Abstract
A 5′ truncated snake venom metalloproteinase was identified from a cDNA library constructed from venom glands of an eastern diamondback rattlesnake (Crotalus adamanteus). The 5′-rapid amplification of cDNA ends (RACE) was used to obtain the 1865 bp full-length cDNA sequence of a snake venom metalloproteinase (CamVMPII). CamVMPII encodes an open reading frame of 488 amino acids, which includes a signal peptide, a pro-domain, a metalloproteinase domain, a spacer, and an RGD-disintegrin domain. The predicted amino acid sequence of CamVMPII showed a 91%, 90%, 83%, and 82% sequence homology to the P-II class enzymes of C. adamanteus metalloproteinase 2, C. atrox CaVMP-II, Gloydius halys agkistin, and Protobothrops jerdonii jerdonitin, respectively. Disintegrins are potent inhibitors of both platelet aggregation and integrin-dependent cell adhesion. Therefore, the disintegrin domain (Cam-dis) of CamVMPII was amplified by PCR, cloned into a pET-43.1a vector, and expressed in Escherichia coli BL21. Affinity purified recombinantly modified Cam-dis (r-Cam-dis) with a yield of 8.5 mg/L culture medium was cleaved from the fusion tags by enterokinase cleavage. r-Cam-dis was further purified by two-step chromatography consisting of HiTrap™ Benzamidine FF column, followed by Talon Metal affinity column with a final yield of 1 mg/L culture. r-Cam-dis was able to inhibit all three processes of platelet thrombus formation including platelet adhesion with an estimated IC50 of 1 nM, collagen- and ADP-induced platelet aggregation with the estimated IC50s of 18 and 6 nM, respectively, and platelet function on clot retraction. It is a potent anti-platelet inhibitor, which should be further investigated for drug discovery to treat stroke patients or patients with thrombotic disorders.
Keywords: Snake venom metalloproteinase, Disintegrin, Crotalus adamanteus, cDNA library, Platelet activity
1. Introduction
Crotalid venoms are rich sources of components that affect the hemostatic system. Snake venom metalloproteinases are zinc-dependent enzymes responsible for hemorrhage that also interfere with hemostasis (Bjarnason and Fox, 1994; Fox and Serrano, 2005). They are divided into three major classes, P-I, P-II, and P-III, based on their general domain organization (Fox and Serrano, 2008). The P-I class contains a pro-domain and a single metalloproteinase domain. The P-II class consists of a pro-domain, a metalloproteinase and a disintegrin domain. The P-III proteinases are composed of a pro-domain, a metalloproteinase, a disintegrin-like domain, and a cysteine-rich domain. The former P-IV class, a P-III structure with C-type lectin-like domains was re-classified into the P-III group as a P-IIId subclass (Fox and Serrano, 2008). Disintegrins may be released by the proteolysis of the P-II or P-III class of snake venom proteins (Jia et al., 1996; Kini and Evans, 1992; Yamada et al., 1999).
Disintegrins are a non-enzymatic group of small molecules (4–14 kDa) that bind selectively to different integrin receptors and are well known to be integrin antagonists. Most disintegrins contain a tripeptide, the RGD motif, which binds to the integrin αIIbβ3 on the platelet surface and inhibit platelet aggregation (Saudek et al., 1991; Williams et al., 1990). However, other disintegrins contain different amino acid sequences including the KGD, RTS, KTS, MGD, MVD, WGD, VGD, MLD, and ECD motifs (Calvete, 2005; Calvete et al., 2003; 2002; McLane et al., 2004). Two antiplatelet drugs, Eptifibatide (a cyclic KGD peptide, Integrilin™) and Tirofiban (an RGD-mimetic, Aggrastat®) were designed based on snake venom disintegrins and have already been approved for clinical use (De Vita et al., 2012; Ji and Hou, 2011; Kristensen et al., 2012). Some RGD-disintegrins also inhibit tumor cell migration, tumor angiogenesis, and tumor metastasis (Jang et al., 2007; Nakamura et al., 1998; Oliva et al., 2007; Sánchez et al., 2009; Yang et al., 2005). In addition to their low molecular weight and blocking integrin activity, disintegrins are of great interest for drug discovery for a variety of disorders such as cancer, osteoporosis, and inflammatory diseases (Walsh and Marcinkiewicz, 2011).
In the present study, we report on a P-II class snake venom metalloproteinase cloned from glands of a C. adamanteus. Its disintegrin domain was subcloned, expressed in Escherichia coli BL21 cells, and tested for its biological activities. This is the first report demonstrating that a recombinantly modified disintegrin from C. adamanteus can inhibit platelet adhesion to fibrinogen, inhibit collagen- and ADP-induced platelet aggregation, and also inhibit platelet function on clot retraction.
2. Materials and Methods
2.1 Snake venom gland extraction and cDNA library construction
Venom glands were dissected from a euthanized adult C. adamanteus snake and immediately frozen in liquid nitrogen. Approximately 30 μg of the venom glands were used to extract the total RNA using the NucleoSpin® RNAII kit (Clontech Laboratories, Inc., CA, USA). A directional cDNA library using 1 μg of total RNA was constructed using the In-Fusion® SMARTer™ cDNA Library Construction Kit (Clontech Laboratories) according to the manufacturer’s instructions. Briefly, 1 μg of total RNA from the venom gland was reverse transcribed into the first-strand cDNA using the SMARTScript™ Reverse Transcriptase (Clontech Laboratories) and the In-Fusion SMARTer CDS primer (Clontech Laboratories) at 42°C for 90 min. Then, double-stranded cDNA (ds cDNA) synthesis was performed on an iCycler Thermal Cycler (Bio-Rad Laboratories, Inc., CA, USA) by LD PCR reaction containing 80 μL of deionized H2O, 10 μL of 10 × advantage 2 PCR buffer, and 2 μL of first-strand cDNA, 50 × dNTP Mix, 5′ PCR primer II A, 3′ In-Fusion SMARTer PCR Primer, and 50 × Advantage 2 Polymerase Mix. The Thermal cycler (Bio-Rad Laboratories) was programmed with an initial denaturation step at 95°C for 1 min followed by 18 cycles for 95°C for 15 s, 65°C for 30 s, and 68°C for 6 min. Lastly, the ds cDNA was purified using CHROMA SPIN™ + TE-1000 size exclusion column chromatography (Clontech Laboratories). Three microliters of each fraction was electrophoresed on a 1.1% agarose/EtBr gel to determine the peak fractions by visualizing the intensity of the bands under UV light. Fractions containing large-, medium-, and small-sized cDNA were pooled, which were then ligated into the pSMART2IFD vector (Clontech Laboratories). The resulting ligation reactions were transformed into Electrocompotent E. coli (Clontech Laboratories). The final resulting plasmid library had over 1 million independent clones having inserts with an approximate average size of 1,000 bp by checking over 2,130 clones using PCR. The 887 clones from the cDNA library were then randomly digested with AluI to determine the cleavage pattern. Based on their sizes and cleavage patterns, the 576 unique plasmid DNAs were selected and sequenced at the DNA Facility, Office of Biotechnology, Iowa State University, Iowa.
2.2 Obtaining the full-length cDNA of the P-II metalloproteinase (CamVMPII)
The full-length cDNA was obtained using the SMARTer™ RACE cDNA amplification kit (Clontech Laboratories) according to the manufacturer’s instructions. The first-strand cDNA was synthesized by a modified oligo (dT) primer (5′-RACE CDS primer A) and the SMARTer II A oligonucleotide. The gene-specific antisense primer (5′-CCTACAGAATGCTTCGGGTTGCACATACCACT-3′) was designed based on the sequence of the metalloproteinase domain of the 5′-truncated PII metalloproteinase gene obtained from cDNA library. The cDNA was amplified using the Advantage 2 PCR kit (Clontech Laboratories), priming by the gene-specific antisense primer and the Universal primer A Mix in the SMARTer™ RACE kit. The RACE product was electrophoresed, purified, and ligated to the pGEM-T Easy vector (Promega, WI, USA). The ligation was transformed into E. coli JM109 competent cells (Promega). Plasmids were extracted from bacterial cell suspensions using the GenElute plasmid miniprep kit (Sigma-Aldrich, MO, USA), and sequenced at the DNA Facility, Office of Biotechnology, Iowa State University, Iowa. The full-length cDNA of CamVMPII was deposited into GenBank with the accession number JX457344.
