Abstract
Burst firing of neurons optimizes neurotransmitter release. GnRH neurons exhibit burst firing activity and T-type calcium channels, which are vital for burst firing activity, are regulated by 17β-estradiol (E2) in GnRH neurons. To further elucidate ion channel expression and E2 regulation during positive and negative feedback on GnRH neurosecretion, we used single cell RT-PCR and real-time qPCR to quantify channel mRNA expression in GnRH neurons. GFP-GnRH neurons expressed numerous ion channels important for burst firing activity. E2-treatment sufficient to induce an LH surge increased mRNA expression of HCN1 channels, which underlie the pacemaker current, the calcium-permeable CaV1.3, CaV2.2, CaV2.3 channels, and TRPC4 channels, which mediate the kisspeptin excitatory response. E2 also decreased mRNA expression of SK3 channels underlying the medium AHP current. Therefore, E2 exerts fundamental changes in ion channel expression in GnRH neurons, to prime them to respond to incoming stimuli with increased excitability at the time of the surge.
Keywords: GnRH neurons, channel subunits, mRNA quantification, LH surge
1. Introduction
As demonstrated in a number of species including the rat, sheep and rhesus monkey, the preovulatory luteinizing hormone (LH) surge is accompanied by a surge in gonadotropin-releasing hormone (GnRH) (Caraty et al., 1989; Chappell and Levine, 2000; Levine and Ramirez, 1982; Pau et al., 1993), suggesting increased activity of GnRH neurons at the time of the GnRH surge. This increased activity is primarily due to increased estradiol levels, because treatment with 17β-estradiol (E2) in ovariectomized (OVX) females can mimic the positive feedback regulation of GnRH and LH secretion (Caraty et al., 1989; Chappell and Levine, 2000). Cell-attached single-unit extracellular recording to evaluate GnRH neuronal firing activity during negative as compared to positive feedback has revealed that the GnRH neuronal firing rate is low during the morning negative feedback period and significantly higher during the evening positive feedback period (Christian et al., 2005). Collectively, these findings would indicate that there are fundamental changes in GnRH neuronal firing activity during the different stages of the ovulatory cycle and during E2-induced negative and positive feedback. The timing of the GnRH (LH) surge and thus the increased activity of GnRH neurons at the time of the surge is E2-dependent, but it is also entrained to a circadian input from the suprachiasmatic nucleus (SCN) at least in some rodent species (Chappell et al., 2009; Christian et al., 2005; Christian and Moenter, 2008; Legan et al., 1975). The mechanism by which the SCN affects GnRH neuronal activity is not known, but circadian cues may involve vasopressin input to kisspeptin neurons and vasoactive intestinal peptide input to GnRH neurons (Christian and Moenter, 2008; Vida et al., 2010; Ward et al., 2009).
In addition to GnRH neurons, kisspeptin neurons are essential for reproductive development and reproductive competence (Oakley et al., 2009). These hypothalamic neurons express ERα, are affected by E2 feedback, and are strongly excitatory to GnRH neurons (Oakley et al., 2009). Kisspeptin excites GnRH neurons by actions on the G protein-coupled receptor 54 (GPR54), also called kisspeptin receptor (Zhang et al., 2008). Evidence from GT1–7 GnRH neuronal cells suggests that GPR54 exhibits an E2-dependent diurnal variation in mRNA expression (Tonsfeldt et al., 2011). These findings, however, have not been confirmed in native GnRH neurons.
Based on a model similar to that described and validated for thalamocortical relay neurons and hypothalamic neurosecretory neurons (Chemin et al., 2002; Erickson et al., 1993b; Kelly and Rønnekleiv, 1994; Kim et al., 2001), we had predicted that T-type calcium channels together with the hyperpolarization-activated, cyclic nucleotide-gated channels (HCN) are essential for induction of burst firing in GnRH neurons, and that the calcium-dependent, small-conductance calcium-activated potassium channels (SK) -type channels, which underlie afterhyperpolarization (AHP) are crucial for allowing repetitive cycles of burst firing (Kelly and Rønnekleiv, 1994; Kelly and Wagner, 2002). All of these channels are active in GnRH neurons and contribute significantly to their signaling pattern (Bosch et al., 2002; Chu et al., 2009; Chu et al., 2010; Kato et al., 2006; Lee et al., 2010; Liu and Herbison, 2008; Spergel, 2007; Zhang et al., 2007; Zhang et al., 2009). Additional channels important for GnRH neuronal firing include canonical transient receptor potential (TRPC) channels, which are activated by kisspeptin, and high voltage activated (HVA) calcium channels, which are important for calcium homeostasis and peptide release (Sun et al., 2010; Zhang et al., 2008).
While previous studies have demonstrated that E2 regulates the expression and/or function of a number of channels in GnRH neurons including T-type and L-type calcium channels (Sun et al., 2010; Zhang et al., 2009), little is known about the channel subtype expression and the E2 and diurnal regulation of the majority of ion channels in GnRH neurons. To begin to understand the E2-induced changes in GnRH neurons, we have explored the mRNA expression of HCN, TRPC, SK, and HVA calcium channels in the morning (negative feedback) and the expression in the evening (positive feedback) in oil- and E2-treated females. Indeed, we have found an E2-induced increased mRNA expression of HCN1, TRPC4, CaV1.3 (L), CaV2.2 (N) and CaV2.3 (R)-type calcium channels in GnRH neurons. In contrast, SK3 mRNA was decreased in GnRH neurons, whereas GPR54 mRNA was not altered at any time-points. These findings indicate that the rising E2-levels exert specific fundamental changes in ion channels expression in GnRH neurons, to prime these neurons for altered responsiveness to incoming stimuli leading to changes in excitability in an E2-dependent manner.
2. Materials and Methods
2.1 Animals
Adult female CBB6 mice (GnRH-GFP) (Suter et al., 2000) were maintained under constant temperature and lights. Two different lighting cycles were used, where lights were on between 0600 hours (zeitgeber time (ZT) 0) and 1800 hours (ZT 12) or where lights were on between 0200 hours (ZT 0) and 1400 hours (ZT 12) local time. The breeders and most of the research animals were kept permanently under reversed lighting schedule, and these animals were used for the majority of evening experiments with some exceptions as noted below. At time of weaning additional research animals were moved to an adjacent room with a regular lighting schedule (0600h–1800h) and were kept there until adulthood (at least 60 days of age). These animals were used for all of the morning experiments and selected evening experiments. We found no evidence for differences between the two lighting schedules. Food and water were provided ad libitum. The females were exposed to male bedding to establish normal estrous cycle prior to bilateral ovariectomy (OVX) and increased response to E2 afterwards (Bronson and Whitten, 1968; Dalal et al., 2001). The animals were OVXed under isofluorane inhalant anesthesia 5–7 days before experimentation, and were given a dose of 4 mg/kg carprofen (Rimadyl, Pfizer Animal Health, New York) immediately following surgery for analgesia. All animal procedures were according to NIH standards and were approved by the Institutional (Oregon Health and Science University) Animal Care and Use Committee.
2.2 Experimental Design
2.2a. Induction of the LH surge
For induction of positive feedback regulation of LH by ovarian steroids in mice, different models have been developed. One model, based on studies by Bronson and colleagues, used E2 implants at the time of OVX combined with a surge-inducing E2 injection 6 days later (Bronson and Vom Saal, 1979). Another similar model (Gee et al., 1984) used an E2 priming implant and an E2 surge implant of differing doses. Both of these models are critically dependent on the “appropriate” concentrations of E2, such that too little or too high levels of E2 will reduce or prevent the LH surge (Bronson and Vom Saal, 1979). We found that, with such sensitivity, we had too many inconsistencies inducing an LH surge in mice. Therefore, we have developed a two-step E2-injection procedure utilizing a priming E2 dose followed by a surge-inducing E2 dose. On day 5 following OVX, the animals were given a subcutaneous injection of a priming dose of 17β-estradiol benzoate (0.25 µg in 50 µl oil) or oil vehicle (50 µl) at ZT 4–5. On day 6, the animals were given a surge dose of 17β-estradiol benzoate (1.0–1.5 µg in 50 µl oil) or oil-vehicle (50 µl) at ZT 4–5. The animals were used for experimentation the following day. The LH surge was induced under both lighting conditions with similar results. When intact animals were used, estrous stage was confirmed by vaginal smears.
2.2b. mRNA quantification in GnRH neuronal pools
In initial experiments we tested the linearity of mRNA expression in single cells compared to pools of GnRH neurons. Since the mRNA expressions of GnRH and GPR54 are quite high in GnRH neurons and these RNAs can be quantified even in single cells, we compared the expression in single cells versus pools of 2, 4 and 8 cells in two intact animals killed during the second day of diestrus. The expression of β-actin was used as control as described below. In addition, we also tested the linearity of one of the high-expressing ion channels, SK3. Since this transcript could only be quantified in pools consisting of at least 5-cells, we compared the expression in pools of 5 and 10 GnRH neurons. For this experiment we used OVX animals, since the SK current (medium IAHP) is reduced acutely with E2 application in GnRH neurons (Chu et al., 2009) and E2-treatment in vivo leads to inhibition of the mIAHP in POA GABAergic neurons (Wagner et al., 2001).
2.2c. Distribution of mRNAs in single GnRH neurons and quantitative mRNA measurements in GnRH neuronal pools
These studies were aimed at investigating the mRNA expression in GnRH neurons of a number of ion channels of which there exist several subunits. In order to determine which subtype is expressed in GnRH neurons, individual GnRH neurons were acutely dispersed, harvested and subjected to single cell RT-PCR using primers selective for each channel subtype (see Table 1). In addition, pools of 5 or 10 neurons were harvested and subjected to real-time qPCR in order to determine the quantitative expression of channel subtypes in GnRH neurons.
