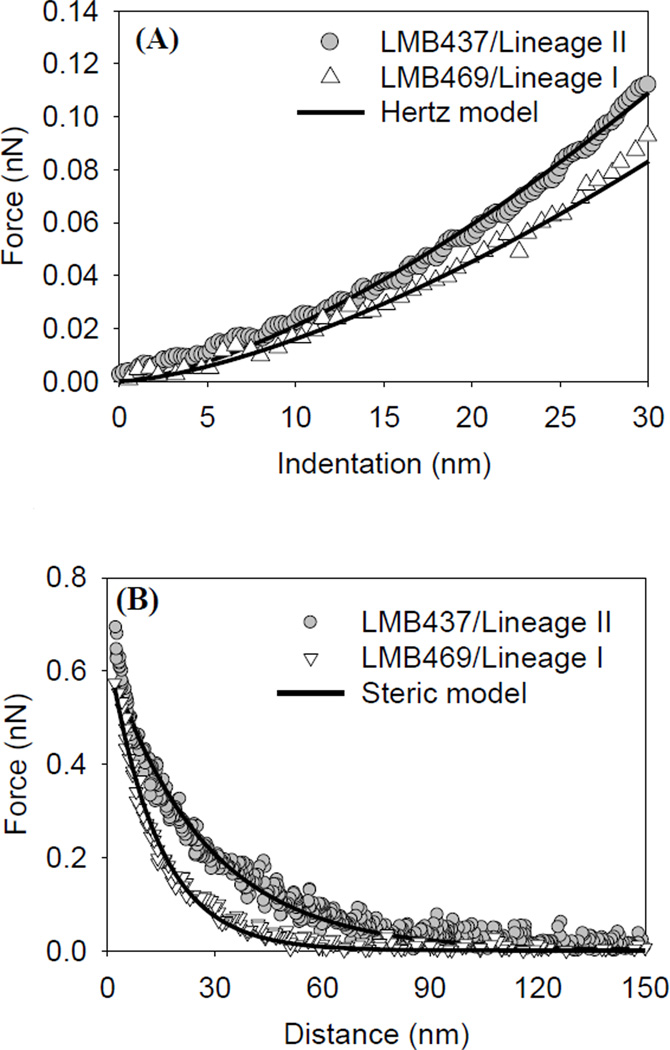
AFM approach raw piezo position-deflection data files were converted to force-indentation files as we described previously [21]. All data were collected under a constant applied load of 1 nN. Quantitative information on bacterial elasticity can be obtained by modeling the measured force-indentation data using the classic Hertz model of contact mechanics [22] (Figure 1A). The model describes the indentation force (F) applied by a non-deformable spherical indenter (the AFM Si3N4 tip) required to indent a distance (δ) in an infinitely deformable elastic half space (the bacterial surface) by:
| (2) |
where E is the bacterial Young’s modulus in kPa and represents the only fitting parameter in eq. 2 R is the indenter radius taken as 40 nm according to the manufacturer value and ν is Poisson ratio of the bacterium taken as 0.5 [22]. For all bacterial strains tested, Hertz model was applied for the first 30 nm indented in the bacterial biopolymer brush. Classic Hertz model ignores tipsurface adhesion. However, since our approach curves displayed only repulsive interactions, Hertz model could be applied to the data. Finally, we have shown previously that the thickness of L. monocytogenes cells is large enough to ignore substrate effects on the elastic properties of bacterial cells [23].
Figure 1.
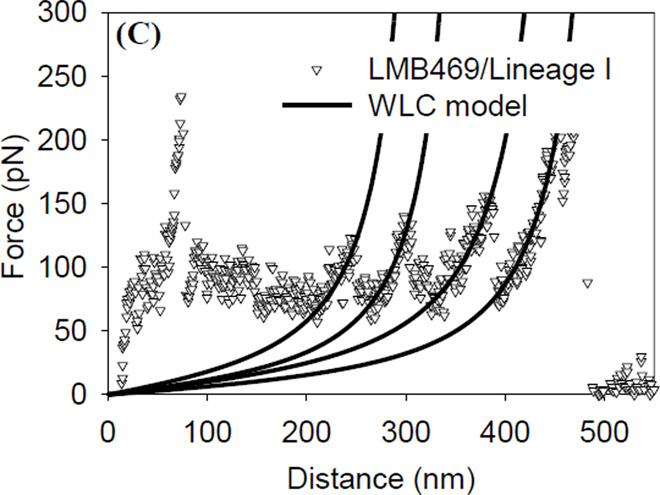
A) Representative force-indentation curves measured on lineages’ I and II strains. Solid lines represent the Hertz model fits to the data with Young’s moduli of 146.7 kPa (r2 = 0.95) and 157.8 kPa (r2 = 0.92) for lineages’ I and II strains, respectively. B) Representative force-distance curves measured on lineages’ I and II strains. Solid lines are the steric model fits to the data with the brush thicknesses and the grafting densities being 86.2 nm and 0.95 × 1016 biopolymers/m2 (r2 = 0.98), and 168.6 nm and 0.59 × 1016 biopolymers/m2 (r2 = 0.98) for lineages’ I and II strains, respectively. C) Solid lines represent WLC model fits to the force-distance data (symbols) measured on L. monocytogenes lineage’ I strain. The persistence lengths and contour lengths that were used to fit the solid line were: (Lp = 0.12, 0.16, 0.14, 0.2, Lc = 350, 393, 500, 540, and r2 = 0.75, 0.82, 0.85, 0.78).
Determination of the thickness and grafting density of the bacterial surface biopolymer brush
The steric force per unit area between the AFM tip and L. monocytogenes surface biopolymers, F, was modeled according to [24–26]:
| (3) |
where kB is Boltzmann constant, T is the absolute temperature taken as 298K, R is the tip radius taken as 40 nm, Γ is the grafted biopolymer density in m−2, D is the distance between the bacterial biopolymers and the AFM tip, and L is the equilibrium thickness of the biopolymer brush layer (Figure 1B).
Elasticity of bacterial surface biopolymers
The entropic elasticities of bacterial surface biopolymers were quantified using the wormlike chain (WLC) statistical thermodynamic model of polymer elasticity [27, 28]. The WLC model describes a polymer chain as an irregular curved filament that has a length of Lc and is linear on the persistence length scale (Lp). LP thus represents the elasticity of the molecule [29]. The force (Fchain) required to stretch a WLC chain to a length D is given by:
| (4) |
The WLC model is usually applied to a portion of the retraction data (Figure 1C) collected on bacterial surfaces. In fitting the force-extension data, Lp values were limited to those larger than 0.154 nm (C–C bond length) [23, 30]. We have chosen the WLC model here over other statistical thermodynamic polymer elasticity models such as the freely jointed chain model because it is better suited to predict the elasticities of the peptidoglycan chains which largely cover the surface of the Gram-positive L. monocytogenes [23].
Colorimetric protein measurements
Bacterial cells of the strain of interest were harvested at the late exponential growth phase as described in the supplementary methods and washed twice by centrifugation at 5525g for ten minutes each round. The collected bacterial pellet was then diluted with 0.2 µm filtered deionized (DI) water to make a suspension with an optical density of ~0.5 at a wavelength of 600 nm. The bacterial suspension was then assayed via colorimetric standard Lowry assay to determine the amounts of bacterial proteins. Bovine serum albumin (BSA) was used as the standard [31].
Statistical description of AFM data
Non-parametric group comparison Mann Whitney Rank Sum test or Dunn test available in Sigma Stat 2.03 (Jandel Scientific) were applied to data to determine if statistical variations were present in adherence or mechanics among two or multiple groups, respectively. The abilities of the Poisson model to fit the adhesion force histograms or the WLC model to fit the force-distance data were judged based on the estimated values of r2 using Microsoft Excel 2010. Likewise, the ability of the steric model to fit force-distance data was judged based on the estimated values of r2 using the Table Curve fitting program (Windows, version 1.11, Jandel Scientific). Force-indentation data were fit to Hertz model using an in-house written C# code that minimizes the differences between experimental forces and theoretical predictions of forces.