2.3 Sequence analysis
The cDNA sequences and predicted amino acid sequences were compared to the sequences in the GenBank database using GenBank BLASTN and BLASTX programs (Altschul et al., 1997). Multiple alignments of the amino acid sequences were performed with Clustal W program (Thompson et al., 1994).
2.4 Construction of expression plasmid
The disintegrin domain of CamVMPII was used as a template for PCR. The cDNA encoding the disintegrin domain, Cam-dis, was amplified by PCR with a forward primer 5′-CGCGAATTCGAGGTGGGAGAAGATTGTGACTG-3′ and a reverse primer 5′-GACTCGAGTTAGCCATAGAGGCCATTTCTGGGA-3′. Two restriction enzyme sites: EcoRI in the forward primer and XhoI in the reverse primer were introduced (underlined). The PCR product was digested with EcoRI and XhoI and gel purified. The PCR product was subcloned into EcoRI and XhoI sites of pET-43.1a expression vector (Novagen, NJ, USA), which contains the Nus tag for cloning and the high-level expression of soluble and active recombinant proteins in E. coli and the histidine tag (His tag) for affinity purification (Ermolova et al., 2003). The ligation was transformed into the E. coli Top10 competent cells (Invitrogen, CA, USA). Plasmids were extracted using the GenElute plasmid miniprep kit (Sigma-Aldrich). Plasmids containing inserts of the predicted size for Cam-dis were performed by PCR and further sequenced to verify that the coding sequence was in-frame with the vector sequence that encodes the Nus-His tag.
2.5 Expression and purification of recombinantly modified Cam-dis (r-Cam-dis)
The confirmed recombinant plasmid DNA (pET-43.1a-Cam-dis) was further transformed into E. coli BL21 (DE3) star cells (Invitrogen). BL21 cells harboring recombinant plasmid DNA were first cultured overnight in shaking flasks containing Luria-Bertani (LB) medium. After inoculation of the overnight culture into fresh LB medium, the cultured cells were grown at 37°C with shaking at 225 rpm until the absorbance at 600 nm (OD600) reached 0.6. The culture was induced with a final concentration of 0.1 mM isopropyl β-d-thiogalactoside (IPTG) for 3 h to induce the expression of recombinant proteins. Bacterial cells were collected by centrifugation and resuspended in 50 mM sodium phosphate, 300 mM NaCl, pH 7.0. Cells were lysed on ice with a Branson Sonifier 450 (Branson, Danbury, CT) with the output control setting at 1, a duty cycle setting of constant, and 6 sonication pulses of 30 s per pulse. The lysate was centrifuged at 10,000 × g for 20 min at 4°C. The soluble supernatant was purified using BD TALON metal affinity resins (BD Biosciences Clontech, CA, USA) in an Econo-Column chromatography column (BIO-RAD, CA, USA), which was previously equilibrated with 50 mM sodium phosphate, 300 mM NaCl, pH 7.0. The His-tagged proteins were eluted using an imidazole elution buffer (50 mM sodium phosphate, 300 mM NaCl containing 150 mM imidazole, pH 7.0) at a flow rate 0.3 mL/min. To generate recombinantly modified Cam-dis (r-Cam-dis) free of vector-encoded sequences, the N-terminal Nus-His-tag fusion proteins were cleaved by recombinant enterokinase (rEK, EMD Chemicals Inc., CA, USA) in rEK cleavage buffer (50 mM NaCl, 20 mM Tris-HCl, 2 mM CaCl2, pH 7.4) for 24 h. rEK was first removed from the reaction using a 1 mL HiTrap™ Benzamidine FF (high sub) column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) according to the manufacturer’s instructions. To remove the N-terminal His-tag fusion proteins from r-Cam-dis, the cleaved sample was re-applied to the Talon Metal affinity column, which was pre-equilibrated with an rEK cleavage buffer. r-Cam-dis was then obtained by washing the column with an rEK cleavage buffer. The column was finally washed with an imidazole elution buffer to remove the N-terminal His-tag fusion proteins bound to the column. r-Cam-dis was dialyzed against 1 × phosphate buffer saline (PBS), pH 7.4 and concentrated using a 3 kDa Amicon Ultra-15 centrifugal filter (Millipore, Carrigtwohill, Ireland), electrophoresed on SDS-PAGE under non-reducing condition.
2.6 N-terminal sequencing
r-Cam-dis (4 μg) was transferred from an SDS-PAGE onto a PVDF membrane (Milipore Corporation, MA, USA) using a Semi-Dry Transblot Cell (BIO-RAD) at 125 mA for 1 h. The membrane was stained with Coomassie blue R-250 for 5 min and distained with 50% methanol for 5 min. The sample membrane was sent out for N-terminal amino acid sequencing at the Protein Facility, Office of Biotechnology, Iowa State University, Iowa.
2.7 Protein concentration determination
UV absorbance at 280 nm was used to accurately quantitate the amount of protein if using a purified protein with a known extinction coefficient (εpercent). The partially purified r-Cam-dis contained the N-terminal part of the cleaved Nus tag (Fig. 3a). The estimated εpercent values of r-Cam-dis and the N-terminal part of the cleaved Nus tag calculated by Protein Identification and Analysis Tools on the Expasy Server were 9 and 7, respectively. Therefore, the average εpercent of the mixture of these two proteins was 8. The concentration of this protein mixture was determined from the absorbance at 280 nm and further calculated by the Beer-Lambert Law using the following equation: , where c is the protein concentration in mg/mL, A280 is the absorbance at 280 nm, εpercent is the percent solution extinction coefficient in the unit of (g/100 mL)−1 cm−1, and l is the pathlength of the cuvette (cm). Purified bovine serum albumin standard (BIO-RAD) was used to calibrate the absorbance at 280 nm. The partially purified r-Cam-dis was diluted in 1 × PBS to a stock concentration of 0.5 mg/mL. A stock solution was divided into aliquots and stored at −20°C. The partially purified r-Cam-dis was diluted serially in 1 × PBS and used for platelet activities. For simplicity, the partially purified r-Cam-dis was noted as r-Cam-dis throughout the manuscript.
Fig 3.
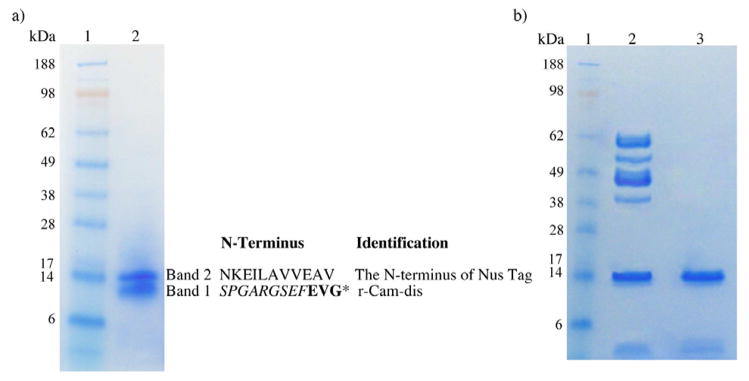
SDS-PAGE of the partially purified r-Cam-dis (a) and cleaved empty pET-43.1a vector (b) under non-reducing condition. Samples were run on 4–12% Bis-Tris Gel using an Xcell SureLock Mini-Cell at 200 V for 30 min. The gel was stained with RapidStain. a) Lane 1: SeeBlue Plus2 Markers; lane 2: the partially purified r-Cam-dis (4 μg). b) Lane 1: SeeBlue Plus2 Markers; lane 2: cleaved empty vector pET-43.1a (3 μg); lane 3: purified cleaved Nus tag (1 μg). The sequence for each band and its identification are on the right. An asterisk (*) represents the N-terminal amino acid sequence of purified r-Cam-dis containing the nine amino acids from the vector (italicized) before the disintegrin sequence (bold).