Table 1.
Primer sequences for single-cell RT-PCR and qPCR.
| Name | Accession Number |
Number (bp) |
Product Length (bp) |
Slope | Efficiency (%) |
(r2) |
|---|---|---|---|---|---|---|
| HCN1 | NM_010408 | 1527-1546 | 136 | −3.098 | 100 | 0.965 |
| 1662-1641 | ||||||
| HCN2 | NM_008226 | 1122-1143 | 97 | −3.247 | 100 | 0.967 |
| 1199-1218 | ||||||
| HCN3 | NM_008227 | 1664-1682 | 118 | −3.210 | 100 | 0.940 |
| 1763-1781 | ||||||
| HCN4 | NM_001081192 | 1929-1950 | 123 | −3.168 | 100 | 0.940 |
| 2030-2051 | ||||||
| TRPC1 | NM_011643 | 1387-1503 | 117 | −3.255 | 100 | 0.930 |
| 1503-1483 | ||||||
| TRPC4 | NM_016984 | 1841-1860 | 116 | −3.032 | 100 | 0.940 |
| 1956-1937 | ||||||
| TRPC5 | NM_009428 | 734-753 | 118 | −3.16 | 100 | 0.952 |
| 832-851 | ||||||
| SK1 | NM_032397 | 1035-1056 | 181 | |||
| 1194-1215 | ||||||
| SK2 | NM_080465 | 1310-1327 | 124 | −3.159 | 100 | 0.940 |
| 1414-1433 | ||||||
| SK3 | NM_080466 | 2182-2203 | 111 | −3.460 | 95 | 0.976 |
| 2292-2273 | ||||||
| Cav 1.2 | NM_009781 | 1642-1659 | 126 | −3.274 | 100 | 0.975 |
| 1750-1767 | ||||||
| Cav 1.3 | NM_028981 | 775-796 | 116 | −3.147 | 100 | 0.980 |
| 890-869 | ||||||
| Cav 2.1 | NM_007578 | 6035-6054 | 75 | −3.345 | 99 | 0.940 |
| 6109-6090 | ||||||
| Cav 2.2 * | NM_001042528 | 5134-5154 | 169 | |||
| 5284-5302 | ||||||
| Cav 2.2# | NM_001042528 | 5281-5302 | 84 | −3.319 | 100 | 0.987 |
| 5343-5364 | ||||||
| Cav 2.3 | NM_009782 | 530-459 | 107 | −3.341 | 99 | 0.990 |
| 636-617 | ||||||
| GPR54* | NM_053244 | 1900-1917 | 245 | |||
| 2125-2144 | ||||||
| GPR54# | NM_053244 | 1916-1934 | 73 | −3.237 | 100 | 0.980 |
| 1988-1968 | ||||||
| GnRH* | NM_008145 | 21-40 | 239 | |||
| 259-278 | ||||||
| GnRH # | NM_008145 | 125-142 | 127 | −3.394 | 97 | 0.969 |
| 251-234 | ||||||
| β-actin | NM_007393 | 849-867 | 63 | −3.302 | 100 | 0.990 |
| 911-890 |
The sense primer is listed first with the antisense primer below.
Primers used for single-cell PCR.
Primers used for qPCR.
2.2d. Effects of oil- and E2-treatment on mRNA expression in GnRH neurons
In initial experiments, we tested whether channel mRNA expression in GnRH neurons was different between morning and evening in ovariectomized (OVX) oil-treated animals. The analysis of 4 neuronal pools (5 cells each) from 3 animals revealed that there were no differences in mRNA expression between the two time-points (Table 2). Since oil-treatment did not change the mRNA expression irrespective of time of day, E2 treatment was compared to oil-treatment during the morning and in separate experiments also during the evening. Pools of 5 (or 10) GnRH neurons were harvested during the morning in one set of experiments (oil- and E2-treated animals) and during the evening in another set of experiments (oil- and E2-treated animals) and channel mRNA expression quantified using real-time qPCR.
Table 2.
Ion channel mRNA expression in GnRH neurons from oil-treated animals.
| Channel | (n) | Morning (ZT 4) | Evening (ZT 11) | Significance |
|---|---|---|---|---|
| CaV 1.3 (L-type) | 3 | 1.098 ± 0.043 | 1.244 ± 0.171 | p = 0.454 |
| CaV 2.3 (R-type) | 3 | 1.215 ± 0.175 | 1.243 ± 0.383 | p= 0.951 |
| TRPC4 | 3 | 1.122 ± 0.116 | 0.898 ± 0.147 | p= 0.298 |
| SK3 | 3 | 1.157 ± 0.206 | 1.382 ± 0.097 | p= 0.378 |
| HCN1 | 3 | 1.093 ± 0.102 | 1.599 ± 0.275 | p= 0.160 |
Four pools of 5 cells were harvested from each of 3 animals at each time point. There were no differences in channel expression comparing evening versus morning samples.
2.3 Tissue preparation
On the day of experimentation, each animal was given an intraperitoneal dose of 15 mg ketamine for sedation purposes. The animal was then rapidly decapitated, trunk blood collected, the brain removed from the skull, and the brain stem removed. The resulting block was mounted on a cutting platform that was then secured in a vibratome well filled with ice-cold, oxygenated (95% O2, 5% CO2) high sucrose artificial cerebral spinal fluid (aCSF) (in mM: 208 sucrose, 2 KCl, 1 MgCl, 1.25 NaH2PO4, 10 HEPES, 26 NaHCO3, 10 dextrose, 2 MgSO4, and 1 CaCl2). Three to four coronal slices (240 µm) were cut through the diagonal band-preoptic area (DB-POA). The slices were transferred to a multiwell auxiliary chamber containing oxygenated aCSF (in mM: 124 NaCl, 5 KCl, 1.44 NaH2PO4, 5 HEPES, 10 Dextrose, 25.99 NaHCO3, 2 CaCl2) and kept there until recovery. In all cases, the uterine weights were recorded as a measure of serum E2 levels.
2.4 GnRH neuronal harvesting and reverse transcription
Each slice was visualized under a Leitz inverted microscope to confirm the exact area of fluorescence GnRH cells and then the medial DB-POA was microdissected under a dissecting microscope. The tissue was incubated in protease (1 mg/ml aCSF) (protease from streptomyces griseus, Sigma) for 15–17 minutes at 37°C then washed four times in low Ca2+ aCSF (1 mM Ca2+) and two times in aCSF. Gentle trituration with flamed Pasteur pipettes of decreasing size were used to disperse the neurons on to a glass bottom dish (60 mm dish). We designed a glass-bottomed 60 mm dish using a 22 × 50 mm glass coverslip to increase the surface area of the plate, to allow greater segregation among cells. The cells settle on the glass bottom of the dish after approximately 12 minutes. For the 15 minutes prior to and during harvesting, a constant flow (2 ml/minute) of oxygenated aCSF circulated into the plate while the effluent circulated out using a peristaltic pump. aCSF flow helped insure fresh, oxygenated media was reaching the cells, and assisted in clearing out unhealthy cells and debris from tituration. The cells harvested were those observed to be fully intact, with long processes extending from at least one axis of the cell. Some cells exhibited tiny hair-like projections from the membrane. The cell membrane was smooth, and the cell body shape was fusiform bipolar or unipolar.
The dispersed cells were visualized, patched with a standard glass pipette (1.5 mm OD/0.84 mm ID, World Precision Instruments, Sarasota, FL) that had been pulled to a 10 µm tip and then harvested with gentle suction using the XenoWorks microinjector system (Sutter Instruments, Navato, CA). This microinjector system allows harvesting of cells with negligible uptake of perfusion fluid into the pipette. The contents of the pipette were expelled into a siliconized 0.5 ml tube containing a solution of 1X Invitrogen Superscript III Buffer, 15U of RNasin (Promega), 10 mM of dithiothreitol (DTT) and diethylpyrocarbonate (DEPC)-treated water in a total of 5 µl for single cells and 8 µl for pooled cells. Cells were harvested as single cells, or as pools of 5 or 10 individual cells. In each case, the tube containing the harvested cells was kept on a chilled (~1°C) metal block during the harvesting procedure (< 10 min for pools of 5 cells and <20 min for pools of 10 cells) and then immediately frozen on dry ice. Each harvested cell and pool of cells were reverse transcribed by first adding dNTPs (0.5 mM, Promega) random primers (100 ng/per tube, Promega) and anchored oligo(dT)20 primer (400 ng/tube, Invitrogen) to each tube, incubated in a thermocycler at 65°C for 5 minutes, and then cooled on ice for 5 minutes. Next, Superscript III reverse-transcriptase (100 U/per tube, Invitrogen), RNAsin (15 U), DTT (6 mM) and DEPC-treated water were added to a final 20 µl volume (25 µl for pooled cells). The reverse transcription (RT) reaction was as follows: 25°C for 5 minutes, 50°C for 60 minutes, 70°C for 15 minutes and 4°C for 5 minutes. Samples were stored at −20°C. In addition, harvested aCSF in the vicinity of the dispersed cells also underwent RT and was used as a control. Cells and tissue RNA used as negative controls were processed as described above but without reverse transcriptase (−RT). Each cell, or pool of cells, was then evaluated using PCR or real-time PCR, respectively.
2.5 Primer design and development
Primer pairs for single cell PCR (scPCR) and quantitative real-time PCR (qPCR) were developed using the mRNA sequence of the respective genes from the National Center for Biotechnology Information (see Table 1). Primers were designed using the Clone Manager software (Sci Ed Software, Cary, NC), and each primer pair crossed intron/exon boundaries to distinguish cDNA amplification products from genomic DNA amplification. In order to measure ion channels and receptor mRNA expression in GnRH neurons using PCR, we designed the most efficient primers possible that were further optimized by adjusting the reaction temperature and magnesium concentrations. For real-time PCR analysis, the amplification efficiency and melting curve were determined for each pair (Table 1). Only primers that exhibited 95–100% efficiency and a single product melting curve were used for the real-time PCR quantification. Most primers could be used for both single cell and real-time PCR analyses, whereas for others (CaV2.2, GPR54 and GnRH) we designed different primer pairs for the two different determinations (Table 1).