Results and Discussion
Cellular mechanics of L. monocytogenes strains
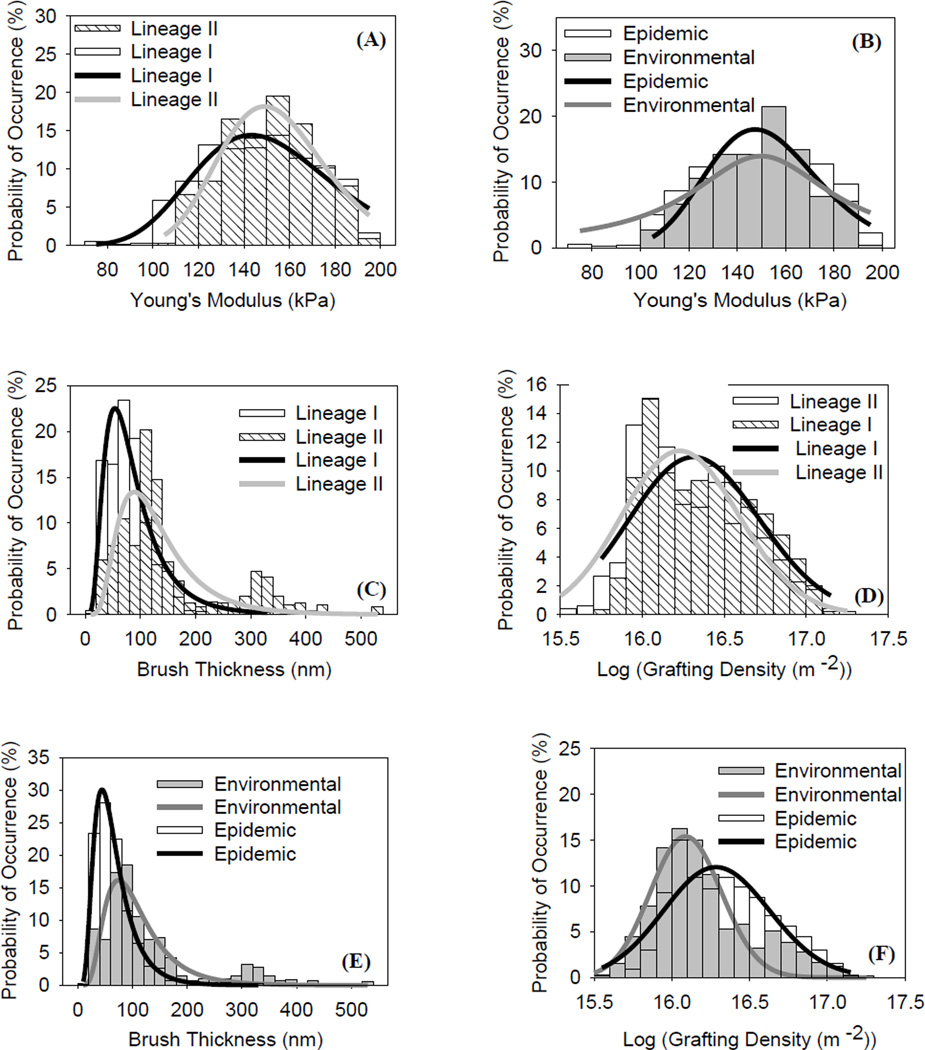
The ability of Hertz model to fit forceindentation data was confirmed for all strains investigated (Figures 1A and S2 in the supplementary data). Young’s moduli data presented in the histograms shown in Figures 2A, 2B and S3 given in the supplementary data were heterogeneous and spanned a range from 75 kPa to 200 kPa for all investigated strains. To describe these heterogeneities, the mean values of the distributions were quantified (Table 1). In addition, log-normal probability function with three parameters was fit the data. Our results indicated that strains that belonged to lineages’ I and II were similar in their heterogeneity. In comparison, epidemic strains were more heterogeneous compared to environmental strains as evident from the larger standard deviation associated with their data (28.7 and 20.7 KPa, respectively). Heterogeneities in bacterial properties in general and in mechanics specifically reflect the variable composition of bacterial surface [14]. When lumped as a group, the average of Young’s moduli estimated for lineage’ II strains (151.1 ± 1.1 kPa, n= 429) was statically higher than that reported for lineage’ I strains (146.8 ± 1.0 kPa, n= 785) (Figure 2A, Table 1, P ≤ 0.001). In comparison, Young’s moduli for epidemic (147.9 ± 1.1 kPa, n= 497) and environmental (148.8 ± 0.9 kPa, n= 471) strains were not statically different (P > 0.5, Table 1, Figure 2B).
Figure 2.
Histograms that show the distributions of Young’s moduli obtained using Hertz model for the investigated: A) lineages’ I and II strains and B) environmental and epidemic strains. Histograms that show the distributions of brush thicknesses obtained using steric model for the investigated: C) lineages’ I and II strains and E) environmental and epidemic strains. Histograms that show the distributions of brush grafting densities obtained using steric model for the investigated: D) lineages’ I and II strains and F) environmental and epidemic strains. Solid lines represent the log-normal dynamic peak function with three parameters fits to the data.
The Young’s moduli values estimated in this study for L. monocytogenes strains are in good agreement with previously reported values in the literature. For example, we have estimated previously Young’s moduli for L. monocytogenes strains that varied in their virulence to be between 52.2 to 113 kPa with virulent strains being more rigid [23]. In another study, Young’s moduli values of Shewanella putrefaciens CIP 8040 measured in 0.1 M KNO3 solution at two pH values (4 and 10) were found to be 210 and 37 kPa, respectively [32]. In a third study, Young moduli of 186 kPa and 300 kPa were estimated for a wild-type Lactobacillus rhamnosus GG cells and a mutant impaired in adherence, biofilm formation, and polysaccharide production, respectively [33]. Finally, Young’s moduli measured on noninfected Escherichia coli was found to be 365± 122 kPa [34].
Mechanics of the surface biopolymer brush of L. monocytogenes strains
The ability of the steric model to fit the approach force-distance data in order to estimate bacterial surface biopolymer brush thickness and grafting density was confirmed in Figures 1B and S4 in the supplementary data. The distributions of the brush thicknesses obtained for each strain are shown in the histograms given in Figures 2C, 2E and S5 in the supplementary data. The length of the bacterial surface biopolymer brushes was in the range of 10–530 nm for all investigated strains (Table 1). Our results indicated that the biopolymers of L. monocytogenes are heterogeneous in their length, reflecting the expected variability in the composition and conformational properties of the bacterial surface biopolymers [35]. We have shown that lineage’ I strains were characterized by significantly shorter biopolymer brushes (82.4 ± 2.0 nm) compared to lineage’ II strains (145.4 ± 6.4 nm) (Table 1, P ≤ 0.001). Our results also indicated that lineage’ II strains are characterized by wider spread histograms (Figure 2C) and an overall larger standard deviation (146.6 nm) compared to lineage’ I strains (62.1 nm). When epidemic and environmental strains were compared for their brush thicknesses, biopolymer brushes of environmental strains (140.6 ± 4.6 nm) were found to be almost twice as long as those of epidemic strains (70.6 ± 2.3 nm) (Figure 3D, P ≤ 0.001). Our results indicated that the standard deviation in the brush thickness estimated for the epidemic strains was smaller than that estimated for the environmental strains (63.7 nm and 123.6 nm, respectively).
Figure 3.