2.8 Platelet adhesion assay
Human blood was collected from healthy adult volunteers who had not taken any medication for at least two weeks prior to sampling. The platelet-rich plasma (PRP) was obtained by centrifugation at 157 × g for 15 min at room temperature. The PRP was then incubated for 10 min at room temperature with 0.25 μg/mL of prostaglandin E-1 (platelet activation inhibitor) and centrifuged at 1,917 × g for 10 min at 24°C. The PRP was washed 2 times with Tyrode’s buffer (145 mM NaCl, 5 mM KCl, 10 mM Hepes, 0.5 mM Na2HPO4, 6 mM glucose, 0.2 % human serum albumin, 1 mM CaCl2, 1 mM MgSO4 and 0.5 IU/mL apyrase, pH 7.4) and was centrifuged at 1,917 × g for 15 min at 24°C. Platelets were suspended in 1 mL of Tyrode’s buffer. Platelets were counted using a Neubauer chamber and adjusted to the concentration required using Tyrode’s buffer.
Platelet adhesion to fibrinogen was measured by the colorimetric method in 96-well microtiter plates as described by Bellavite et al. (1994). The 96-well polystyrene microplate (Corning, NY, USA) was coated with 100 μL/well of human fibrinogen (2 mg/mL) at 4°C overnight. Platelets (10 × 106 cells) were pre-incubated with various concentrations of r-Cam-dis at 37°C for 1 h. Unbound adhesive proteins were removed from the coated wells by plate aspiration. Two hundred microliters of 1 × PBS containing 1% bovine serum albumin were added and incubated at 37°C for 1 h. After two washes with PBS, 100 μL/well of the treated platelets were added to the wells and incubated at 37°C for 2 h. After the washing step, the adherent platelets were allowed to react for 1 h at 37°C with 150 μL of substrate solution (5 mM p-nitrophenyl phosphate dissolved in 0.1 M citrate buffer, pH 5.4 containing 0.1% Triton X-100). The reaction was stopped by the addition of 100 μL of 2N NaOH. The p-nitrophenol produced by the reaction was measured at 405 nm using a Beckman Coulter AD340 microplate reader. The percentage of platelet adhesion was determined by assigning 100% to the number of platelets, which adhered without being pre-treated with r-Cam-dis. As a negative control to adhesion, the wells were coated with bovine serum albumin (2 mg/mL). r-Mojastin-1 at a final concentration of 7 μg/mL (880 nM) was used as the disintegrin control. The cleaved Nus tag at a final concentration of 1 μg/mL was used as the vector control.
2.9 Platelet aggregation assay
The platelet aggregation assay was performed with human whole blood as previously described (Sánchez et al., 2010). Four hundred and fifty microliters of 10% citrated human blood was incubated at 37°C at least 5 min prior to use with equal amounts of 0.15 M sodium chloride. r-Cam-dis at various concentrations was incubated with the blood samples in a Chrono-log Whole Blood Aggregometer (Chrono-log, PA, USA) at 37°C for 1 min. Platelet aggregation was initiated by a 5 μg/mL of collagen or a final concentration of 10 μM ADP. r-Mojastin-1 was used for comparison (Sánchez et al., 2010). The cleaved Nus tag (0.2 mg/mL) was used as the vector control. The percentage of impedance was measured. The percent inhibition of platelet aggregation was calculated using the following equation: , where C is the units of platelet aggregation (ohms) for the control, and E is the unit of platelet aggregation (ohms) for the experimental fraction. The extent of the inhibition of platelet aggregation was assessed by comparison with the maximal aggregation induced by the control dose of agonists (collagen and ADP). The median inhibitory concentration (IC50) was calculated from a dose-dependent curve using Microsoft Excel 2007.
2.10 Clot retraction assay
Platelet function as a function of clot retraction was measured using human whole blood on a Sonoclot Coagulation and Platelet Function Analyzer (SIENCO, Inc.) as described previously in detail (Hett et al., 1995; Miyashita and Kuro, 1998; Sánchez et al., 2010; Tucci et al., 2006). The Sonoclot analyzer measures the activated clot time (ACT), clot rate, and platelet function on clot retraction by measuring the viscosity changes of a whole blood or plasma sample (von Kaulla et al., 1975). The time that plasma remains a liquid is reported as the ACT. The clot rate is defined as the change in clot signal with a change in time. The platelet function is obtained from the timing and quality of the clot retraction. Briefly, a cuvette containing glass beads (gbACT + KIT) was placed into the cuvette holder, which maintains the temperature at 37°C. A pre-warmed 10 μL sample (37°C) of a 0.3 M CaCl2 was added to one side of the cuvette. Ten microliters of r-Cam-dis at various concentrations (0.01, 0.05, and 0.2 mg/mL) was added to the other side of the cuvette. Three hundred and thirty microliters of citrated blood was added to the cuvette. The platelet function was measured and the data was downloaded from Signature Viewer™ program v. 3.1, software provided by Sienco, Inc. on an iMac computer. The control was citrated blood that was activated with glass beads contained in the cuvette and 0.3 M CaCl2. r-Mojastin-1 (1 mg/mL) was used for comparison. The cleaved Nus tag (0.2 mg/mL) was used as the vector control. The significance was analyzed by the student’s t-test. The level of significance was at P < 0.05. P value was compared with the control.
3. Results
3.1 Amino acid sequence of CamVMPII
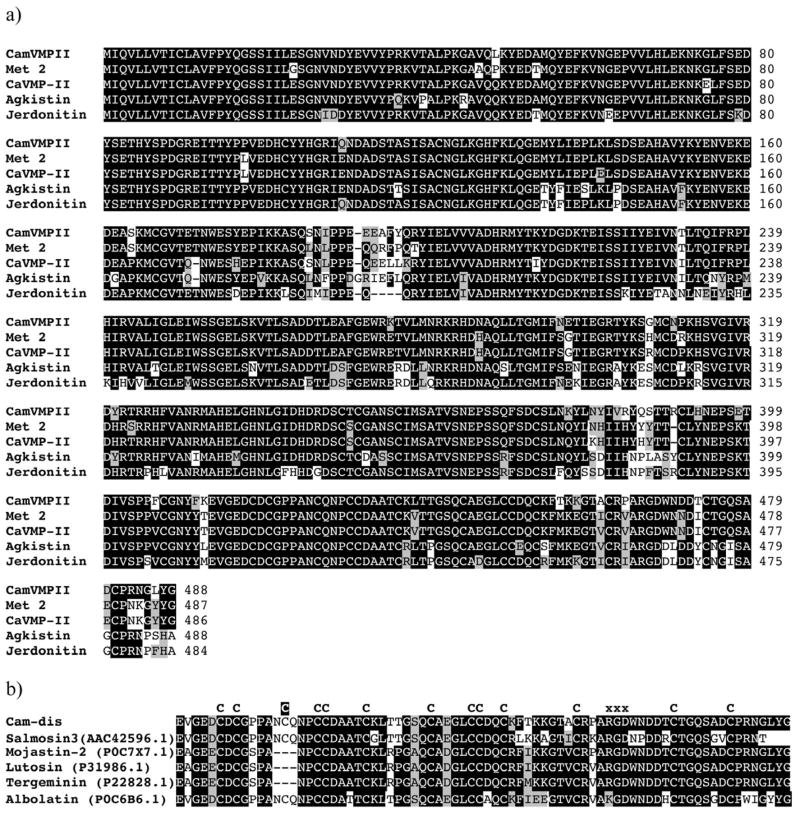
Using a partial sequence obtained from the cDNA library, a P-II metalloproteinase-specific antisense primer was designed for the 5′-RACE to obtain the full-length cDNA. This P-II metalloproteinase from C. adamanteus was designated as CamVMPII. The cDNA sequence of CamVMPII was 1865 bp in length, including 15 nucleotides of a 5′-end untranslated region (UTR), an open reading frame of 1464 nucleotides, and 386 nucleotides of 3′-end UTR with termination codon. The deduced amino acid sequence consisted of a 20-residue signal sequence with the initial ATG codon, a 177-residue pro-domain containing a cysteine-switch domain (SKMCGVT), a 199-residue metalloproteinase domain with a zinc-binding motif (HELGHNLGIDH), a 16-residue spacer region, and a 76-residue disintegrin domain containing the integrin receptor site RGD with 13 cysteine amino acids (Fig. 1). The protein BLAST search and CLUSTAL W multiple sequence alignments revealed that the amino acid sequence of CamVMPII was homologous to C. adamanteus metalloproteinase 2 (AEJ31985.1), C. atrox CaVMP-II (ACV83932.1), Gloydius halys agkistin (AAL60587.1), and Protobothrops jerdonii jerdonitin (P83912.1) with 91%, 90%, 83%, and 82% identity, respectively (Fig. 2a).