2.6 Single cell PCR
Single cell PCR (sc-PCR) was performed using 2 to 3 µl of cDNA template from each RT reaction in a 30 µl PCR mix. Forty to fifty cycles of amplification were performed using a Bio-Rad C1000 Thermal Cycler (Bio-Rad, Hercules, CA) according to established protocols (Qiu et al., 2011; Zhang et al., 2009). PCR products were visualized with ethidium bromide on a 2% agarose gel. The PCR product in single cells for each primer pair was sequenced and confirmed to be identical to each respective mRNA sequence.
2.7 Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was performed on an Applied Biosystems (ABI) 7500 Fast Real-time PCR System using the Power SybrGreen mastermix method according to established protocols (Zhang et al., 2009). In all cases, real-time PCR assays were tested to determine and compare the efficiencies of the target and control gene amplifications. Serially diluted cDNAs (1:50–1:12,800 for most primers; 1:1–1:12,800 for GnRH) from mouse POA, assayed in triplicate, were used to construct standard curves and the efficiency was calculated as follows: E = 10(−1/m) − 1, where m is slope (Livak and Schmittgen, 2001; Pfaffl, 2001). Amplification efficiencies are listed in Table 1. The ABI sequence detection software system (version 1.3) was used to generate the standard curves. The comparative ΔΔCT method was used to calculate the individual values for each sample as described previously (Zhang et al., 2009). Briefly, cDNA samples from GnRH pools were run in duplicates and the mean ΔCT of the oil-treated pool values was used as a calibrator when comparing the mRNA quantities to the E2-treated samples. The relative linear quantity of the target gene was calculated using the formula 2−ΔΔCT (Livak and Schmittgen, 2001). Therefore, the data were expressed as an n-fold change in gene expression normalized to a reference gene (β-actin) and relative to the oil-control values. The comparative ΔΔCT method was also used in Experiment 2b to compare mRNA quantities between a single cell, and pools of cells. Here the mean ΔCT of all the samples was used as calibrator and the relative quantity was calculated using the 2−ΔΔCT formula. When comparing mRNA expression levels between different genes, the mean ΔCT of one of the gene values was used as a calibrator to which the other genes were compared. In all instances, the PCR efficiencies were similar between the different primer pairs (see table 1). The data are reported as relative mRNA expression. Data were expressed as mean ± SEM and differences in gene expression were analyzed by two-tailed Student’s t-test or one-way ANOVA followed by Newman-Keuls multiple comparison post hoc test.
2.8 Radioimmunoassay for luteinizing hormone
Mouse LH radioimmunoassay (RIA) was performed by the Endocrine Technology and Support Lab, Oregon National Primate Research Center (Oregon Health and Science University, Beaverton, OR) using a traditional double-antibody RIA procedure similar to that described previously (Pau et al., 1986). The mouse LH RIA kit was purchased from Dr. Parlow (NHPP, Harbor-UCLA Medical Center, Los Angeles) that includes rat LH (NIDDK-rLH-I-8, a.k.a. AFP-12066B; 100 mg) for iodination, mouse LH (AFP-5306A; 2.5 mg) for standards and rabbit anti-rat LH serum (NIDDK-anti-rLH-S-10) for use at a final dilution of 1:750,000. The detection limit of the assay was 0.2 ng/ml. A mouse serum pool (ET-mouse # 4) was used in triplicate in each assay as quality control. The inter-assay variation was 14.7% and the intra-assay variation was 3.8%.
3. Results
3.1. E2-induced LH release
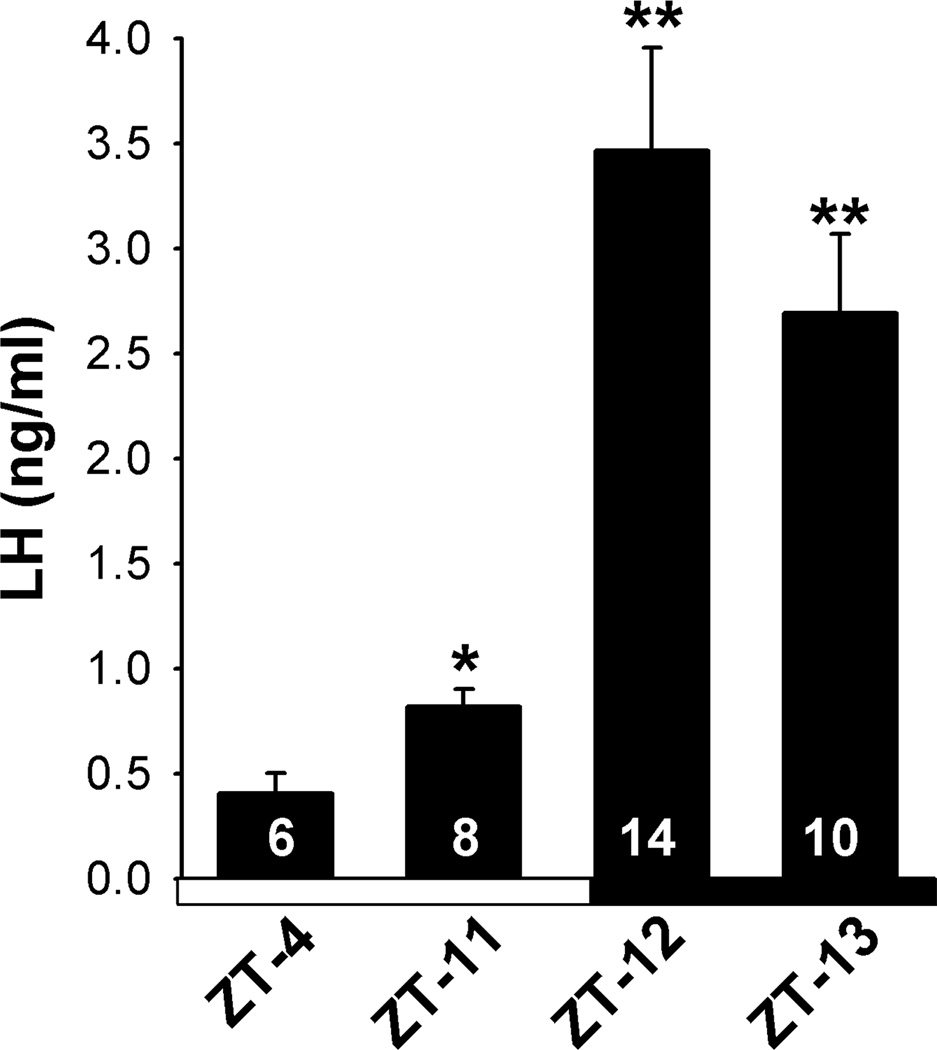
OVX animals were injected with a low priming dose followed by a LH surge-inducing dose of E2. With this procedure, we consistently induced a LH surge in CBB6 GnRH mice (Fig. 1). The timing of the surge was similar to that reported previously in mice kept under 12h light and 12h dark conditions (Gee et al., 1984; Wintermantel et al., 2006). All animals had low serum LH levels in the morning at ZT 4–5 (n=6) following E2-treatment. The mean serum LH levels were slightly, but significantly elevated in animals euthanized at ZT11 (p<0.05) (n=8) and highly elevated in animals euthanized at ZT12 (p<0.001; n=14) or ZT 13 (p<0.01; n=10) (Fig.1). LH levels were also measured in OVX oil-treated animals and as expected found to be elevated during the morning (ZT 4–5) and evening (ZT 11–13) (Data not shown). The uterine weights (fluid expelled and fat trimmed) in OVX E2-treated mice were 104.5 ± 4.1 mg at ZT 4–5 (n=6) and 112.7 ± 5.3 mg (n=8) at ZT 11, 108.0 ± 3.1 mg (n=14) at ZT 12, 111.6 ± 4.5 mg (n=10) at ZT 13. As expected, OVX oil-treated mice had significantly lower uterine weights of 22.8 ± 1.2 mg (n=10) and 22.1 ± 0.9 mg (n=10) at ZT 4–5 and ZT 11–13, respectively.
Figure 1.
Mean serum levels of LH in OVX, E2-treated mice housed under 12 h light, 12 h dark lighting condition and killed at one of four time points, ZT4, ZT11, ZT12, ZT13. Lights were turned on at ZT0 and turned off at ZT12. Serum LH levels at ZT11, 12 and 13 were elevated compared to LH levels at ZT4; *, p<0.05, **, p<0.01, Student’s t-test. The number of animals at each time point is indicated.