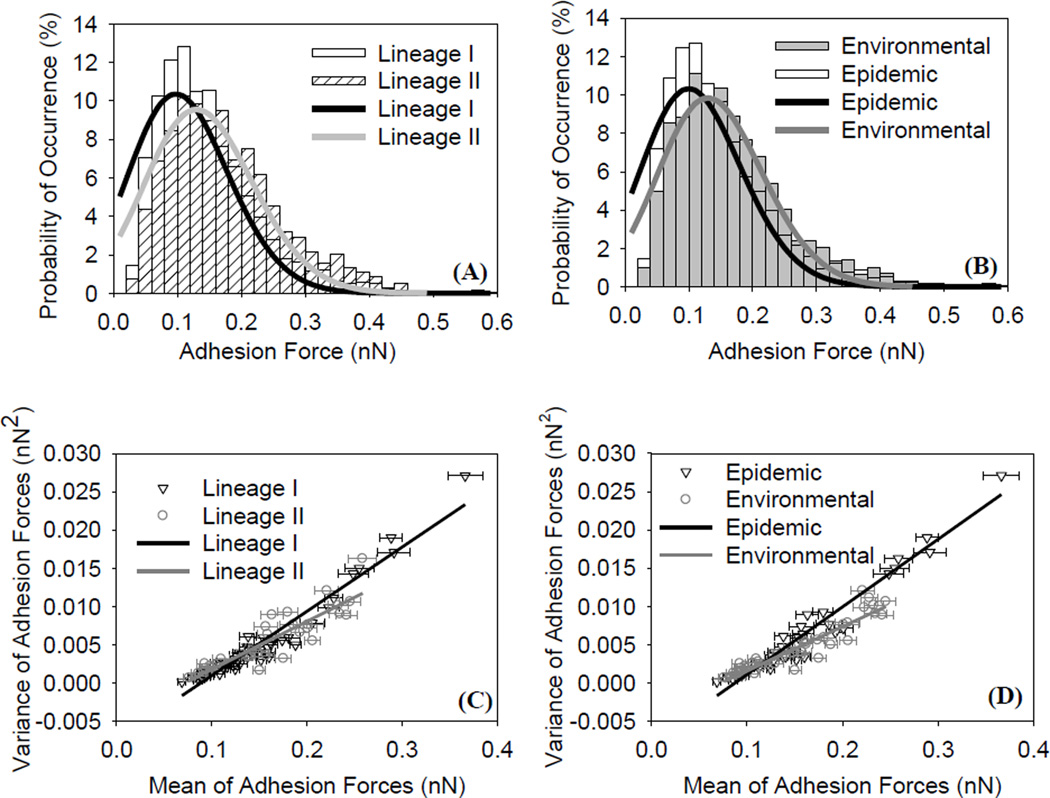
Histograms that show the distributions of the overall AFM adhesion forces measured between the investigated L. monocytogenes: A) lineages’ I and II strains and B) environmental and epidemic strains and Si3N4 in water. Solid lines represent the Poisson model fits to the data. Scatter plots of the variance (σF2) versus the mean(µF) of the adhesion force data for: C) lineages I strains and II and D) environmental and epidemic strains. Solid lines are linear regression fits to the data.
Histograms of the grafting densities estimated from the steric model for all L. monocytogenes strains are shown in Figures 2D, 2F and S6 in the supplementary data. The grafting densities were heterogeneous and ranged between 1015 and 1017 (molecules/m2) for all strains investigated. The standard deviation in the grafting densities data was reported to be 1.9 × 1016 (molecules/m2). Heterogeneities in the estimated grafting densities reflect the variations in the surface composition of the bacterial surface biopolymers on the strains investigated. When averaged, the means of the grafting densities of the bacterial surface biopolymer brushes of lineages’ I and II strains were not statically different (P = 0.254, Table 1). The grafting densities of the bacterial surface biopolymer brushes were statically larger on average for the epidemic strains (2.52×1016 ± 0.07 ×1016 biopolymers/m2) compared to the environmental strains (1.95 ×1016 ± 0.07 ×1016 biopolymers/m2) (Table1, Figures 2D and 2F, P ≤ 0.001).
Our steric model results agree well with bacterial surface biopolymer brush thicknesses and grafting densities previously reported in the literature. We have shown previously that the thicknesses and grafting densities of bacterial surface biopolymer brushes of different L. monocytogenes strains were estimated to be between 134 and 178 nm and 1.32 ×1016 molecules/m2 to 2.71 ×1016 molecules/m2 [23]. Likewise, the thickness of Escherichia coli biopolymer brush was found to be 93 nm in water [21]. In another study, the length of the lipopolysaccharides of Pseudomonas aeruginosa was estimated to be 171 ± 5 nm [36]. To our knowledge, we present here the first study that reports heterogeneities in the thicknesses and grafting densities estimated at different locations on bacterial surfaces. Contrary to what has largely been accepted in the literature about the homogeneity of the approach data [37], our results indicate that approach curves registered at different locations on bacterial cell surfaces are quite different and thus should be considered individually upon modeling.
Conformations of the surface biopolymers of L. monocytogenes strains
Elastic properties of individual chains of bacterial surface biopolymers were estimated using WLC model [30] (Table 1, Figures 1C and S7 in the supplementary data). Only fitting parameters that were obtained with r2 values higher than or equal to 0.7 were reported in this study. On average, persistence lengths estimated for biopolymers of lineage’ I strains were found to be significantly lower (0.33 ± 0.01 nm) compared to those estimated for lineage’ II strains (0.37 ± 0.02 nm) (P= 0.017, Table 1). In comparison, the estimated contour lengths were found not to be statically different for lineage’ I (827.2 ± 32.5) and lineage’ II strains (825.5 ± 38.6) (P = 0.798, Table 1). Persistence lengths estimated for the epidemic strains were found to be significantly lower (0.31 ± 0.01 nm) compared to those estimated for the environmental strains (0.38 ± 0.02 nm) (P= 0.002, Table 1). Additionally, estimated contour lengths were found to be statically lower for the epidemic strains (720.9 ± 31.3) compared to the environmental strains (943.2 ± 37.2) (P ≤ 0.001, Table 1).
Note that contour lengths are expected to be directly proportional to the stretched lengths of the biopolymer chains. However, since we have no definite information on the types of biopolymers being stretched on bacterial surface, the actual lengths of the biopolymer chains were not known [30]. In addition, it has to be noted that the AFM tip can contact the biopolymer chain anywhere along its contour and thus the contour lengths estimated from the WLC modeling of the retraction AFM generally reflect the length of a portion of the biopolymer chain [30]. The large contour lengths shown here can be explained in part by unfolding of proteins which largely cover the surface of L. monocytogenes strains [38].
The persistence lengths obtained for the investigated strains of L. monocytogenes compare well with previously reported persistence lengths for other bacteria in the literature. We have shown previously that the virulent strains of L. monocytogenes are characterized by more flexible surface biopolymers (0.21 ± 0.08 nm) compared to the avirulent strains (0.24 ± 0.14 nm) [23]. Similarly, the persistence lengths estimated for Pseudomonas putida KT2442 in water were flexible and ranged between 0.154–0.28 nm [30]. Comparatively, the persistence length value of 0.38 nm and contour length values that ranged from 228 nm to 353 nm predicted using the WLC model were found to be in good agreement with physical and structural dimensions of OmpA-AmCyan protein expressed on the surface of Escherichia coli cells [39]. Finally, WLC model was successfully applied to polypeptides stretching curves obtained with Pseudomonas aeruginosa coated tips over mica surfaces. The persistence length used in the fitting data was around 0.5 nm [40].