Fig. 1.
cDNA and deduced amino acid sequences of CamVMPII with the translated open reading frame from the start codon ATG to the termination codon TAA (shaded in gray). The cysteine-switch motif (SKMCGVT) is bolded. The zinc-binding sequences (HELGHNLGIDH) and Met-turn motif (CIM) are underlined and boxed, respectively. CamVMPII has an RGD motif (black box) within the disintegrin domain. Each domain is indicated by arrows.
Fig. 2.
Multiple alignment of predicted amino acid sequence of CamVMPII with other homologous venom proteins. The alignment was generated with the Clustal W multiple sequence alignment program with manual adjustment and displayed with box shaded. a) CamVMPII is closely homologous to class P-II snake venom metalloproteinases, metalloproteinase 2 (Met 2), CaVMP-II, agkistin, and jerdonitin. b) The disintegrin domain of CamVMPII (Cam-dis) is compared with other medium-sized disintegrins. The numbers in parenthesis are the NCBI accession numbers. The tripeptide binding motif are marked by the letter “X” above the sequences. All cysteine residues (letter “C” above the sequences) are conserved except for an extra cysteine residue (the letter “C” highlighted in black) in Cam-dis, salmosin 3, and albolatin.
Analysis of the predicted amino acid sequence revealed that Cam-dis is a medium-sized disintegrin with an RGD motif. Fig. 2b shows amino acid sequences in comparison with Cam-dis and other closely related medium-sized disintegrins. NCBI protein BLAST analysis showed that the deduced amino acid sequence of the cloned Cam-dis had an 83% sequence identity with salmosin-3 (AAC42596.1) from Gloydius brevicaudus and the native disintegrin mojastin 2 (P0C7X7.1) isolated from the venom of C. scutulatus scutulatus, and was 82% identical to lutosin (P31986.1) from C. viridis, tergeminin (P22828.1) from Sistrurus catenatus tergeminus, and albolatin from Cryptelytrops albolabris (Fig. 2b).
3.2 Expression and purification of r-Cam-dis
The cDNA-coding region for the disintegrin domain was amplified by PCR and subcloned into the EcoRI and XhoI sites of the pET-43.1a expression vector. The yield of affinity r-Cam-dis secreted in the culture supernatant was 8.5 mg/L of culture medium. Following rEK cleavage of the fusion protein, and further purification by two-step chromatography as described above, a yield of 1 mg of protein was obtained. SDS-PAGE analysis revealed two protein bands at ~14 kDa and ~9 kDa, corresponding to the N-terminal part of cleaved Nus tag and r-Cam-dis, respectively (Fig 3a, lane 2). The purity of r-Cam-dis was approximately 54%, as calculated by ImageJ 4.16r software (http://imagej.nih.gov/ij). As shown in Fig. 2, Cam-dis consisted of 76 amino acids and had a calculated molecular weight of ~8 kDa. The N-terminal end contained an extra nine amino acids derived from the vector (SPGARGSEF), which added an additional molecular weight of ~1 kDa, making r-Cam-dis ~9 kDa with a pI 4.51 by Protein Identification and Analysis Tools on the Expasy Server. The N-terminal sequence analysis of the product confirmed that the rEK cleavage occurred at the expected recognition sequence (DDDDK) of the fusion protein and the unexpected secondary sites at other basic amino acids in the Nus tag (Fig. 3). The empty vector was also expressed, purified, and digested with enterokinase. The protein band at about 14 kDa was observed and further purified using the same purification procedure as described above (Fig 3b, lane 3) and used as the vector control (cleaved Nus tag) for platelet activities including platelet adhesion, platelet aggregation, and platelet function on clot retraction.
3.3 Platelet adhesion inhibition
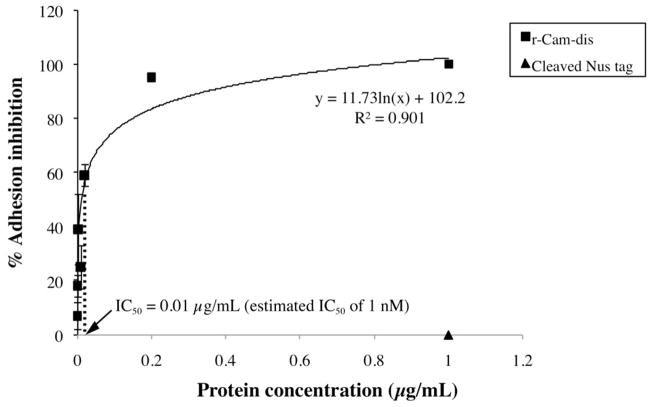
r-Cam-dis inhibited platelet adhesion to fibrinogen with an IC50 of 0.01 μg/mL (the estimated IC50 of 1 nM) (Fig. 4). r-Mojastin-1 used as a disintegrin control (880 nM) inhibited platelet adhesion by 97%.
Fig. 4.
Inhibition of platelet adhesion to fibrinogen by r-Cam-dis. Platelet (10 × 106 cells) were preincubated with various concentrations of r-Cam-dis at 37°C for 1 h prior to adding to fibrinogen-coated wells. p-nitrophenyl phosphate containing 0.1% Triton X-100 was used as the substrate solution. The reaction product p-nitrophenol was measured at 405 nm. The IC50 was 0.01 μg/mL (an estimated IC50 value of 1 nM). The cleaved Nus-tag (~14 kDa; ▲) had no effect on platelet adhesion. The vertical bars represent the standard deviation. n = 3.
3.4 Platelet aggregation inhibition
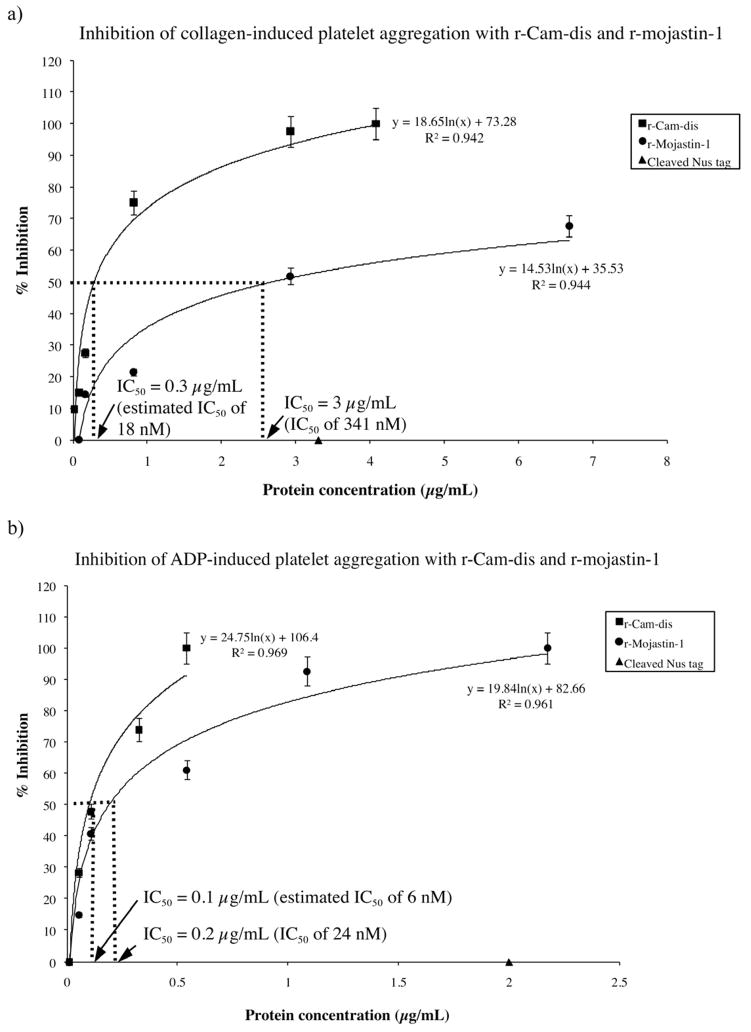
To determine the inhibitory effect of r-Cam-dis on human platelets, in vitro collagen-, and ADP-induced platelet aggregation was carried out using human whole blood. r-Mojastin-1, a recombinant RGD-disintegrin, which is a potent inhibitor of ADP-induced platelet aggregation (Sánchez et al., 2010) was also tested for comparison. r-Cam-dis inhibited the collagen- and ADP-induced platelet aggregation in a dose-dependent manner with IC50 values of 0.3 μg/mL and 0.1 μg/mL, respectively (Fig. 5). To consider the molar ratio of platelet aggregation inhibition induced by collagen and ADP, the IC50 values would be 18 nM and 6 nM, respectively. The IC50 values, for inhibiting collagen- and ADP-induced platelet aggregation, of r-mojastin-1 were 3 μg/mL (341 nM) and 0.2 μg/mL (24 nM), respectively. It was evident that the inhibitory platelet aggregation effects of r-Cam-dis were more potent than that of r-mojastin-1 induced by both agonists, suggesting that r-Cam-dis is a strong platelet aggregation inhibitor. The cleaved Nus tag (vector control, 14 kDa) was incapable of inhibiting collagen- and ADP-induced platelet aggregation.