3.2 Linear regression analysis of mRNA expression in GnRH neurons
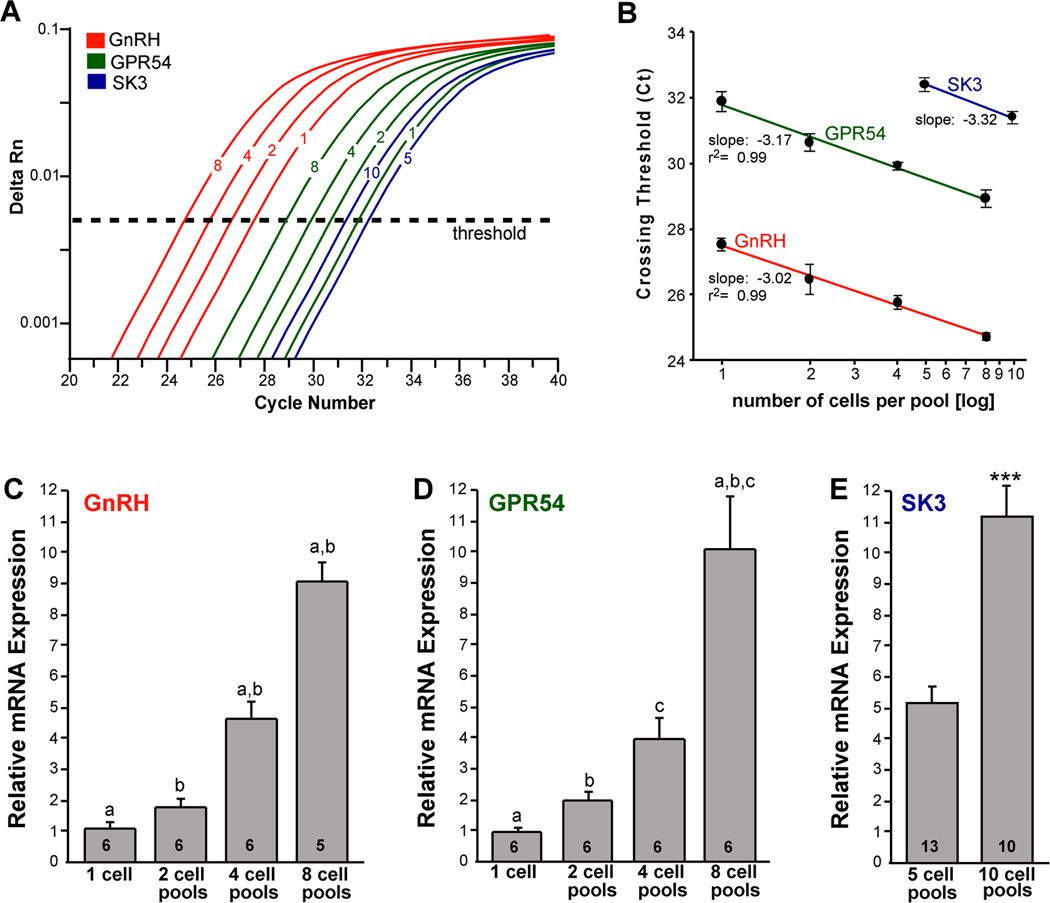
In initial experiments, we analyzed the quantitative mRNA expression of highly expressed genes in individual neurons (n = 6 cells from 2 animals) and pools of 2 neurons (n=6 pools from 2 animals), 4 neurons (n= 6 pools from 2 animals) and 8 GnRH neurons (n=5–6 pools from 2 animals). For analysis of GnRH, the cell content was diluted 1:10 to accommodate for the high expression of GnRH mRNA. Linear regression analysis revealed that increasing the number of cells resulted in a linear decrease in the CT values (r2 = 0.99 for both GnRH and GPR54; Fig. 2A, B), which resulted in a near linear increase in mRNA expression (Fig. 2C, D). We also measured the mRNA expression of small conductance calcium-activated potassium (SK3) channels in pools of 5 GnRH neurons (13 pools from 5 animals) and 10 GnRH neurons (10 pools from 5 animals). (SK3 could not be quantified by real-time PCR in single cells or in pools of 2–4 individual GnRH neurons). The analysis revealed a slope of −3.32 (Fig. 2B), and a doubling of SK3 mRNA levels in pools of 10 as compared to 5 neurons (Fig. 2B, E).
Figure 2.
qPCR amplification for GnRH, GPR54 and SK3 in GnRH neurons. Amplification of cDNA synthesized from differing amounts of RNA in individual GnRH neurons and pools of 2, 4, and 8 cells (GnRH and GPR54) as well as pools of 5 and 10 cells for SK3. cDNA from GnRH single cells and pools was diluted 1:10 for GnRH qPCR measurements. GPR54 and SK3 were quantified in full strength cDNA. A. Cycle number was plotted against normalized fluorescence intensity (delta Rn) to visualize the PCR amplification. Cycle threshold (CT; dashed line) is the point in the amplification at which the sample values were calculated. B. Linear regression analysis plotting crossing threshold (CT) versus the log scale of number of cells collected in each pool (mean ± SEM; n = number of individual cells or pools of cells, which were 5–6 for GnRH and GPR54, and 10–13 for SK3. GnRH, GPR54 and SK3 produced slope values of −3.02, −3.17, and −3.23 respectively, which corresponds to a doubling between each cycle with high linearity (Pearson’s coefficient r2= 0.99). β-actin showed a similar linear relationship between individual cells and cell pools (slope −3.00, r2 = 0.98; data not shown). The amplification efficiency for each primer pair is listed in Table 1. These efficiencies allowed us to use the ΔΔCT method for quantification. C. Bar graphs illustrating the quantitative analysis of relative mRNA expression of GnRH in individual GnRH neurons and in pools of 2, 4 and 8 cells (one-way ANOVA; a-a, b-b, p<0.001; n=6 individual cells and 5–6 pools). D. Bar graphs illustrating the quantitative analysis of relative mRNA expression of GPR54 in individual GnRH neurons and in pools of 2, 4 and 8 cells (one-way ANOVA; a-a, b-b, c-c, p<0.001; n = 6 individual cells and 6 pools). E. Bar graphs illustrating the quantitative analysis of relative mRNA expression of SK3 in GnRH neuronal pools containing 5 and 10 cells (Student’s t-test; ***, p<0.001, n = 10–13 pools).
3.3 mRNA expression in GnRH neurons
These studies were designed to explore whether ion channels (hyperpolarization-activated cyclic nucleotide-gated (HCN), high voltage-activated (HVA) calcium, SK and canonical transient receptor potential (TRPC) channels) important for burst firing activity (Kelly and Rønnekleiv, 1994; Lee et al., 2010) are expressed in mouse GnRH neurons, and which subtypes are the most prevalent. We also explored the expression of GPR54, which is required for activation of GnRH neurons by kisspeptin at the time of the LH surge (Clarkson et al., 2008). For these experiments we used intact female mice, killed at ZT 4–5 during the proestrus/estrus phase of their cycle with a mean uterine weight of 97.6 ± 8.2 mg (n=6), confirming that E2 levels were high. For a more quantitative analysis of mRNA expression in GnRH neurons, we used real-time quantitative PCR (qPCR) in pools of GnRH neurons.
HCN channel subtypes
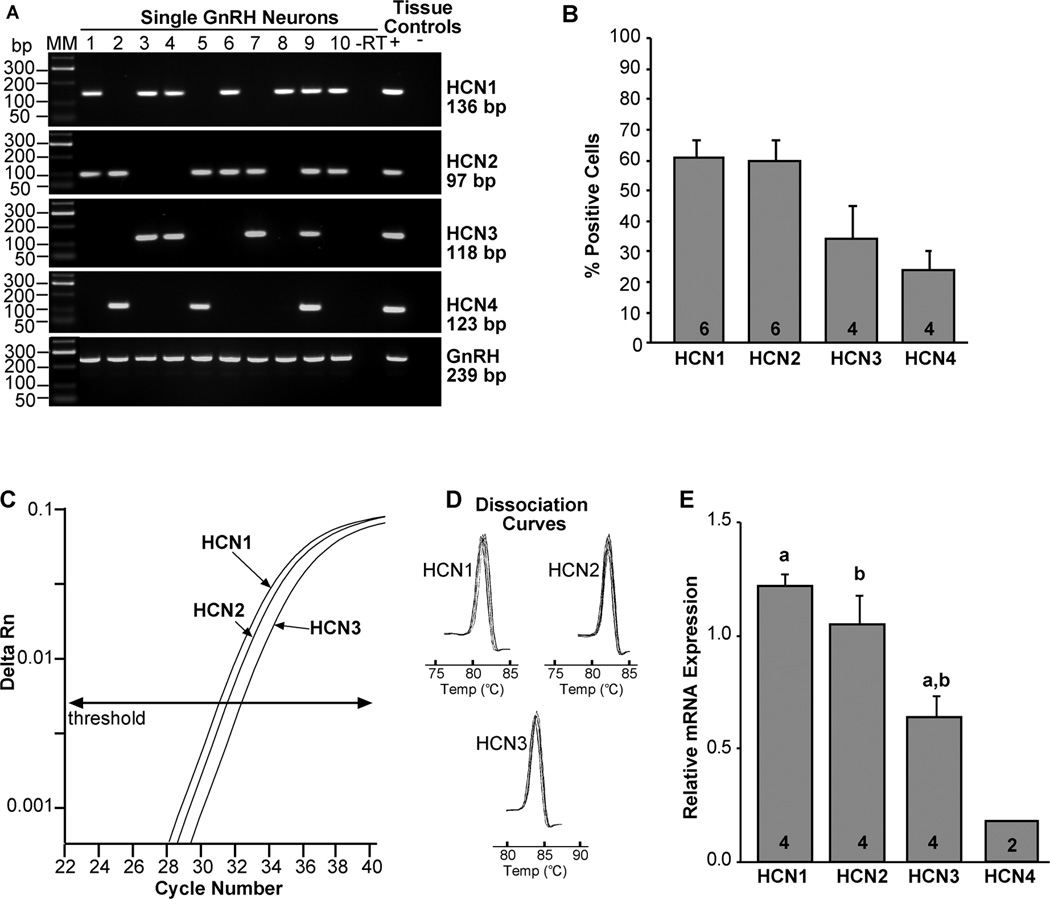
It has been reported previously in immature mouse GnRH neurons that HCN3 is the primary transcript, HCN4 was barely detectable, and HCN1 and HCN2 transcripts were not expressed (Constantin and Wray, 2008). Our analysis of 48–72 cells from four animals revealed that HCN1 and HCN2 transcripts were the most prevalent and were about equally expressed in adult mouse GnRH neurons, 61 ± 6% and 60 ± 7%, respectively (Fig 2A, B). Moreover, either HCN1 and/or HCN2 subtypes were expressed in at least 90% of the cells. HCN3 mRNA was detected in 34 ± 11% of GnRH neurons and HCN4 in 24 ± 6% (Fig. 3A, B).