The mechanical properties of the bacterial cells are expected to be affected by the rigidity of the cellular membrane [23], the osmotic pressure of the cell [41] and the conformational properties of bacterial surface biopolymers [30]. Since membranes of bacterial cells are only few nanometers in thickness [42], their contribution to the mechanics can be ignored. In addition, since cells are grown in nutrient media that have constant ionic composition and strains tested have very similar zeta potentials (Figure S8 in supplementary data), their osmotic pressures are expected to be similar [43]. The only remaining influencing factor that is expected to largely affect the cellular mechanics is the conformational properties of the bacterial surface biopolymers. Since bacterial cells generally display a wide array of biopolymers on their surfaces, spatial variations in the elastic properties of biopolymers spread on the cell surface and thus of cells as well are to be expected [22]. In fact, the cell wall of L. monocytogenes is composed mainly of polysaccharides and peptidoglycans [38]. The range of the persistence lengths estimated using WLC reflects the different elastic properties of the bacterial surface biopolymers including peptidoglycans and polysaccharides (Table 1). The Young’s moduli of the bacterial cells will be largely influenced by the elastic properties of these biopolymers. Combined, the more rigid individual biopolymer chains, the larger the number of biopolymer chains and the longer biopolymer chains observed for lineage’ II strains compared to those observed for lineage’ I strains are potentially responsible for the higher Young’s moduli estimated for the later strains (Table 1).
In a recent study in the literature [44], mechanical properties of two strains of streptococcus salivarius HB7 and HBV51 that varied in the length of their fibrils from 91 nm to 63 nm were investigated as the ionic strength of the interactions media was decreased from 57 mM to 5.7 mM using an array of techniques including quartz crystal microbalance with dissipation (QCM-D), AFM, and total-internal-reflection microscopy (TIRM). Their results showed that upon decrease in ionic strength, hydrophilic and negatively charged fibrils of HBV51 remained partially stretched resulting in a decreased overall stiffness of the adhesive bond. In comparison, the more hydrophobic and less negatively charged fibrils of the HB7 strain were suggested to collapse on the cell surface. Although the authors used QCM-D and TIRM to arrive to their conclusions, their findings agree with our modeling results which pointed to the importance of investigating the roles of the mechanical and physiochemical properties of the bacterial surface fibrils in affecting the overall Young’s moduli of bacterial cells [44].
Cellular adhesion of L. monocytogenes to Si3N4 in water
The quantified nanoscale adhesion forces of L. monocytogenes strains to Si3N4 in water were distributed over a wide range (0.025 – 0.620 nN) (Table 1, Figures 3A, 3B and S9 in the supplementary data). Such heterogeneity is largely attributed to variations in the composition, structure and physicochemical properties of the bacterial surface biopolymers [35, 45, 46]. To describe the distribution of adhesion forces, the average of all data points represented in each histogram in Figure S9 was calculated and compared among various strains (Table 1). Our results showed that lineage’ II strains have statistically higher overall adhesion to Si3N4 (0.175 ± 0.002 nN, n = 1495) compared to lineage’ I strains (0.149 ± 0.001 nN, n = 3258) (Figure 3A, P ≤ 0.001). Similarly, environmental strains were characterized by significantly higher adhesion to Si3N4 (0.165 ± 0.002 nN, n = 2203) compared to epidemic strains (0.150 ± 0.002 nN, n = 2550) (Figure 3B, P ≤ 0.001). The quantified adhesion forces between the investigated strains of L. monocytogenes and Si3N4 in water were comparable to reported adhesion forces quantified between Gram-positive bacteria and Si3N4 in the literature. We have shown previously that the adhesion forces of different strains of L. monocytogenes to Si3N4 in water ranged from 0.227 ± 0.005 nN to 0.515 ± 0.01 nN [20]. In a second study, the adhesion forces measured between Staphylococcus epidermidis and Si3N4 in water were found to be 0.203 ± 0.066 nN for biofilm positive strains and 0.181 ± 0.064 nN for biofilm negative strains [47].
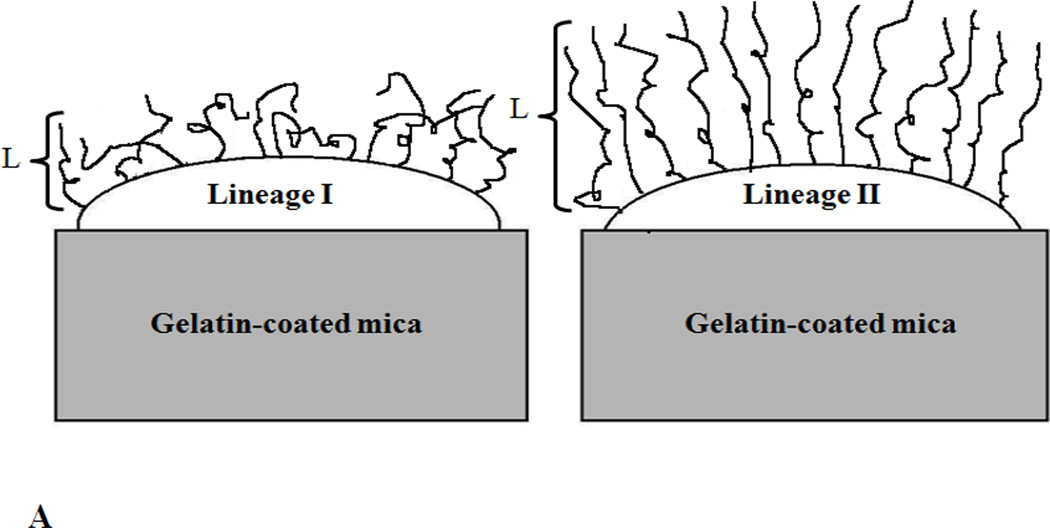
Our results showed that Lineage’ II strains were characterized by a higher overall cellular adhesion compared to lineage’ I strains. This can be in part attributed to the longer biopolymers present on the surfaces of lineage’ II strains compared to lineage’ I strains as was estimated from the steric model fits to the force-distance approach data (Table 1). Longer biopolymer brushes are expected to furnish longer arms for the bacterial cells to reach to inert surfaces more easily and thus enhance their adhesion to surfaces [48]. In addition, we have shown that lineage’ II strains have larger persistence lengths for their biopolymers. This suggests that on average, longer segments of the biopolymer chains are available for interactions compared to those on lineage’ I strains, explaining in part the higher adhesion forces observed for lineage’ II strains. Finally, although the grafting densities estimated for both lineages were similar, we have to remember that the grafting densities estimated from the steric model reflect the apparent grafting densities [25]. Thus, since the biopolymers of lineage’ I strains have shorter persistence lengths compared to the biopolymers of lineage’ II strains, they can adapt more flexible conformations, potentially resulting in single chains contacting the surfaces more than once and thus getting counted as more than one chain in the grafting density estimates (Figure 4A). The larger number of biopolymers available for interactions is expected to lead to higher adhesion forces as well [49].
Figure 4.