Fig. 5.
Inhibition of platelet aggregation induced by collagen (a) and ADP (b) with r-Cam-dis and r-mojastin-1. The inhibition assay was performed with human whole blood as described in the Materials and Methods. Platelet aggregation was initiated by a 5 μg/mL of collagen or a final concentration of 10 μM ADP. The maximal aggregation in the absence of recombinant disintegrin was given as 100% aggregation. The IC50s values for inhibiting collagen-induced platelet aggregation were 0.3 μg/mL (an estimated IC50 of 18 nM) for r-Cam-dis and 3 μg/mL (an IC50 of 341 nM) for r-mojastin-1. The IC50s values for inhibiting ADP-induced platelet aggregation were 0.1 μg/mL (an estimated IC50 of 6 nM) for r-Cam-dis and 0.2 μg/mL (an IC50 of 24 nM) for r-mojastin-1. The cleaved Nus-tag (~14 kDa; ▲) did not inhibit collagen- and ADP-induced platelet aggregation. The vertical bars represent the standard deviation. n = 3.
3.5 Clot retraction inhibition
The effect of r-Cam-dis on platelet function was determined by measuring the clot retraction, a simple measurement of platelet interacting with fibrinogen/fibrin, using the Sonoclot analyzer. A platelet function greater than 1 represents normal clot retraction. A platelet function of less than 1 represents no clot retraction and a flat Sonoclot Signature after fibrin formation. Sonoclot signatures from representative samples are shown in Fig. 6. r-Cam-dis had no effects on the ACT and clot rate but significantly inhibited platelet function in a dose-dependent manner (Fig. 6 and Table 1). r-Mojastin-1 did not affect the ACT, clot rate, nor platelet function, which was consistent with previous results (Sánchez et al., 2010). The cleaved Nus tag (vector control) did not have an effect on the ACT, clot rate, and platelet function.
Fig. 6.
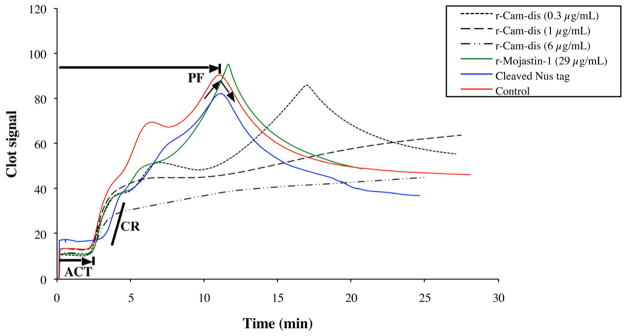
Data from Sonoclot signature and further analyzed by Microsoft Excel 2007. r-Cam-dis were added to the final concentrations of 0.3, 1, and 6 μg/mL in whole blood using glass bead activated cuvettes (gbACT + KIT) on a Sonoclot Analyzer System. The activated clot time (ACT) is the time (min) in which whole blood begins to clot. The clot rate is defined as the rate of fibrin polymerization, which is the slope in the linear part of the curves and is defined as the change clot signal with change in time (U = Δsignal/Δtime). The platelet function (PF, the timing and quality of the clot retraction) is a calculated value, derived by using an automated numeric integration of changes in the Sonoclot signature after fibrin formation has completed. The control was citrated blood that was activated with glass beads contained in the cuvette and 0.3 M CaCl2. r-Mojastin-1 at a final concentration of 29 μg/mL (4 μM) was used for comparison.
Table 1.
Sonoclot analysis of whole blood platelet function on clot retraction by r-Cam-dis and r-mojastin-1.
| Sample | Platelet functiona Mean ± SD (n=3) | P value |
|---|---|---|
| Controlb | 2.63 ± 0.31 | - |
| r-Cam-dis (0.3 μg/mL) | 2.63 ± 0.38 | 1.00 |
| r-Cam-dis (1 μg/mL) | 0.23 ± 0.15 | 0.0003 |
| r-Cam-dis (6 μg/mL) | 0.17 ± 0.06 | 0.0002 |
| r-Mojastin-1 (29 μg/mL)c | 3.00 ± 0.44 | 0.30 |
| Cleaved Nus tag | 2.90 ± 0.89 | 0.65 |
Bold letter indicates the significant reduction of the platelet function by r-Cam-dis compared with the control at P < 0.05.
Platelet function is defined as the function of clot retraction. A platelet function greater than 1 represents normal clot retraction. A platelet function of less than 1 represents no clot retraction.
The control was citrated blood that was activated with glass beads contained in the cuvette and 0.3 M CaCl2.
r-Mojastin-1 at a final concentration of 29 μg/mL (4 μM) was used for comparison.
4. Discussion
Snake venom metalloproteinases are widely distributed in Viperid and Crotalid snake venoms and are involved in several pathological and biological effects, such as hemorrhage, inflammation, necrosis, hypotension, pro-coagulant, anticoagulant, and antiplatelet activities (Bjarnason and Fox, 1994; Fox and Serrano, 2009; 2005; Kamiguti, 2005; Kini, 2006). They are multifunctional-domain proteins with a large number of variability in snake venoms and are grouped into several subclasses depending on the organization of their domains (Fox and Serrano, 2008). The P-II and P-III metalloproteinases are further divided into P-IIa to P-IIe and P-IIIa to P-IIId subclasses, respectively, based on their variable post-translation modifications (Fox and Serrano, 2009; 2008).
In this study, we have characterized a metalloproteinase, CamVMPII, from the cDNA library of C. adamanteus. Its deduced amino acid sequence consists of a signal peptide, a pro-domain, a metalloproteinase domain, a spacer, and a disintegrin domain and belongs to the non-proteolytic processed subclass P-IIb snake venom metalloproteinase. CamVMPII shares 91% identity with the previously reported metalloproteinase 2 from the same snake (accession # AEJ31985.1), 90% identity with CaVMP-II from C. atrox (accession # ACV83932.1), 83% identity with agkistin from Gloydius halys (accession # AAL60587.1), and 82% identity with jerdonitin from Protobothrops jerdonii (accession # P83912.1). The cysteinyl residues are conserved in all of these sequences. Interestingly, a number of different amino acids in coding regions were observed between the predicted amino acid sequences of C. adamanteus CamVMPII (in this study) and metalloproteinase 2 (Rokyta et al., 2011), illustrating the diversity of metalloproteinases in this snake. A great diversity of the class P-II has been reported in Bothrops neuwiedi snake venom transcripts that could be reflected in the generation of new functional proteins in the venoms (Moura-da-Silva et al., 2011).
The amino acid sequence comparison of the disintegrin domain (Cam-dis) with other closely related medium-sized disintegrins revealed that conserved cysteine residues were observed among disintegrins (Fig. 2b). Cam-dis, salmosin 3 (Park et al., 1998), and albolatin (Singhamatr and Rojnuckarin, 2007) shared an additional cysteine located in the N-terminal end. An additional cysteine residue may form a disulfide bond with another residue located in the spacer region that is believed to protect the proteolysis of the disintegrin domain from the metalloproteinase domain (Fox and Serrano, 2009; 2005).