Figure 3.
Expression of HCN channels in GnRH neurons from intact proestrus/estrus EGFP-GnRH mice. A, Representative gels illustrating the mRNA expression of HCN channel subtypes 1–4. The expected sizes for the PCR products are as follows: for HCN1, 136 bp; for HCN2, 97 bp; for HCN3, 118 bp; for HCN4, 123 bp; and for GnRH, 239 bp. As a negative control, a cell reacted without reverse transcriptase (−RT) did not express any of the transcripts. POA tissue RNA was also included as positive control (+, with RT) and negative control (−, without RT). MM, molecular markers. B, Summary bar graphs of the percentage expression of HCN1, HCN2, HCN3, and HCN4. 12 cells/animal from 4–6 animals were analyzed by sc-PCR, and the mean number of neurons expressing HCN channel subtypes from each animal was determined. Bar graphs represent the mean ± SEM of the percentage of GnRH neurons expressing each HCN subtype per animal. C, Quantitative real-time PCR assay with amplification curves for HCN1, HCN2 and HCN3 subunits. Cycle number was plotted against the normalized fluorescence intensity (delta Rn) to visualize the PCR amplification. The cycle threshold (CT, arrow) is the point in the amplification at which the sample values were calculated. The amplification efficiencies for each primer pair is listed in Table 1. These efficiencies allowed us to use the ΔΔCT method for quantification. D, The melting curves depict single-product melting at 82, 83 and 84°C for HCN1, HCN2 and HCN3, respectively, illustrating that only one product was formed for each transcript in GnRH 5-cell pools. E, Bar-graphs illustrating the relative mRNA expression of HCN1, HCN2, HCN3 and HCN4. a-a, p<0.01, b-b, p<0.05; Student’s t-test. HCN4 was measured in 10-cell pools (n=2) and was often below the level of detectability and was, therefore, not included in the statistical analysis. The number of animals is indicated.
For a more quantitative analysis of HCN channel expression in GnRH neurons, we used real-time quantitative PCR (qPCR) in pools of 5 or 10 cells. HCN1, HCN2 and HCN3 mRNAs were measured in 5-cell pools (3 pools from each of 4 animals), whereas HCN4 mRNA was not detected in 5-cell pools and borderline or below detectability in 10 cell pools (2 pools from each of 2 animals) by real-time PCR in GnRH neurons (Fig. 2C–E). The quantification revealed that HCN1 ≥ HCN2 > HCN3 >> HCN4 (Fig. 3E; one-way-ANOVA; a-a, p<0.005, b-b, p<0.05; n = 4).
HVA calcium channel subtypes
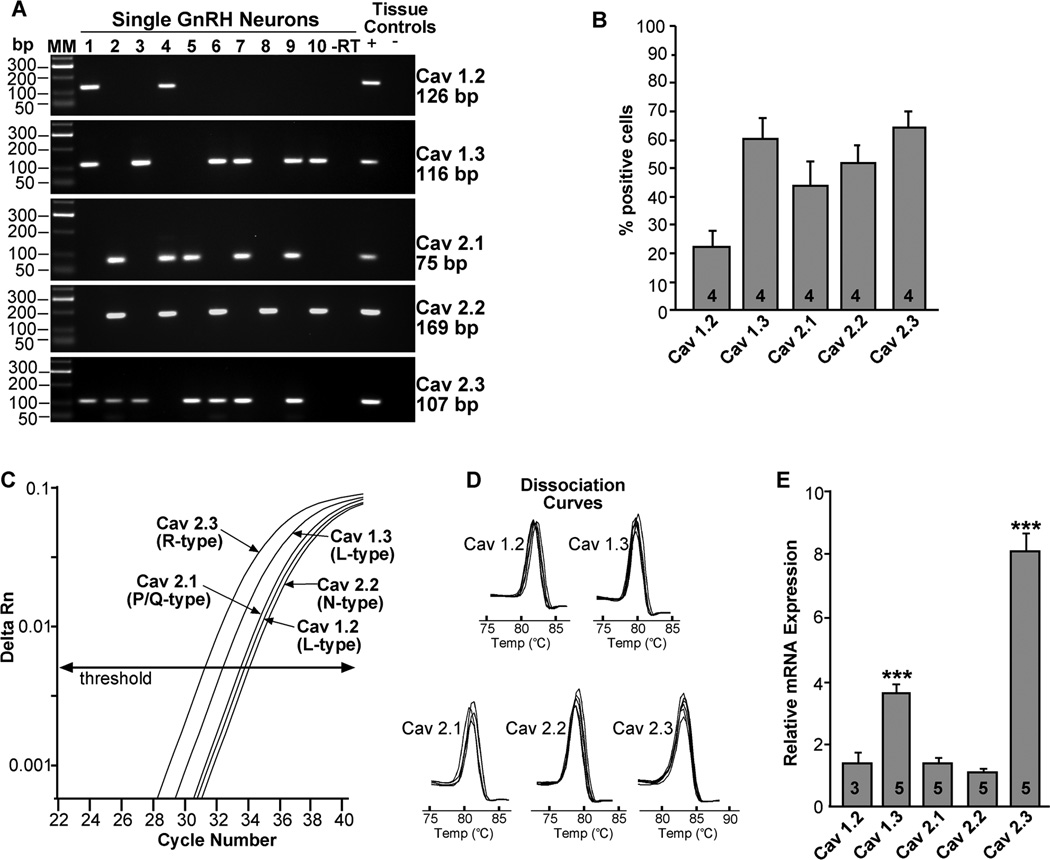
There are four L-type channel isoforms (CaV1.1, 1.2, 1.3, 1.4), but only CaV1.2 and CaV1.3 have been reported in rat GnRH neurons (Tanaka et al., 2010). We confirmed that CaV1.2 and CaV1.3, but not CaV1.4, were expressed in mouse GnRH neurons. Since CaV1.1 is expressed primarily in muscle (Catterall et al., 2003), we did not investigate this L-type subunit and instead focused our analysis on CaV1.2 and 1.3 L-type calcium channel isoforms. Based on the analysis of 48 cells from 4 animals, CaV1.2 was expressed in 22 ± 6 %, and CaV1.3 was expressed in 60 ± 7% of GnRH neurons. P/Q-type channels, CaV2.1, and N-type channels, CaV2.2, were expressed in 44 ± 9% and 52 ± 6% of GnRH neurons, respectively, while the R-type channel, CaV2.3, was detected in 65 ± 5% of GnRH neurons (Fig. 4A, B).
Figure 4.
Expression of HVA channels in GnRH neurons from intact proestrus/estrus EGFP-GnRH mice. A, Representative gels illustrating the mRNA expression of HVA channel subtypes. The expected sizes for the PCR products are as follows: for CaV1.2 (L), 126 bp; CaV1.3 (L), 116 bp; CaV2.1 (P/Q), 75 bp; CaV2.2 (N), 169 bp; CaV2.3 (R), 107 bp. As a negative control, a cell reacted without reverse transcriptase (−RT) did not express any of the transcripts. POA tissue RNA was also included as positive control (+, with RT) and negative control (−, without RT). MM, molecular markers. B, Summary bar graphs of the percentage expression of HVA channel subtypes in GnRH neurons. 12 cells/animal from 4 animals were analyzed by sc-PCR, and the mean number of neurons expressing HVA channel subtypes from each animal was determined. Bar graphs represent the mean ± SEM of the percentage of GnRH neurons expressing each HVA subtype per animal. C, Quantitative real-time PCR assay with amplification curves for CaV2.3, CaV1.3, CaV2.1, CaV1.2, and CaV2.2. Cycle number was plotted against the normalized fluorescence intensity (delta Rn) to visualize the PCR amplification. The cycle threshold (CT, arrow) is the point in the amplification at which the sample values were calculated. The amplification efficiencies for each primer pair is listed in Table 1. D, The melting curves depict single-product melting at 82, 80, 81, 80 and 83°C for CaV1.2, 1.3, 2.1, 2.2 and 2.3, respectively, illustrating that only one product was formed for each transcript in GnRH 5- or 10-cell pools. E, Bar-graphs illustrating the relative mRNA expression of the various CaV channel subtypes. R-type (CaV2.3) mRNA expression was significantly higher than that of the other channel subtypes (***, p<0.001; one-way ANOVA). L-type (CaV1.3) mRNA expression was significantly higher than CaV1.2, 2.1 and 2.2 mRNA levels (**, p<0.01; one way ANOVA). The number of animals is indicated.
For quantification of HVA channel subtypes expression in GnRH neurons, the CaV1.3 (L), CaV2.1(P/Q), and CaV2.3 (R) subtypes were quantified by qPCR in 5-cell pools (3 pools from each of 5 animals) and CaV1.2 and CaV2.2 in 10-cell pools (2 pools from each of 3–5 animals). The relative mRNA expression of CaV2.3 was significantly higher than the expression of the other HVA channel subtypes (Fig. 4C–E; p<0.001). In addition, CaV1.3 mRNA expression was significantly higher than the expression of CaV1.2, 2.1 and 2.2 subtypes (Fig. 4C–E; p<0.01). There were no significant differences between CaV1.2, 2.1 and 2.2 mRNA expressions.