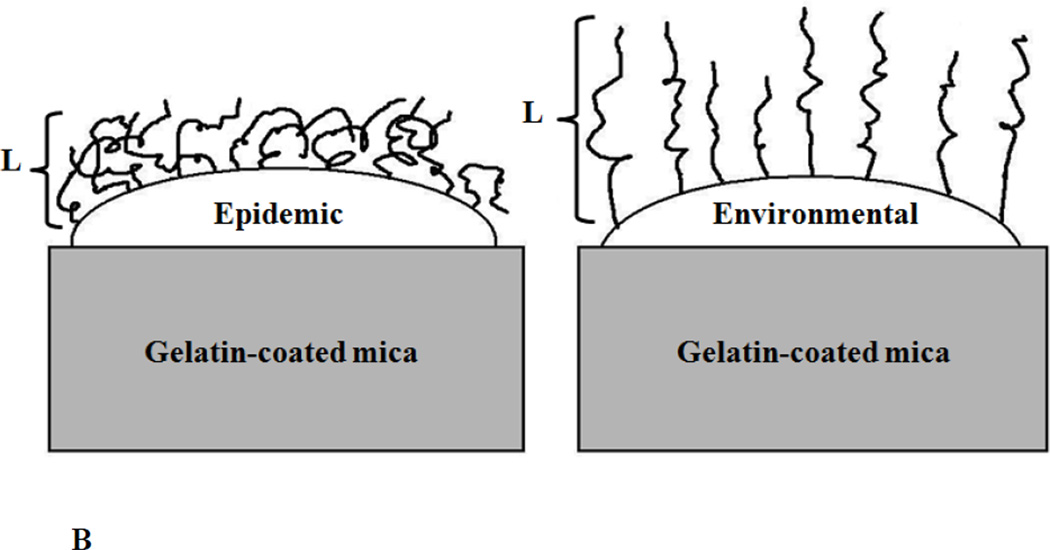
Hypothetical schematics that show how the biopolymers of L. monocytogenes: A) lineages’ I and II strains and B) environmental and epidemic strains are distributed on their surfaces. L is the bacterial surface polymer brush thickness. Schematics are drawn not to scale for clarity purposes.
Molecular components of the adhesion forces measured between L. monocytogenes and Si3N4 in water
The ability of Poisson model to fit the distributions of adhesion affinities was confirmed (r2= 0.87 on average, Figures 3A, 3B and S9 in the supplementary data). As a result, linear regressions of the force variance (σF2) versus the average force (µF) data were used to compute the specific and nonspecific components of the interaction forces for each strain investigated (Table 1, Figures 3C, 3D and S9 in the supplementary data).Our results indicated that, on average, the nonspecific forces were slightly (~4%) higher than the specific forces for lineage’ I strains (Table 1), indicating that both specific forces and nonspecific forces play important roles in governing the adhesion of these strains to Si3N4. In comparison, the nonspecific forces for lineage’ II strains were ~ 15% higher than the specific forces (Table 1), indicating that the adhesion of lineage’ II strains to Si3N4 is dominated by nonspecific forces. When compared, the specific and nonspecific forces for lineage’ I strains were higher than their counter parts calculated for lineage’ II strains. When individual strains were considered, the nonspecific forces were larger than the specific forces for all of them except for the epidemic, lineage’ I LMB496 strain. This indicated that nonspecific forces in general dominate the adhesion of L. monocytogenes strains to Si3N4 (Table 1).
Similarly, nonspecific forces were 1.1% to 8.6% higher than the specific forces for individual epidemic strains indicating that both specific forces and nonspecific forces have important roles in the adhesion of these strains to Si3N4 (Table 1). In comparison, nonspecific forces for all individual environmental strains investigated were 8.5% to 24.6% higher than specific forces estimated for the same strains indicating that nonspecific forces dominate the adhesion of the environmental strains to Si3N4 (Table 1). Our results indicated that both specific forces as well as nonspecific forces were important players in governing the overall adhesion of lineage’ I strains to Si3N4. In comparison, nonspecific forces dominated the interactions of lineage’ II strains to Si3N4.
When compared, the magnitudes of the specific forces obtained for lineage’ I strains were higher than those obtained for lineage’ II’ strains. It has to be noted that the specific forces calculated here using Poisson statistical approach represent the average strength of a single hydrogen bond for this particular system. This can possibly be due to the presence of functional groups such as carboxyl, amide, phosphate and carbohydrate moieties [50] on the surfaces of lineage’ I strains that can form stronger hydrogen bonds with Si3N4 compared to lineage’ II strains. Similarly, the magnitude of the total nonspecific forces obtained for lineage I’ strains was larger than that reported for lineage’ II strains. Note that the sum of the specific and nonspecific forces calculated from Poisson model does not add to the total adhesion force measured by AFM. This can be justified by the fact that not all the strains investigated can form specific hydrogen bonds with Si3N4. The overall higher adhesion observed for lineage’ II strains in comparison to that of lineage I’ strains is due to the ability of the former to establish ~1.63 hydrogen bonds on average with Si3N4 compared to only 0.73 hydrogen bonds for the later (Table 1). This is reasonable given that lineage’ II strains have longer biopolymers with more exposed segments available for interactions with Si3N4 compared to the flexible collapsed biopolymers shown for lineage’ I strains (Figure 4A). The strength of the hydrogen bond is very sensitive to the structure of the interacting molecules where the geometries and the directionality vary [51]. The typical strength of a hydrogen bond lies between 0.1 nN and 0.4 nN [52], and there is not a lower limit for the strengths of hydrogen bonds [53]. According to the recently proposed description of “hydrogen bond” by IUPAC [53]; electrostatic forces play a significant role in the directionality of the interacting molecules and hence the strength of hydrogen bonding [54].
The higher adhesion observed for the environmental strains compared to the epidemic strains can be attributed in part to their longer surface biopolymer brushes (Table 1). In addition, we have shown that environmental strains have larger persistence length values for their biopolymers. That indicates that on average, longer segments of the biopolymer chains are available for interactions. The higher grafting densities observed for the epidemic strains can be in part the result of the shorter persistence lengths for their biopolymers resulting in more flexible biopolymers that can collapse on the bacterial surface (Figure 4B) and in part due to the expression of full length proteins on their surfaces compared to environmental strains. In one study, epidemic L. monocytogenes strains were found to express full-length internalin, a virulence surface protein, more frequently compared to environmental food isolates [55]. The study above indicates that although similar proteins may be present on both environmental and epidemic strains of L. monocytogenes, these proteins may still be characterized by different conformational properties.
Our results agree well with macroscale adhesion experiments performed using a microtiter plate method between L. monocytogenes strains isolated from seafood and cheese industry samples (industrial or environmental strains) and from patients with listeriosis (clinical or epidemic strains) and polystyrene or stainless steel surfaces [56]. The results of the study above indicated that the clinical strains were less adherent to inert surfaces compared to the environmental strains [56].
However, although our results indicated that the environmental strains have higher overall adhesion, epidemic strains were characterized by higher specific and nonspecific forces (Table 1). This can be explained in part by the larger grafting densities observed for the epidemic strains. However, this also indicates that the number of hydrogen bonds that can form between environmental strains and Si3N4 (n= 1.59) are larger compared to those that can form between epidemic strains and Si3N4 (n= 0.72). This is reasonable given that environmental strains have biopolymers that are approximately twice as long as those of the epidemic strains. This indicates that larger segments of biopolymer chains are exposed and thus more side groups are potentially available for hydrogen bonding. In addition, our results indicated that both specific and nonspecific forces are important in establishing interactions between epidemic strains and Si3N4. In comparison, nonspecific forces dominated the interactions between environmental strains and Si3N4 (Table 1). Our results thus indicate that although both strains have relatively close overall adhesion forces, they varied significantly in the molecular mechanisms that govern their interactions with surfaces. The higher adhesion of environmental strains was accompanied with longer and more extended surface biopolymers regardless of their surface protein contents as was shown from our colorimetric standard Lowry assays where the epidemic strains were characterized by higher total protein content (0.068 g/L) compared to environmental strains (0.058 g/L) (Table 1). Prior literature studies have indicated that certain individual proteins on bacterial surfaces may play a significant role in various bacterial functions such as pathogenicity, intracellular motility or adhesion to surfaces. For example, a study has pointed to the significant relation between the sequence of fibronectin binding protein A on Staphylococcus aureusbinding strain and the actual activity of this protein, and hence its effect on the strain binding strength where strains with different polymorphisms of this protein showed differences in their adhesion to fibronectin coated surfaces [57]. Several studies have shown the important role of the surface protein listeriolysin O (LLO) in L. monocytogenes virulence [58, 59]. Other studies have also indicated the role of Act A protein in listeria monocytogenes’ actin-based intracellular motility [60].