The disintegrin domain is a part of snake venom metalloproteinases, which involves the binding of integrin receptors. Disintegrins typically contain the RGD or other tripeptide motifs that can inhibit integrin-mediated platelet aggregation and block many biological functions of integrins on cell surfaces (Calvete et al., 2005; Huang et al., 1987; Scarborough et al., 1991). Functional recombinant disintegrins have been expressed in E. coli, such as rhodostomin (Chang et al., 1993), jararhajin ECD-disintegrin (Moura-da-Silva et al., 1999), r-mojastin-1 (Sánchez et al., 2010), r-rubistatin (Carey et al., 2012), DisBa-01 (Montenegro et al., 2012; Ramos et al., 2008), and r-viridistatin-2 (Lucena et al., 2012).
In this study, the disintegrin domain of CamVMPII, Cam-dis, was expressed in E. coli, purified, and cleaved from the Nus-His fusion tags using recombinant enterokinase. r-Cam-dis showed two major protein bands with molecular weights of about 9 and 14 kDa under non-reducing condition. The N-terminal amino acid sequences of 9-kDa and 14-kDa proteins revealed that the 9 kDa protein was r-Cam-dis containing an additional nine amino acids from the vector at the N-terminus end. Structural-functional studies of disintegrins revealed that disulfide bonding is essential for the binding of disintegrins to integrin receptors, whereas the integrin binding specificity and affinity of disintegrins depends on the tripeptide and amino acid residues in the C-terminal end (Calvete et al., 2005; McLane et al., 2001; Wierzbicka-Patynowski et al., 1999; Yahalom et al., 2002). Moreover, the NMR structure of RGD-disintegrins including salmosin (PDB code 1iq2), kistrin (PDB code 1kst), flavoridin (PDB code 1fvl), and echistatin (PDB code 2ech) revealed that the conformational uniqueness of a disintegrin is a major contributing factor for the integrin binding selectivity, which is necessary for biological activity (Shin et al., 2003). Therefore, the additional nine amino acids with several hydrophobic amino acids could introduce a conformational change in the protein structure that may alter its biological activity. The native disintegrin, if found in the venom, should be further isolated from C. adamanteus venom for the purpose of comparability. Further investigation on the three-dimensional structures of the native disintegrin and r-Cam-dis will be required to understand and identify their biological activities. The 14 kDa protein was identical to the N-terminal end of the pET-43.1a vector, which might be derived from the enterokinase cleavage at the secondary sites located towards the amino terminal end on the pET-43.1a vector. We tested several dilutions of recombinant enterokinase to r-Cam-dis (1:20, 1:50, 1:100, and 1:200) and incubation times (2, 4, 8, 16, 24, and 32 h) to optimize the efficiency of the cleavage; however, the unexpected proteolytic protein (~14 kDa) released during the enterokinase cleavage of the Nus-His-tagged r-Cam-dis fusion protein was observed in all conditions. This non-targeted cleavage product at about 14 kDa corresponded to the cleavage of the N-terminal end of Nus tag after its -ER124 sequence, which is one of three different secondary cleavage sites (-MK134, -ER143, and -DR156 sites) as previously reported (Liew et al., 2005). We expressed the empty vector, purified, and cleaved with recombinant enterokinase with the same protocol for purification of r-Cam-dis. The cleaved Nus tag protein (~14 kDa) was then isolated and used as the vector control for platelet activities.
Platelets play an important role in hemostasis, clot stability, and clot retraction. Upon vascular injury, platelets adhere to the damage sites, spread, aggregate, secrete several activating agents to further promote aggregation, which lead to a platelet thrombus. However, inappropriate platelet adhesion and activation can lead to thrombosis, myocardial infarctions, and strokes. Since disintegrins affect the mechanisms of platelet plug formation, they could be a drug candidate for arterial thrombosis and heart disease. Therefore, the effects of r-Cam-dis on platelet adhesion, platelet aggregation, and platelet function on clot retraction were characterized. To exclude the cleaved Nus-tag from being involved in the activity, the cleaved Nus-tag (~14 kDa) was used as the vector control and had no effect on platelet adhesion, aggregation, or platelet function on clot retraction (Fig. 4 – 6, Table 1).
Platelet adhesion is the first step in the initiation phase of platelet plug formation for inhibiting blood loss after vessel wall injury. Previous studies reported that disintegrins inhibit platelet adhesion to immobilized extracellular matrix including fibrinogen, fibronectin, and collagen (Da Silva et al., 2009; Sánchez et al. 2010; You et al., 2003). In our study, r-Cam-dis inhibited platelet adhesion to fibrinogen with an IC50 of 0.01 μg/mL (an estimated IC50 of 1 nM). In addition, r-Cam-dis dose-dependently inhibited collagen- and ADP-induced platelet aggregation using whole blood with the IC50s of 0.3 μg/mL (an estimated IC50 of 18 nM) and 0.1 μg/mL (an estimated IC50 of 6 nM), respectively. Based on the molar concentration, the inhibition of collagen-induced platelet aggregation of r-Cam-dis was about 19 times more efficient than that of r-mojastin-1 (Fig. 5a). Similarly, the IC50 value for inhibiting ADP-induced platelet aggregation was 4 times more efficient than r-mojastin-1 (Fig. 5b). Recently, two potent recombinant RGD-disintegrins, r-mojastin-1 (Sánchez et al., 2010) and r-viridistatin 2 (Lucena et al., 2012), have been reported to inhibit ADP-induced platelet aggregation in whole blood with IC50s of 46 nM and 34 nM, respectively. The IC50, for inhibiting ADP-induced platelet aggregation in PRP, of r-mojastin-1 was 122 nM, which was more efficient than other recombinant disintegrins with IC50s ranging from 126 to 6000 nM (Sánchez et al., 2010). Platelet aggregation studies using PRP with recombinant non-RGD-disintegrins, recombinant barbourin (a KGD monomeric disintegrin), could inhibit ADP-induced platelet aggregation with IC50 values ranging from 330 to 370 nM (Marques et al., 2001). In addition, Singhamatr et al. (2007) reported that recombinant albolatin, a KGD homodimeric disintegrin, inhibits collagen-induced platelet aggregation with IC50 of 976 nM, but is incapable of inhibiting ADP-induced platelet aggregation. This indicated that r-Cam-dis is a potent platelet aggregation inhibitor.
In addition, r-Cam-dis had anti-platelet function activity demonstrated by inhibiting platelet interaction with fibrinogen/fibrin (clot retraction), but did not interfere with activated clotting time and clot rate (Fig. 6). The ability of integrin αIIbβ3 to bind to fibrinogen plays a crucial role in platelet aggregation, adhesion, and hemostasis, suggesting that r-Cam-dis binds to the platelet integrin αIIbβ3. However, other integrin specificity for r-Cam-dis should be further investigated.
In summary, a full-length cDNA of snake venom metalloproteinase class P-II containing the disintegrin domain was characterized from the venom glands of C. adamanteus. Its disintegrin domain was cloned and expressed with activity. This is the first study to report that a new recombinantly modified RGD-disintegrin from C. adamanteus was shown to have strong anti-platelet activities. It was able to inhibit three mechanisms of platelet plug formation including platelet adhesion, platelet aggregation, and platelet function on clot retraction. Due to its potent anti-platelet activity and medium size, r-Cam-dis could be used to develop new agents for the treatment of thrombosis and heart disease.
Highlights.
A novel snake venom metalloproteinase, CamVMPII was identified from cDNA library of venom glands of Crotalus adamanteus.
The disintegrin domain (r-Cam-dis) of CamVMPII is expressed in Escherichia coli.
r-Cam-dis inhibits platelet adhesion, platelet aggregation, and platelet function on clot retraction.
r-Cam-dis is a potent anti-platelet inhibitor.
Acknowledgments
This work was funded by the NNTRC, Texas A&M University-Kingsville: NCRR/Viper Resource Center P40 OD01960-10 and Texas A&M University-Kingsville-Research Development Funds (Acct. # 160302 and 160315-00022). We would like to thank Barret S. Henriquez and Rolando Barrientes, Jr., students at Texas A&M University-Kingsville for their assistance. We are grateful to Nora Diaz De Leon, the NNTRC administrative officer for technical assistance. We thank Angela Wyro for her technical review of the manuscript. We gratefully acknowledge all of the NNTRC staff.