TRPC channel subtypes
Using sc-PCR, we had determined previously that most TRPC channel subtypes are expressed in GnRH neurons (Zhang et al., 2008). However, based on the electrophysiological and pharmacological analysis, we had evidence that the TRPC1,4,5 channel subfamily was the primary effector of kisspeptin’s actions (Zhang et al., 2008). Therefore, we focused our quantitative analysis on TRPC1, TRPC4 and TRPC5 channel subtypes in GnRH neurons. The qPCR analysis revealed that TRPC4 was the most highly expressed channel subtype in GnRH neurons compared to TRPC1 and TRPC5 mRNAs (Fig. 5: p<0.001, n=3–5). The mRNA expression of TRPC5 (10-cell pools) was quite low in GnRH neurons and was only detected in 3 of 5 animals.
Figure 5.
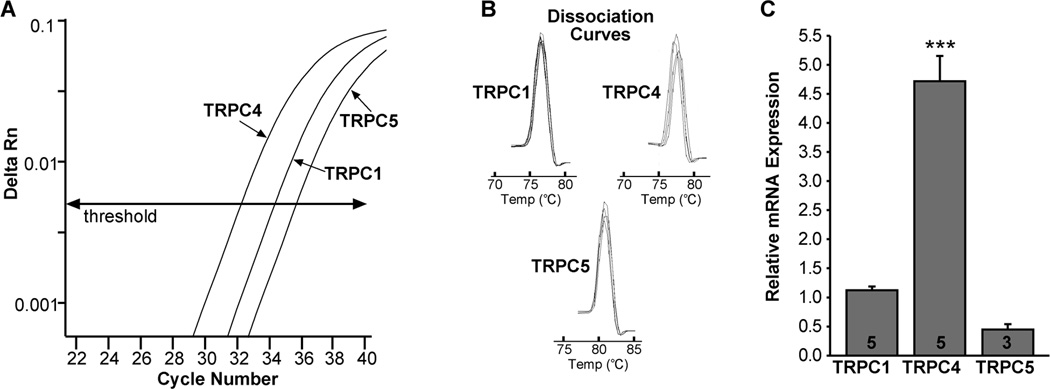
TRPC channel subtype distribution by real-time PCR. A, qPCR assay with amplification curves for TRPC1, TRPC4 and TRPC5 subunits. (TRPC1 and TRPC4 were analyzed in 5-cell pools and TRPC5 in 10-cell pools). Cycle number was plotted against the normalized fluorescence intensity (delta Rn) to visualize the PCR amplification. The cycle threshold (CT, arrow) is the point in the amplification at which the sample values were calculated. The amplification efficiencies for each primer pair is listed in Table 1. B, Melting curves depict single-product melting at 77, 78, and 81°C for TRPC1, TRPC4 and TRPC5, respectively, illustrating that only one product was formed for each transcript in GnRH neuronal pools. C, Bar-graphs illustrating the relative mRNA expression of TRPC1, TRPC4 and TRPC5 (*** p<0.001 TRPC4 compared to TRPC1 and TRPC5). The number of animals is indicated.
SK channel subtypes
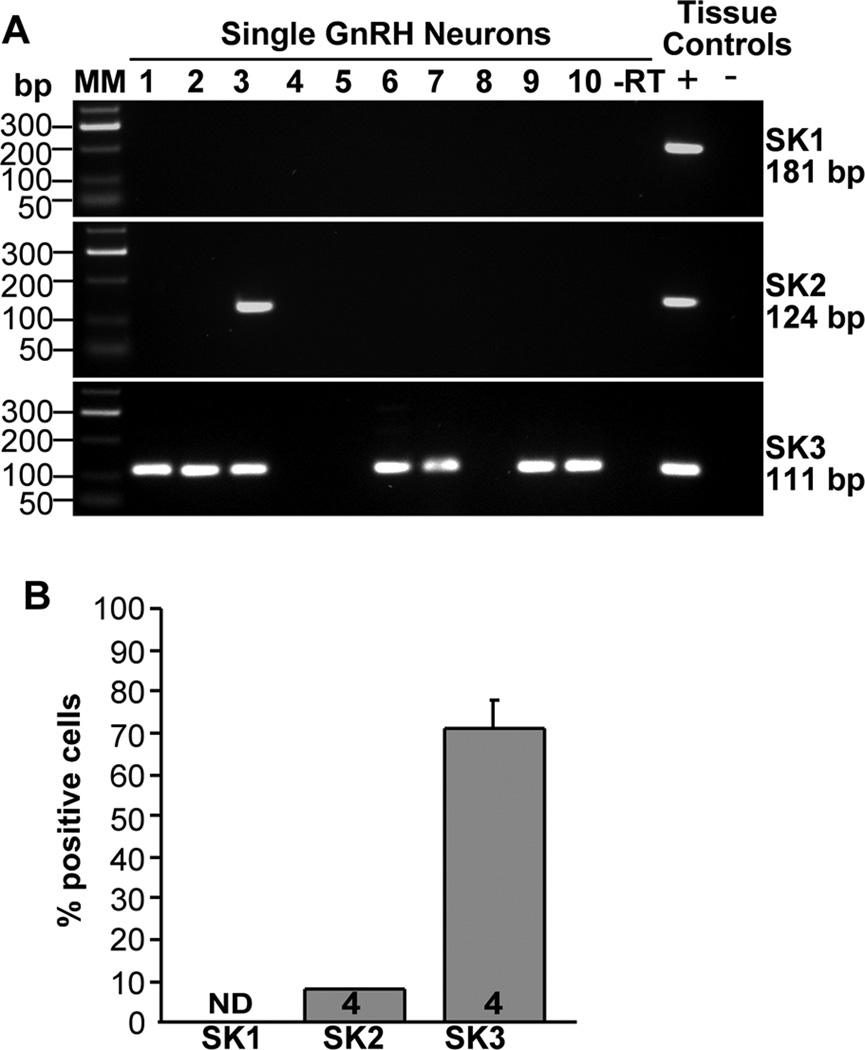
We had previously determined that SK3 is expressed in guinea pig GnRH neurons (Bosch et al., 2002). In addition, Kato and colleagues (2006) reported that SK1, 2 and 3 are all expressed in rat GnRH neurons. However, based on analysis of 48 cells from 4 animals, we determined that SK3 is the major subtype in mouse GnRH neurons and that this SK subtype was found in 71 ± 7% of the cells (Fig. 6A, B). In contrast, SK 2 was found in only one cell in each animal (8%) and SK1 was not expressed in mouse GnRH single neurons.
Figure 6.
SK channel subtype distribution in GnRH neurons. A, Representative gels illustrating the mRNA expression of SK channel subtypes 1–3. The expected sizes for the PCR products are as follows: for SK1, 181 bp; for SK2, 124 bp; for SK3, 111 bp. As a negative control, a cell reacted without reverse transcriptase (−RT) did not express any of the transcripts. POA tissue RNA was also included as positive control (+, with RT) and negative control (−, without RT). MM, molecular markers. B, Summary bar graphs of the percentage expression of SK1, SK2 and SK3 subunits. 12 cells/animal from 4 animals were analyzed by sc-PCR, and the mean number of neurons expressing SK channel subtypes from each animal was determined. Bar graphs represent the mean ± SEM of the percentage of GnRH neurons expressing each SK subtype per animal. Note SK1 was not detected in mouse GnRH neurons.
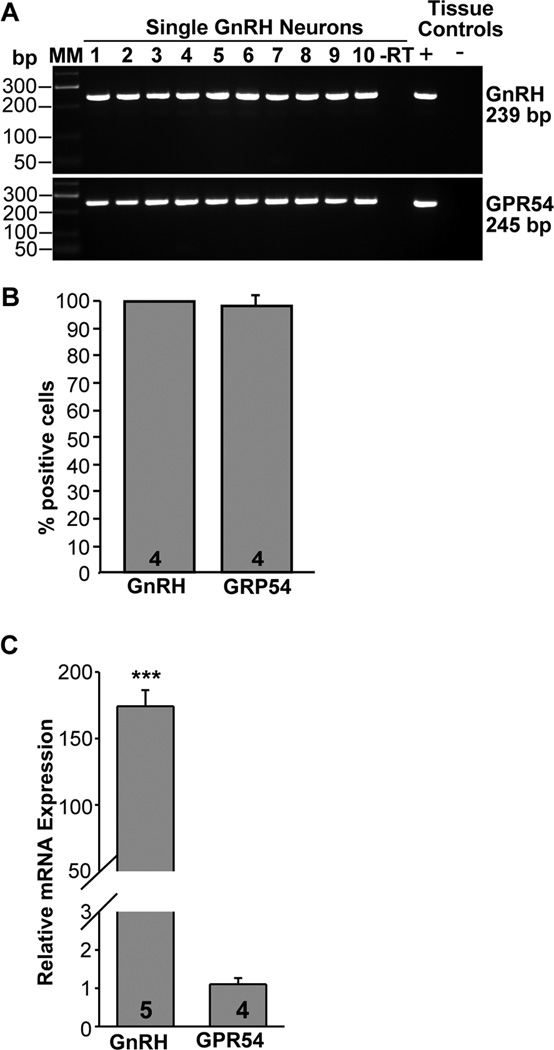
GnRH and kisspeptin receptor, GPR54
Based on in situ hybridization analysis in mice, approximately 90% of GnRH neurons express the GPR54 receptor (Han et al., 2005). Currently, we analyzed 48 GnRH neurons from 4 female mice using sc-PCR. This analysis revealed that GPR54 mRNA was expressed in 97 % of GnRH neurons; only one cell in one of the animals did not express GPR54 mRNA (Fig. 7A, B). Based on qPCR analysis, GnRH mRNA was highly expressed in GnRH neurons as revealed by the very low CT value (see Fig. 2). GPR54 mRNA was also robustly expressed (see Fig. 2), but still more than 150-fold lower than GnRH mRNA expression (Fig. 7C).
Figure 7.