Relationship between mechanics and adherence
Nanomechanical properties of bacterial cells are expected to be influenced by the bacterial surface composition of biomolecules which were also shown to affect bacterial adhesion to surfaces. For example in a recent study, the presence of the virulence surface proteins on the bacterial surface was correlated with an increase in the bacterial membrane rigidity [61]. Although the study above did not correlate adherence and mechanics, we have shown that adhesion is positively correlated with the number of bacterial surface biopolymers (data not shown). The relationship between adherence and mechanics can be investigated using an array of adhesion models such as the Johnson-Kendall and Roberts (JKR) and Derjaguin, Muller and Toporov (DMT) [62]. These models assume that the surface forces such as loads applied by indenters are a function of the separation distance between the two surfaces in contact and they follow the force derived from the Lennard-Jones potential:
| (5) |
where F is the surface force, zo is the equilibrium separation distance, w is the work of adhesion and z is separation distance. The rigidity of the interacting surfaces will influence the separation distance and the contact area between the two surfaces and thus will affect adhesion per the equation above. Our results provide direct evidence that mechanics and adherence are related. We have shown that the conformational properties of the surface biopolymers were critical to determining the ability of these molecules to form specific bonds with the surfaces they interact with. The higher adhesion observed for lineage’ II strains compared to lineage’ I strains was accompanied with longer and more extended surface biopolymers regardless of their surface protein contents as was shown from our colorimetric standard Lowry (Table 1). Our findings are in sync with prior findings in the literature. For example, the strength of the adhesion force of E. coli to Si3N4 was shown to be affected by the length of the core lipopolysaccharides (LPS) molecules present on the cell surface. Bacterial LPS are considered as important virulence factors and significant determinants in the adhesion of bacteria to nonbiological surfaces [63]. In the future, we will quantify the effects of adhesion on cellular mechanics according to JKR and DMT models of contact mechanics.
Importance of molecular details in probing bacterial adherence and mechanics at the cellular-level
Our results showed that lineage’ I strains were characterized by lower overall adhesion forces and higher specific and nonspecific forces compared to lineage’ II strains. In addition, lineage’ II strains were characterized by higher Young’s moduli and longer and stiffer biopolymers compared to lineage’ I strains. Strains of both lineages were similar in their grafting densities. Our results thus point to the potency of using nanomechanical cellular and molecular properties of bacterial cells as additional means that can aid in distinguishing between lineages’ I and II strains. Our results also indicated that epidemic and environmental strains of L. monocytogenes and irrespective of their lineage group were characterized by similar Young’s moduli of elasticties and adhesion forces at the cellular level. However, at a molecular level, epidemic strains were characterized by higher specific and nonspecific forces, shorter, denser and more flexible biopolymers compared to the environmental strains. Differences in molecular scale forces can be attributed to differences in the structure, conformations, charge, hydrophobicity and the types of side groups available on the biopolymer chains [49, 64], all properties which were shown to be largely dependent on the type of environment in which bacterial strains survive and adhere in [65]. Our results thus point to the importance of considering the molecular details when comparing the functions of bacterial cells. Therefore, although the cells investigated here were structurally similar at the cellular level, they were inherently different at the molecular level in their mechanics, adherence and pathogenicity.
In addition, our results indicated that lineage’ II strains were more heterogeneous in their adhesion, biopolymer brush thicknesses, grafting densities and persistence lengths compared to lineage’ I strains. Lineages’ I and II strains were comparable in the heterogeneities of their contour lengths and Young’s moduli of elasticties. The heterogeneities were quantified based on the magnitudes of standard deviations as well as based on statistical variations between individual strains that belonged to each lineage. Such heterogeneity is expected as literature studies indicate that lineage’ II strains are capable of horizontal gene transfer, leading to a higher generic heterogeneity among strains as well as allows for a better ability for the strains to survive and multiply in different environments [14]. Our results also indicated that environmental strains were more heterogeneous in their adhesion, brush thicknesses and persistence and contour lengths compared to epidemic strains. Both environmental and epidemic strains were comparable in the heterogeneities of their Young’s moduli of elasticties and grafting densities. Such heterogeneity is expected as environmental strains reflect bacterial strains that are adaptive to a wide array of environmental conditions compared to epidemic strains which are generally constrained to in vivo environmental conditions. A previous study that has conducted pulsed-field gel electrophoresis analysis on Vibrio vulnificus strains, has similarly reported a higher heterogeneity for the environmental strains compared to the epidemic strains [66]. Our results thus point to the importance of mapping bacterial properties on the entire bacterial surface due to expected heterogeneities in bacterial surface properties.
Supplementary Material
Highlights.
Lineage’ I strains were characterized by shorter and more flexible biopolymers.
Lineage’ II strains were characterized by higher adhesion strengths to Si3N4.
Lineage’ I strains were characterized by higher specific and nonspecific forces.
Bacterial elasticties were similar for epidemic and environmental strains.
Epidemic strains were characterized by short, dense and collapsed biopolymers.
Acknowledgements
The authors would like to thank Prof. Douglas call, a professor at the Department of Veterinary Microbiology & Pathology, the School for Global Animal Health at Washington State University (WSU) for providing us with all bacterial strains used in the current study. We would like to thank the National Institutes of Health (NIH) grants 1R03AI077590–01A1 and 5R03AI077590– 02 for financial support of this work. Finally, we would like to thank Ken Dorrance, a physics student at WSU, for his C# code used in this study to fit the Hertz model to the force-indentation data and Sharon Cox, Ala’ Abu- Lail and Fabiola Quiroa for help in data analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cossart P, Mengaud J. Listeria monocytogenes: A model system for the molecular study of intracellular parasitism. Molecular Biology and Medicine. 1989;6:463–474. [PubMed] [Google Scholar]
- 2.Fenlon DR, Wilson J, Donachie W. The incidence and level of Listeria monocytogenes contamination of food sources at primary production and initial processing. Journal of Applied Bacteriology. 1996;81:641–650. doi: 10.1111/j.1365-2672.1996.tb03559.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, Elmose J, Call DR. Interactions between the environmental pathogen Listeria monocytogenes and a free-living protozoan (Acanthamoeba castellanii) Environmental Microbiology. 2007;9:913–922. doi: 10.1111/j.1462-2920.2006.01213.x. [DOI] [PubMed] [Google Scholar]