Abrreviations
- ACT
activated clotting time
- ADP
adenosine diphosphate
- cDNA
complementary deoxyribonucleic acid
- CR
clot rate
- IPTG
isopropyl β-d-thiogalactoside
- PCR
polymerase chain reaction
- PF
platelet function
- pI
isoelectric point
- RACE
rapid amplification of cDNA ends
- RGD motif
arginine-glycine-aspartic acid motif
- PRP
platelet-rich plasma
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- UTR
untranslated region
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This research was approved by Texas A&M University-Kingsville Human Subjects Committee.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavite P, Andrioli G, Guzzo P, Arigliano P, Chirumbolo S, Manzato F, Santonastaso C. A colorimetric method for the measurement of platelet adhesion in microtiter plates. Anal Biochem. 1994;216:444–450. doi: 10.1006/abio.1994.1066. [DOI] [PubMed] [Google Scholar]
- Bjarnason JB, Fox JW. Hemorrhagic metalloproteinases from snake venoms. Pharmacol Ther. 1994;62:325–372. doi: 10.1016/0163-7258(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Calvete JJ. Structure-function correlations of snake venom disintegrins. Curr Pharm Des. 2005;11:829–835. doi: 10.2174/1381612053381783. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Fox JW, Agelan A, Niewiarowski S, Marcinkiewicz C. The presence of the WGD motif in CC8 heterodimeric disintegrin increases its inhibitory effect on alphaII(b)beta3, alpha(v)beta3, and alpha5beta1 integrins. Biochemistry. 2002;41:2014–2021. doi: 10.1021/bi015627o. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Marcinkiewicz C, Monleón D, Esteve V, Celda B, Juárez P, Sanz L. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005;45:1063–1074. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Moreno-Murciano MP, Theakston RD, Kisiel DG, Marcinkiewicz C. Snake venom disintegrins: novel dimeric disintegrins and structural diversification by disulphide bond engineering. Biochem J. 2003;372(Pt 3):725–734. doi: 10.1042/BJ20021739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey CM, Bueno R, Gutierrez DA, Petro C, Lucena SE, Sánchez EE, Soto JG. Recombinant rubistatin (r-Rub), an MVD disintegrin, inhibits cell migration and proliferation, and is a strong apoptotic inducer of the human melanoma cell line SK-Mel-28. Toxicon. 2012;59:241–248. doi: 10.1016/j.toxicon.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Hu ST, Huang TF, Chen SH, Lee YH, Lo SJ. Rhodostomin, an RGD-containing peptide expressed from a synthetic gene in Escherichia coli, facilitates the attachment of human hepatoma cells. Biochem Biophys Res Commun. 1993;190:242–249. doi: 10.1006/bbrc.1993.1037. [DOI] [PubMed] [Google Scholar]
- Da Silva M, Lucena S, Aguilar I, Rodríguez-Acosta A, Salazar AM, Sánchez EE, Girón ME, Carvajal Z, Arocha-Piñango CL, Guerrero B. Anti-platelet effect of cumanastatin 1, a disintegrin isolated from venom of South American Crotalus rattlesnake. Thromb Res. 2009;123:731–739. doi: 10.1016/j.thromres.2008.08.001. [DOI] [PubMed] [Google Scholar]
- De Vita M, Coluccia V, Burzotta F, Romagnoli E, Trani C. Intracoronary Use of GP IIb/IIIa Inhibitors in Percutaneous Coronary Interventions. Curr Vasc Pharmacol. 2012;10:448–453. doi: 10.2174/157016112800812764. [DOI] [PubMed] [Google Scholar]
- Ermolova NV, Ann Cushman M, Taybi T, Condon SA, Cushman JC, Chollet R. Expression, purification, and initial characterization of a recombinant form of plant PEP-carboxylase kinase from CAM-induced Mesembryanthemum crystallinum with enhanced solubility in Escherichia coli. Protein Expr Purif. 2003;29:123–131. doi: 10.1016/s1046-5928(03)00014-7. [DOI] [PubMed] [Google Scholar]
- Fox JW, Serrano SM. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Fox JW, Serrano SM. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008;275:3016–3030. doi: 10.1111/j.1742-4658.2008.06466.x. [DOI] [PubMed] [Google Scholar]
- Fox JW, Serrano SM. Timeline of key events in snake venom metalloproteinase research. J Proteomics. 2009;72:200–209. doi: 10.1016/j.jprot.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Hett DA, Walker D, Pilkington SN, Smith DC. Sonoclot analysis. Br J Anaesth. 1995;75:771–776. doi: 10.1093/bja/75.6.771. [DOI] [PubMed] [Google Scholar]
- Huang TF, Holt JC, Lukasiewicz H, Niewiarowski S, Trigramin A low molecular weight peptide inhibiting fibrinogen interaction with platelet receptors expressed on glycoprotein IIb-IIIa complex. J Biol Chem. 1987;262:16157–16163. [PubMed] [Google Scholar]
- Jang YJ, Kim DS, Jeon OH, Kim DS. Saxatilin suppresses tumor-induced angiogenesis by regulating VEGF expression in NCI-H460 human lung cancer cells. J Biochem Mol Biol. 2007;40:439–443. doi: 10.5483/bmbrep.2007.40.3.439. [DOI] [PubMed] [Google Scholar]
- Ji X, Hou M. Novel agents for anti-platelet therapy. J Hematol Oncol. 2011;4:44. doi: 10.1186/1756-8722-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia LG, Shimokawa K, Bjarnason JB, Fox JW. Snake venom metalloproteinases: structure, function and relationship to the ADAMs family of proteins. Toxicon. 1996;34:1269–1276. doi: 10.1016/s0041-0101(96)00108-0. [DOI] [PubMed] [Google Scholar]
- Kamiguti AS. Platelets as targets of snake venom metalloproteinases. Toxicon. 2005;45:1041–1049. doi: 10.1016/j.toxicon.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Kini RM. Anticoagulant proteins from snake venoms: structure, function and mechanism. Biochem J. 2006;397:377–387. doi: 10.1042/BJ20060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini RM, Evans HJ. Structural domains in venom proteins: evidence that metalloproteinases and nonenzymatic platelet aggregation inhibitors (disintegrins) from snake venoms are derived by proteolysis from a common precursor. Toxicon. 1992;30:265–293. doi: 10.1016/0041-0101(92)90869-7. [DOI] [PubMed] [Google Scholar]
- Kristensen SD, Würtz M, Grove EL, De Caterina R, Huber K, Moliterno DJ, Neumann FJ. Contemporary use of glycoprotein IIb/IIIa inhibitors. Thromb Haemost. 2012;107:215–224. doi: 10.1160/TH11-07-0468. [DOI] [PubMed] [Google Scholar]
- Liew OW, Ching Chong JP, Yandle TG, Brennan SO. Preparation of recombinant thioredoxin fused N-terminal proCNP: Analysis of enterokinase cleavage products reveals new enterokinase cleavage sites. Protein Expr Purif. 2005;41:332–340. doi: 10.1016/j.pep.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Lucena SE, Jia Y, Soto JG, Parral J, Cantu E, Brannon J, Lardner K, Ramos CJ, Seoane AI, Sánchez EE. Anti-invasive and anti-adhesive activities of a recombinant disintegrin, r-viridistatin 2, derived from the Prairie rattlesnake (Crotalus viridis viridis) Toxicon. 2012;60:31–39. doi: 10.1016/j.toxicon.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JA, George JK, Smith IJ, Bhakta V, Sheffield WP. A barbourin-albumin fusion protein that is slowly cleared in vivo retains the ability to inhibit platelet aggregation in vitro. Thromb Haemost. 2001;86:902–908. [PubMed] [Google Scholar]