Expression of GPR54 mRNA in GnRH neurons from intact proestrus/estrus EGFP-GnRH mice. A, Representative gels illustrating the mRNA expression of GPR54 and GnRH in EGFP-GnRH mice. The expected sizes for the PCR products are as follows: for GnRH, 239 bp; for GPR54,245 bp; As a negative control, a cell reacted without reverse transcriptase (−RT) did not express any of the transcripts. POA tissue RNA was also included as positive control (+, with RT) and negative control (−, without RT). MM, molecular markers. B, Summary bar graphs of the percentage expression of GnRH and GPR54 in EGFP-GnRH neurons. 12 cells/animal from 4 animals were analyzed by sc-PCR, and the mean number of neurons expressing GnRH and GPR54 mRNAs from each animal was determined. Bar graphs represent the mean ± SEM of the percentage of GnRH neurons expressing GnRH and GPR54 per animal. Essentially, all EGFP neurons harvested expressed GnRH and GPR54. C, qPCR analysis of of GnRH and GPR54 mRNA expression in GnRH neurons. Bar-graphs illustrating the relative mRNA expression of GnRH and GPR54 (***, p<0.001; Student’s t-test; the number of animals is indicated).
3.3 qPCR analysis of channel mRNA expression in GnRH neurons and modulation by E2 during negative and positive feedback
These experiments were done in OVX oil-treated control and E2-treated females analyzed during the morning (ZT 4 – 5 negative feedback) and/or during the evening (ZT 11–12 positive feedback. We used real-time PCR (qPCR) to quantify the relative expression of ion channel subtypes in GnRH neurons in E2-treated as compared to oil-treated animals using the primers outlined in Table 1. The quantifications were done in pools of 5 neurons except as noted in the text.
HCN channel subtypes
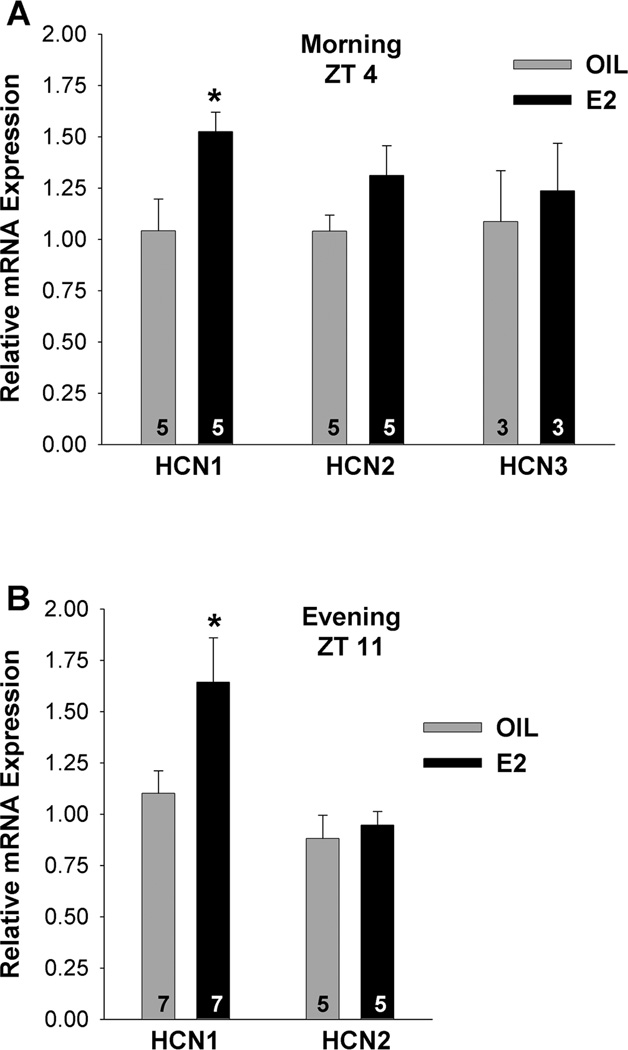
HCN1, HCN2 and HCN3 mRNAs were quantified in cells obtained at ZT4–5, and HCN1 and HCN2 mRNAs were also quantified at ZT11–12 in oil control and E2-treated females. Based on this analysis, HCN1 mRNA expression was significantly increased in GnRH neurons from E2-treated as compared to oil-treated animals at both time points (Fig. 8; p<0.05, n = 5–7). In contrast, there was no difference in HCN2 or HCN3 mRNA expression between oil- and E2-treated females (Fig. 8; n = 3–5). Due to the low mRNA expression of HCN4, we were not able to quantify differences between the two groups of animals, even in 10-cell pools.
Figure 8.
E2 regulates HCN1 channel mRNA expression in GnRH neurons. A,B, Quantitative real-time PCR measurements of HCN1, HCN2 and HCN3 mRNAs in GnRH neuronal pools (3–4 pools per animal) from oil- and E2-treated mice (n=3–7 animals per group) obtained during the morning, ZT4 (A) or during the evening, ZT11 (B). The expression values were calculated via the ΔΔCT method and normalized to the mean ΔCT of the oil-treated samples. Bar graphs represent the mean ± SEM. *, p<0.05, oil versus E2.
HVA calcium channel subtypes
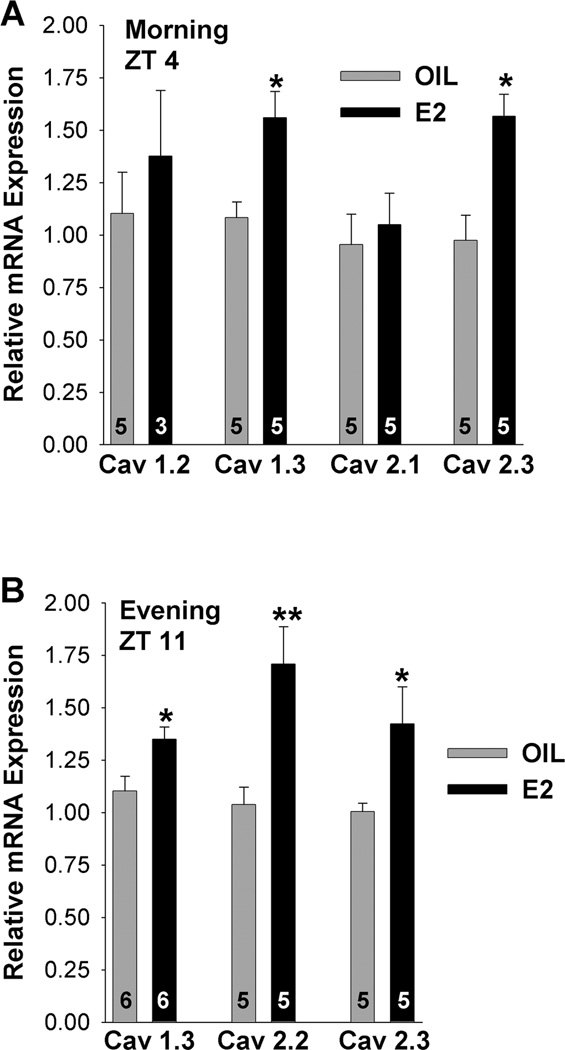
Based on previous electrophysiology findings, HVA L-type and N-type calcium currents are regulated by E2 in GnRH neurons (Sun et al., 2010). We were, therefore, interested in exploring whether the mRNA expression of HVA channel subtypes was also regulated by E2 in GnRH neurons. CaV1.2 (L-type, 10-cell pools), CaV1.3 (another L-type), CaV2.1 (P/Q-type) and CaV2.3 (R-type) mRNAs were all quantified at ZT4–5 in oil- and E2-treated females, but only CaV1.3 and CaV2.3 mRNAs were increased with E2-treatment (Fig. 9A; *p<0.05, n=5). In contrast, CaV1.2 and CaV2.1 mRNA expression was not altered by E2-treatment and were not further analyzed. CaV1.3 and CaV2.3 mRNAs were also quantified at ZT 11–12 and found to be increased in E2-treated as compared to oil-treated females during the evening (Fig. 9B; *p<0.05, n=5–6). CaV2.2 mRNA (N-type; 10-cell pools) was only evaluated during the evening and found to be significantly increased in GnRH neurons from E2-treated as compared to oil-treated animals (Fig. 9B; **p<0.01, n=5).
Figure 9.
E2 regulates HVA channel mRNAs in GnRH neurons. A,B, Quantitative real-time PCR measurements of CaV1.2, CaV1.3, CaV2.1 and CaV2.3 mRNAs during the morning, ZT4 (A) and CaV1.3, CaV2.2 and CaV2.3 mRNAs during the evening, ZT11 (B) in GnRH neuronal pools (3–4 pools of 5 or 10 (CaV1.2, CaV2.2) cells per animal) from oil- and E2-treated mice (n=3–6 animals per group). The expression values were calculated via the ΔΔCT method, normalized to β-actin and relative to the oil control values. Bar graphs represent the mean ± SEM. *, p<0.05, oil versus E2.