- 4.Gellin BG, Broome CV, Bibb WF, Weaver RE, Gaventa S, Mascola L. The epidemiology of Listeriosis in the United-States-1986. American Journal of Epidemiology. 1991;133:392–401. doi: 10.1093/oxfordjournals.aje.a115893. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C, Zhang M, Ju J, Nietfeldt J, Wise J, Terry PM, Olson M, Kachman SD, Wiedmann M, Samadpour M, Benson AK. Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: Identification of segments unique to lineage II populations. Journal of Bacteriology. 2003;185:5573–5584. doi: 10.1128/JB.185.18.5573-5584.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebuffo-Scheer CA, Schmitt J, Scherer S. Differentiation of Listeria monocytogenes serovars by using artificial neural network analysis of Fourier-transformed infrared spectra. Applied and Environmental Microbiology. 2007;73:1036–1040. doi: 10.1128/AEM.02004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiedemann M, Bruce JL, Keating C, Johnson AE, McDonough PL, Batt CA. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infection and Immunity. 1997;65:2707–2716. doi: 10.1128/iai.65.7.2707-2716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nightingale KK, Windham K, Wiedmann M. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 2005;187:5537–5551. doi: 10.1128/JB.187.16.5537-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen OF, Skouboe P, Dons L, Rossen L, Olsen JE. Listeria monocytogenes exists in at least three evolutionary lines: Evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology. 1995;141:2053–2061. doi: 10.1099/13500872-141-9-2053. [DOI] [PubMed] [Google Scholar]
- 10.Nadon CA, Woodward DL, Young C, Rodgers FG, Wiedmann M. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 2001;39:2704–2707. doi: 10.1128/JCM.39.7.2704-2707.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts A, Nightingale K, Jeffers G, Fortes E, Kongo JM, Wiedmann M. Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology. 2006;152:685–693. doi: 10.1099/mic.0.28503-0. [DOI] [PubMed] [Google Scholar]
- 12.Wiedmann M. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 2002;85:524–531. [PubMed] [Google Scholar]
- 13.Ward TJ, Gorski L, Borucki MK, Mandrell RE, Hutchins J, Pupedis K. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. Journal of Bacteriology. 2004;186:4994–5002. doi: 10.1128/JB.186.15.4994-5002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Jesus AJ, Whiting RC. Thermal inactivation, growth, and survival studies of Listeria monocytogenes strains belonging to three distinct genotypic lineages. Journal of Food Protection. 2003;66:1611–1617. doi: 10.4315/0362-028x-66.9.1611. [DOI] [PubMed] [Google Scholar]
- 15.Gray MJ, Zadoks RN, Fortes ED, Dogan B, Cai S, Chen Y, Scott VN, Gombas DE, Boor KJ, Wiedmann M. Food and human isolates of Listeria monocytogenes form distinct but overlapping populations. Appl. Environ. Microbiol. 2004;70:5833–5841. doi: 10.1128/AEM.70.10.5833-5841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clinical Microbiology Reviews. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woolhouse ME, Taylor LH, Haydon DT. Population biology of the multihost pathogen. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- 18.Kim SH, Bakko MK, Knowles D, Borucki MK. Oral inoculation of A/J mice for detection of invasiveness differences between Listeria monocytogenes epidemic and environmental strains. Infection and Immunity. 2004;72:4318–4321. doi: 10.1128/IAI.72.7.4318-4321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doktycz MJ, Sullivan CJ, Hoyt PR, Pelletier DA, Wu S, Allison DP. AFM imaging of bacteria in liquid media immobilized on gelatin coated mica surfaces. Ultramicroscopy. 2003;97:209–16. doi: 10.1016/S0304-3991(03)00045-7. [DOI] [PubMed] [Google Scholar]
- 20.Park BJ, Haines T, Abu-Lail NI. A correlation between the virulence and the adhesion of Listeria monocytogenes to silicon nitride: An atomic force microscopy study. Colloids and Surfaces B: Biointerfaces. 2009;73:237–243. doi: 10.1016/j.colsurfb.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 21.Abu-Lail NI, Camesano TA. The effect of solvent polarity on the molecular surface properties and adhesion of Escherichia coli. Colloids and Surfaces. B, Biointerfaces. 2006;51:62–70. doi: 10.1016/j.colsurfb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Touhami A, Nysten B, Dufrene YF. Nanoscale mapping of the elasticity of microbial cells by AFM. Langmuir. 2003;19:4539–4543. [Google Scholar]
- 23.Park B, Abu-Lail NI. Variations in the nanomechanical properties of virulent and avirulent Listeria monocytogenes. Soft Matter. 2010;6:3898–3909. doi: 10.1039/b927260g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander SJ. Adsorption of chain molecules with a polar head a scaling description. Journal of Physics II (Paris-France) 1977;38:983–987. [Google Scholar]
- 25.de Gennes PG. Polymers at interface: A simplified view. Advances in Colloid and Interface Science. 1987;27:189–209. [Google Scholar]
- 26.Butt H-J, Kappl M, Mueller H, Raiteri R, Meyer W, Ruhe J. Steric forces measured with the atomic force microscope at various temperatures. Langmuir. 1999;15:2559–2565. [Google Scholar]
- 27.Ortiz C, Hadziioannou G. Entropic elasticity of single polymer chains of poly(methacrylic acid) measured by atomic force microscopy. Macromolecules. 1999;32:780–787. [Google Scholar]
- 28.Fisher TE, Oberhauser AF, Carrion-Vazquez M, Marszalek PE, Fernandez JM. The study of protein mechanics with the atomic force microscope. Trends in Biochemical Sciences. 1999;24:379–84. doi: 10.1016/s0968-0004(99)01453-x. [DOI] [PubMed] [Google Scholar]
- 29.van der Aa BC, Michel RM, Asther M, Zamora MT, Rouxhet PG, Dufrene YF. Stretching cell surface macromolecules by atomic force microscopy. Langmuir. 2001;17:3116–3119. [Google Scholar]
- 30.Abu-Lail NI, Camesano TA. Elasticity of Pseudomonas putida KT2442 surface polymers probed with single-molecule force microscopy. Langmuir. 2002;18:4071–4081. [Google Scholar]
- 31.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. The Journal of Biological Chemistry. 1951;193:265–275. [PubMed] [Google Scholar]
- 32.Gaboriaud F, Bailet S, Dague E, Jorand F. Surface sructure and nanomechanical properties of shewanella putrefaciens bacteria at two pH values (4 and 10) determined by atomic force microscopy. Journal of Bacteriology. 2005;187:3864–3868. doi: 10.1128/JB.187.11.3864-3868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francius G, Lebeer S, Alsteens D, Wildling L, Gruber HJ, Hols P, De Keersmaecker S, Vanderleyden J, Dufrene YF. Detection, localization, and conformational analysis of single polysaccharide molecules on live bacteria. ACS Nano. 2008;2:1921–1929. doi: 10.1021/nn800341b. [DOI] [PubMed] [Google Scholar]
- 34.Chen YY, Yew TR, Hsu JL, Chang HY, Wu CC, Peng HL. Surface rigidity change of Escherichia coli after filamentous bacteriophage infection. Langmuir. 2009;25:4607–4614. doi: 10.1021/la8036346. [DOI] [PubMed] [Google Scholar]
- 35.Camesano TA, Abu-Lail NI. Heterogeneity in bacterial surface polysaccharides, probed on a single-molecule basis. Biomacromolecules. 2002;3:661–667. doi: 10.1021/bm015648y. [DOI] [PubMed] [Google Scholar]