- McLane MA, Kuchar MA, Brando C, Santoli D, Paquette-Straub CA, Miele ME. New insights on disintegrin-receptor interactions: eristostatin and melanoma cells. Haemostasis. 2001;31:177–182. doi: 10.1159/000048061. [DOI] [PubMed] [Google Scholar]
- McLane MA, Sánchez EE, Wong A, Paquette-Straub C, Perez JC. Disintegrins. Curr Drug Targets Cardiovasc Haematol Disord. 2004;4:327–355. doi: 10.2174/1568006043335880. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Kuro M. Evaluation of platelet function by Sonoclot analysis compared with other hemostatic variables in cardiac surgery. Anesth Analg. 1998;87:1228–1233. doi: 10.1097/00000539-199812000-00002. [DOI] [PubMed] [Google Scholar]
- Montenegro CF, Salla-Pontes CL, Ribeiro JU, Machado AZ, Ramos RF, Figueiredo CC, Morandi V, Selistre-de-Araujo HS. Blocking αvβ3 integrin by a recombinant RGD disintegrin impairs VEGF signaling in endothelial cells. Biochimie. 2012;94:1812–1820. doi: 10.1016/j.biochi.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Moura-da-Silva AM, Furlan MS, Caporrino MC, Grego KF, Portes-Junior JA, Clissa PB, Valente RH, Magalhães GS. Diversity of metalloproteinases in Bothrops neuwiedi snake venom transcripts: evidences for recombination between different classes of SVMPs. BMC Genet. 2011;12:94. doi: 10.1186/1471-2156-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura-da-Silva AM, Línica A, Della-Casa MS, Kamiguti AS, Ho PL, Crampton JM, Theakston RD. Jararhagin ECD-containing disintegrin domain: expression in escherichia coli and inhibition of the platelet-collagen interaction. Arch Biochem Biophys. 1999;369:295–301. doi: 10.1006/abbi.1999.1372. [DOI] [PubMed] [Google Scholar]
- Nakamura I, Tanaka H, Rodan GA, Duong LT. Echistatin inhibits the migration of murine prefusion osteoclasts and the formation of multinucleated osteoclast-like cells. Endocrinology. 1998;139:5182–5193. doi: 10.1210/endo.139.12.6375. [DOI] [PubMed] [Google Scholar]
- Oliva IB, Coelho RM, Barcellos GG, Saldanha-Gama R, Wermelinger LS, Marcinkiewicz C, Benedeta Zingali R, Barja-Fidalgo C. Effect of RGD-disintegrins on melanoma cell growth and metastasis: involvement of the actin cytoskeleton, FAK and c-Fos. Toxicon. 2007;50:1053–1063. doi: 10.1016/j.toxicon.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Park D, Kang I, Kim H, Chung K, Kim DS, Yun Y. Cloning and characterization of novel disintegrins from Agkistrodon halys venom. Mol Cells. 1998;8:578–584. [PubMed] [Google Scholar]
- Ramos OH, Kauskot A, Cominetti MR, Bechyne I, Salla Pontes CL, Chareyre F, Manent J, Vassy R, Giovannini M, Legrand C, Selistre-de-Araujo HS, Crépin M, Bonnefoy A. A novel alpha(v)beta (3)-blocking disintegrin containing the RGD motive, DisBa-01, inhibits bFGF-induced angiogenesis and melanoma metastasis. Clin Exp Metastasis. 2008;25:53–64. doi: 10.1007/s10585-007-9101-y. [DOI] [PubMed] [Google Scholar]
- Rokyta DR, Wray KP, Lemmon AR, Lemmon EM, Caudle SB. A high-throughput venom-gland transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon. 2011;57:657–671. doi: 10.1016/j.toxicon.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Sánchez EE, Lucena SE, Reyes S, Soto JG, Cantu E, Lopez-Johnston JC, Guerrero B, Salazar AM, Rodríguez-Acosta A, Galán JA, Tao WA, Pérez JC. Cloning, expression, and hemostatic activities of a disintegrin, r-mojastin 1, from the mohave rattlesnake (Crotalus scutulatus scutulatus) Thrombosis Research. 2010;126:e211–e219. doi: 10.1016/j.thromres.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez EE, Rodríguez-Acosta A, Palomar R, Lucena SE, Bashir S, Soto JG, Pérez JC. Colombistatin: a disintegrin isolated from the venom of the South American snake (Bothrops colombiensis) that effectively inhibits platelet aggregation and SK-Mel-28 cell adhesion. Arch Toxicol. 2009;83:271–279. doi: 10.1007/s00204-008-0358-y. [DOI] [PubMed] [Google Scholar]
- Saudek V, Atkinson RA, Pelton JT. Three-dimensional structure of echistatin, the smallest active RGD protein. Biochemistry. 1991;30:7369–7372. doi: 10.1021/bi00244a003. [DOI] [PubMed] [Google Scholar]
- Scarborough RM, Rose JW, Hsu MA, Phillips DR, Fried VA, Campbell AM, Nannizzi L, Charo IF. Barbourin. A GPIIb-IIIa-specific integrin antagonist from the venom of Sistrurus m barbouri. J Biol Chem. 1991;266:9359–9362. [PubMed] [Google Scholar]
- Shin J, Hong SY, Chung K, Kang I, Jang Y, Kim DS, Lee W. Solution structure of a novel disintegrin, salmosin, from Agkistrondon halys venom. Biochemistry. 2003;42:14408–14415. doi: 10.1021/bi0300276. [DOI] [PubMed] [Google Scholar]
- Singhamatr P, Rojnuckarin P. Molecular cloning of albolatin, a novel snake venom metalloprotease from green pit viper (Trimeresurus albolabris), and expression of its disintegrin domain. Toxicon. 2007;50:1192–1200. doi: 10.1016/j.toxicon.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci MA, Ganter MT, Hamiel CR, Klaghofer R, Zollinger A, Hofer CK. Platelet function monitoring with the Sonoclot analyzer after in vitro tirofiban and heparin administration. J Thorac Cardiovasc Surg. 2006;131:1314–1322. doi: 10.1016/j.jtcvs.2006.01.041. [DOI] [PubMed] [Google Scholar]
- von Kaulla KN, Ostendorf P, von Kaulla E. The impedance machine: a new bedside coagulation recording device. J Med. 1975;6:73–88. [PubMed] [Google Scholar]
- Walsh EM, Marcinkiewicz C. Non-RGD-containing snake venom disintegrins, functional and structural relations. Toxicon. 2011;58:355–362. doi: 10.1016/j.toxicon.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Wierzbicka-Patynowski I, Niewiarowski S, Marcinkiewicz C, Calvete JJ, Marcinkiewicz MM, McLane MA. Structural requirements of echistatin for the recognition of alpha(v)beta(3) and alpha(5)beta(1) integrins. J Biol Chem. 1999;274:37809–37814. doi: 10.1074/jbc.274.53.37809. [DOI] [PubMed] [Google Scholar]
- Williams J, Rucinski B, Holt J, Niewiarowski S. Elegantin and albolabrin purified peptides from viper venoms: homologies with the RGDS domain of fibrinogen and von Willebrand factor. Biochim Biophys Acta. 1990;1039:81–89. doi: 10.1016/0167-4838(90)90229-9. [DOI] [PubMed] [Google Scholar]
- Yahalom D, Wittelsberger A, Mierke DF, Rosenblatt M, Alexander JM, Chorev M. Identification of the principal binding site for RGD-containing ligands in the alpha(V)beta(3) integrin: a photoaffinity cross-linking study. Biochemistry. 2002;41:8321–8331. doi: 10.1021/bi025690t. [DOI] [PubMed] [Google Scholar]
- Yamada D, Shin Y, Morita T. Nucleotide sequence of a cDNA encoding a common precursor of disintegrin flavostatin and hemorrhagic factor HR2a from the venom of Trimeresurus flavoviridis. FEBS Lett. 1999;451:299–302. doi: 10.1016/s0014-5793(99)00604-3. [DOI] [PubMed] [Google Scholar]
- Yang RS, Tang CH, Chuang WJ, Huang TH, Peng HC, Huang TF, Fu WM. Inhibition of tumor formation by snake venom disintegrin. Toxicon. 2005;45:661–669. doi: 10.1016/j.toxicon.2005.01.013. [DOI] [PubMed] [Google Scholar]
- You WK, Jang YJ, Chung KH, Kim DS. A novel disintegrin-like domain of a high molecular weight metalloprotease inhibits platelet aggregation. Biochem Biophys Res Commun. 2003;309:637–642. doi: 10.1016/j.bbrc.2003.08.049. [DOI] [PubMed] [Google Scholar]