- 36.Ivanov IE, Kintz EN, Porter LA, Goldberg JB, Burnham NA, Camesano TA. Relating the physical properties of Pseudomonas aeruginosa lipopolysaccharides to virulence by atomic force microscopy. Journal of Bacteriology. 2011;193:1259–1266. doi: 10.1128/JB.01308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eaton P, Fernandes JC, Pereira E, Pintado ME, Malcata FX. Atomic force microscopy study of the antibacterial effects of chitosans on Escherichia coli and Staphylococcus aureus. Ultramicroscopy. 2008;108:1128–1134. doi: 10.1016/j.ultramic.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 38.Saiki I, kamisango K-I, Tanio Y, Okumura H, Yamamura Y, Azuma I. Adjuvant activity of purified peptidoglycan of Listeria monocytogenes in mice and guinea pigs. Infection and Immunity. 1982;38:58–66. doi: 10.1128/iai.38.1.58-65.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lower BH, Yongsunthon R, Vellano FP, III, Lower SK. Simultaneous force and fluorescsence measurements of a protein that forms a bond between a living bacterium and a solid surface. Journal of Bacteriology. 2005;187:2127–2137. doi: 10.1128/JB.187.6.2127-2137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Touhami A, Jericho MH, Boyd JM, Beveridge TJ. Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa Pili by using atomic force microscopy. Journal of Bacteriology. 2006;188:370–377. doi: 10.1128/JB.188.2.370-377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood JM, Bremer E, Csonka LN, Kraemer R, Poolman B, van der Heide T, Smith LT. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comparative Biochemistry and Physiology A-Molecular & Integrative Physiology. 2001;130:437–460. doi: 10.1016/s1095-6433(01)00442-1. [DOI] [PubMed] [Google Scholar]
- 42.Arnoldi M, Kacher CM, bauerlein E, Radmacher M, Fritz M. Elastic properties of the cell wall of Magnetospirillum gryphiswaldense investigated by AFM. Appl. Phys. A: Materials Science and Processing. 1998;66:S613–S617. [Google Scholar]
- 43.Vasseur C, Baverel L, Hebraud M, Labadie J. Effect of osmotic, alkaline, acid or thermal stresses on the growth and inhibition of Listeria monocytogenes. Journal of Applied Microbiology. 1999;86:469–476. doi: 10.1046/j.1365-2672.1999.00686.x. [DOI] [PubMed] [Google Scholar]
- 44.Olsson ALJ, Arun N, Kanger JS, Busscher HJ, Ivanov IE, Camesano TA, Chen Y, Johannsmann D, van der Mei HC, Sharma PK. The influence of ionic strength on the adhesive bond stiffness of oral streptococci possessing different surface appendages as probed using AFM and QCM-D. Soft Matter. 2012;8:9870–9876. [Google Scholar]
- 45.Dorobantu LS, Bhattacharjee S, Foght JM, Gray MR. Atomic force microscopy measurement of heterogeneity in bacterial surface hydrophobicity. Langmuir. 2008;24:4944–4951. doi: 10.1021/la7035295. [DOI] [PubMed] [Google Scholar]
- 46.Boonaert CJP, Rouxhet PG, Dufrene YF. Surface properties of microbial cells probed at the nanometer scale with atomic force microscopy. Surface and interface analysis. 2000;30:32–35. [Google Scholar]
- 47.Hu Y, Ulstrup J, Zhang J, Molin S, Dupres V. Adhesive properties of Staphylococcus epidermidis probed by atomic force microscopy. Physical Chemistry Chemical Physics (Incorporating Faraday Transactions) 2011;13:9995–10003. doi: 10.1039/c0cp02800b. [DOI] [PubMed] [Google Scholar]
- 48.Tsuneda S, Aikawa H, Hayashi H, Yuasa A, Hirata A. Extracellular polymeric substances responsible for bacterial adhesion onto solid surface. FEMS microbiology letters. 2003;223:287–292. doi: 10.1016/S0378-1097(03)00399-9. [DOI] [PubMed] [Google Scholar]
- 49.Atabek A, Liu Y, Pinzon-Arango PA, Camesano TA. Importance of LPS structure on protein interactions with Pseudomonas aeruginosa. Colloids and Surfaces B: Biointerfaces. 2008;67:115–121. doi: 10.1016/j.colsurfb.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 50.Abdalla OM, Christen GL, Davidson PM. Chemical composition of Listeria monocytogenes survival in white pickled cheese. Journal of Food Protection. 1993;56:841–846. doi: 10.4315/0362-028X-56.10.841. [DOI] [PubMed] [Google Scholar]
- 51.Kamya PRN, Muchall HM. Revisiting the effects of sequence and structure on the hydrogen bonding and π-stacking interactions in nucleic acids. J. Phys. Chem. A. 2011;115:12800–12808. doi: 10.1021/jp203918z. [DOI] [PubMed] [Google Scholar]
- 52.Pauling L. The nature of the chemical bond. New York: Cornell University Press; 1960. [Google Scholar]
- 53.Arunan E, Desiraju GR, Klein RA, Sadlej J, Scheiner S, Alkorta I, Clary DC, Crabtree RH, Dannenberg JJ, Hobza P, Kjaergaard HG, Legon AC, Mennucci B, Nesbitt DJ. Defining the hydrogen bond: An account. Pure Appl. Chem. 2011;83:1619–1636. [Google Scholar]
- 54.Gordesli FP, Abu-Lail NI. Combined Poisson and soft-particle DLVO analysis of the specific and nonspecific adhesion forces measured between L. monocytogenes grown at various temperatures and silicon nitride. Environmental Science & Technology. 2012;46:10089–10098. doi: 10.1021/es300653w. [DOI] [PubMed] [Google Scholar]
- 55.Jacquet C, Doumith M, Gordon JI, Martin PMV, Cossart P, Lecuit M. A molecular marker for evaluating the pthogenic potential of foodborne Listeria monocytogenes. Journal of Infectious Diseases. 2004;189:2094–2100. doi: 10.1086/420853. [DOI] [PubMed] [Google Scholar]
- 56.Tresse O, Shannon K, Pinon A, Malle P, Vialette M, Midelet-Bourdin G. Variable adhesion of Listeria monocytogenes isolates from food-processing facilities and clinical cases to inert surfaces. Journal of Food Protection. 2007;70:1569–1578. doi: 10.4315/0362-028x-70.7.1569. [DOI] [PubMed] [Google Scholar]
- 57.Lower SK, Casillas-Ituarte NN, Yongsunthon R, Taylor ES, DiBartola AC, Lower BH, Lamlertthon S, Reller LB, Fowler VG, Jr, Lins RD, Edmonson C, McIntyre LM, Que YA, Ros R. Polymorphisms in fibronectin binding protein A of Staphylococcus aureus are associated with infection of cardiovascular devices. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18372–18377. doi: 10.1073/pnas.1109071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kayal S, Charbit A. Listeriolysin O: A key protein of Listeria monocytogenes with multiple functions. Fems Microbiology Reviews. 2006;30:514–529. doi: 10.1111/j.1574-6976.2006.00021.x. [DOI] [PubMed] [Google Scholar]
- 59.Moors MA, Levitt B, Youngman P, Portnoy DA. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infection and Immunity. 1999;67:131–139. doi: 10.1128/iai.67.1.131-139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shetron-Rama LM, Marquis H, Bouwer HG, Freitag NE. Intracellular induction of Listeria monocytogenes actA expression. Infection and Immunity. 2002;70:1087–1096. doi: 10.1128/IAI.70.3.1087-1096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arnal L, Serra DO, Salvarezza RC, Yantorno OM, Vela ME. Adhesion contribution to Nanomechanical properties of the Virulent Bordetella pertussis envelope. Langmuir. 2012;28:7461–7469. doi: 10.1021/la300811m. [DOI] [PubMed] [Google Scholar]
- 62.Johnson KL, Greenwood JA. An adhesion map for the contact of elasctic spheres. Journal of Colloid and Interface Science. 1997;192:326–333. doi: 10.1006/jcis.1997.4984. [DOI] [PubMed] [Google Scholar]
- 63.Razatos A, ONG Y-L, Sharma MM, Georgiou G. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 1998;95:11059–11064. doi: 10.1073/pnas.95.19.11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y, Busscher HJ, van der Mei HC, Norde W. Statistical analysis of long- and short-range forces involved in bacterial adhesion to substratum surfaces as measured using atomic force microscopy. Applied and Environmental Microbiology. 2011;77:5065–5070. doi: 10.1128/AEM.00502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Briandet R, Meylheuc T, Maher C, Bellon-Fontaine MN. Listeria monocytogenes Scott A: Cell surface charge, hydrophobicity, and electron donor and acceptor characteristics under different environmental growth conditions. Applied and Environmental Microbiology. 1999;65:5328–5333. doi: 10.1128/aem.65.12.5328-5333.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong H-C, Shau-Yan Chen S-Y, Chen MY, Oliver JD, Hor L-I, Tsai W-C. Pulsed-field gel electrophoresis analysis of Vibrio vulnificus strains isolated from Taiwan and the United States. Appl Environ Microbiol. 2004;70:5153–5158. doi: 10.1128/AEM.70.9.5153-5158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.