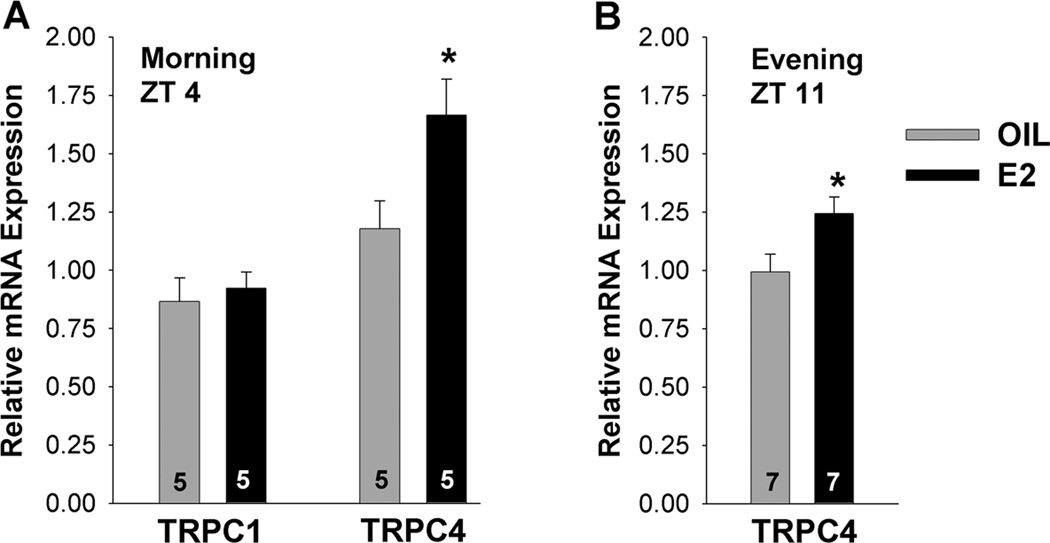
TRPC channel subtypes
qPCR analysis of TRPC 1 and TRPC4 channel subtypes in GnRH neurons at ZT 4–5 in oil- and E2-treated OVX female mice revealed that the E2 treatment increased TRPC4 mRNA levels (Fig. 10A; *p<0.05, n=5). In contrast, TRPC1 mRNA was not altered with E2 treatment and was not further analyzed (Fig. 10A). The mRNA expression of TRPC5 was quite low in GnRH neurons, even in 10-cell pools (see Fig. 4). Therefore, it was not possible to evaluate the effects of E2 on TRPC5 mRNA expression. Similar to the morning data, TRPC4 mRNA expression was increased with E2 treatment relative to oil-treatment at ZT 11–12 (p<0.05; n=7) (Fig. 10B).
Figure 10.
E2 regulates TRPC4 mRNA in GnRH neurons. A, B, Quantitative real-time PCR measurements of TRPC1 and TRPC4 mRNAs during the morning, ZT4 (A) and TRPC4 during the evening, ZT11 (B) in GnRH neuronal pools (3–4 pools of 5 cells per animal) from oil- and E2-treated mice (n=5–7 animals per group). The expression values were calculated via the ΔΔCT method, normalized to β-actin and relative to the oil control values. Bar graphs represent the mean ± SEM. *, p<0.05, oil versus E2.
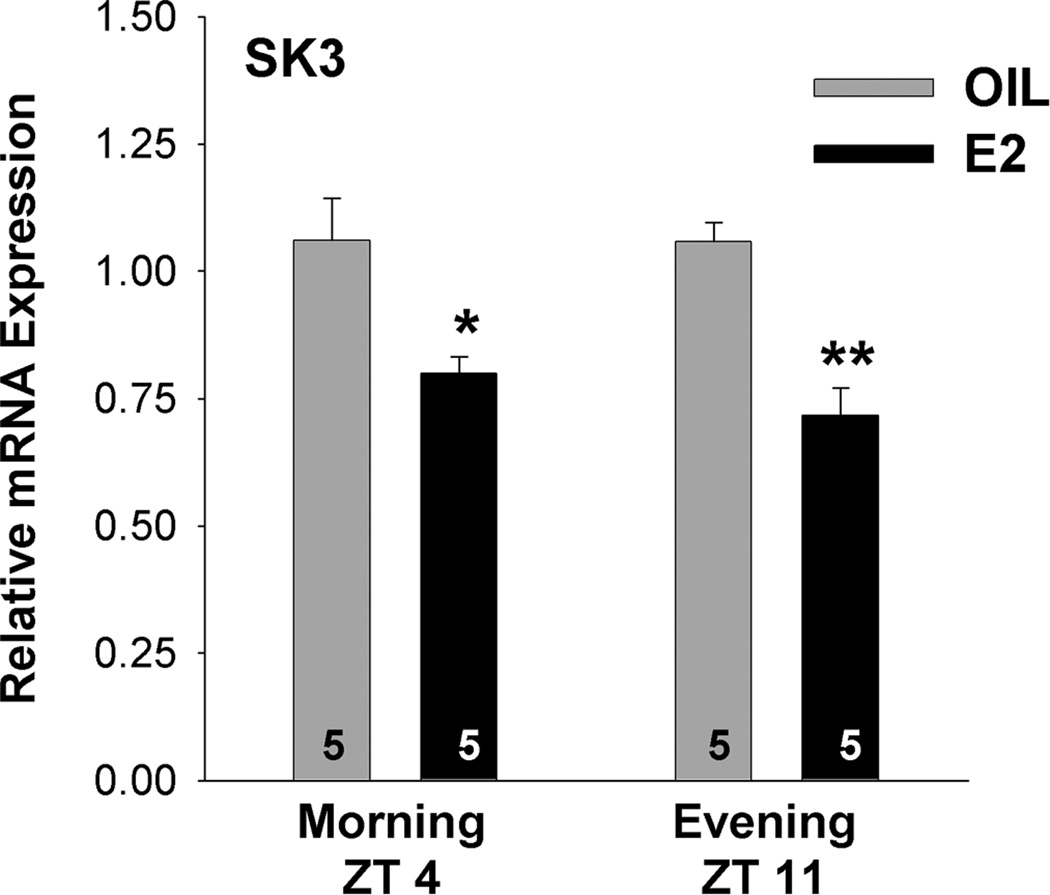
SK channel subtypes
Since SK1 was not expressed in mouse GnRH neurons and SK2 was expressed in < 8% of the cells, we focused our analysis on the SK3 subtype. The E2-treatment decreased SK3 mRNA at ZT 4–5 (p<0.05; n=5) and further decreased the expression of this channel subtype at ZT 11–12 (p<0.01; n=5) (Fig. 11).
Figure 11.
E2 decreases SK3 mRNA in GnRH neurons. Quantitative real-time PCR measurements of SK3 mRNA during the morning, ZT4 (A) and SK3 mRNA during the evening, ZT11 in GnRH neuronal pools (3–4 pools of 5 cells per animal) from oil- and E2-treated mice (n=5 animals per group). The expression values were calculated via the ΔΔCT method, normalized to β-actin and each E2 value was calculated relative to the respective oil control value. Bar graphs represent the mean ± SEM. *, p<0.05, **, p<0.01, oil versus E2.
3.4 qPCR analysis of GnRH and GPR54 mRNA expression in GnRH neurons and modulation by E2 during negative and positive feedback
Prior to ovulation, it has been established that a surge of GnRH is released into the portal vasculature, which prompts a similar surge of LH (Caraty et al., 1989; Chappell and Levine, 2000; Levine and Ramirez, 1982; Pau et al., 1993). This bolus release of GnRH occurs during positive E2 feedback, but data describing changes in GnRH mRNA levels have been equivocal (Gore and Roberts, 1997; Sagrillo et al., 1996). The lack of an effect of E2 on GnRH mRNA was confirmed in the current study. Thus, an E2 regime that induces a LH surge had no significant effect on GnRH mRNA concentration (relative expression) at ZT 4–5 in comparison to mRNA expression at ZT 11–12 on the day of induced proestrus (E2-treated morning, relative expression (RE) 1.02 ± 0.07; E2-treated evening, RE 0.84 ± 0.08 p>0.05; n=5). In addition, the GnRH mRNA expression in E2-treated as compared to oil-treated animals was not different at any time-point (oil morning, RE 1.03 ± 0.11, E2 morning, RE 1.05 ± 0.07; oil evening, RE 1.02 ± 0.06, E2 evening, RE 0.98 ± 0.09, p>0.05, n=5 in each group).
Similarly, the mRNA expression of GPR54 was analyzed at ZT 4–5 (morning) and ZT 11–12 (evening) in oil- and E2-treated animals. The analysis showed that the relative mRNA expression of GPR54 in GnRH neurons was not altered with E2-treatment at either time-point (oil morning, RE 1.02 ± 0.05, E2 morning, RE 0.91 ± 0.14; oil evening, RE 1.04 ± 0.08, E2 evening, RE 0.96 ± 0.12, p>0.05, n=5 in each group).
4. Discussion
Presently we evaluated the mRNA expression and dominant subunits of numerous ion channels important for burst firing and transmitter (neuropeptide) release, including HCN, HVA, TRPC and SK channels, as well as their expression during E2 negative and positive feedback regulation of LH secretion. The salient findings were the following: 1) both HCN1 and HCN2 channel mRNAs were highly expressed in mouse GnRH neurons, but only the mRNA expression of HCN1 was regulated by E2. 2) The majority of HVA channel subtypes were expressed in GnRH neurons, and CaV1.3 (L), CaV2.2 (N) and CaV2.3 (R) transcripts were increased by E2 treatment. 3) TRPC4 mRNA in comparison to TRPC1 and TRPC5 was the most highly expressed in mouse GnRH neurons, and TRPC4 mRNA was increased in E2-treated mice. 4) Of the SK channel subtypes, only SK3 mRNA exhibited significant expression in GnRH neurons, and this channel subtype was decreased in E2-treated animals. The increased expression of ion channels important for maintaining burst firing activity and decreased expression of channels attenuating this activity would help augment robust firing critical for the GnRH super secretion responsible for the LH surge.
For induction of the LH surge by ovarian steroids in mice, different models have been developed in which the animals are given E2-implants with and without surge-inducing treatment of E2 (Bronson and Vom Saal, 1979; Christian et al., 2005; Gee et al., 1984). We have developed a two-step E2 injection procedure in which we used a priming dose followed by a surge-inducing dose of E2 in OVX mice. With this procedure we were consistently able to induce a LH surge in the CBB6 GFP-GnRH mice (Fig. 1). With a reliable model of E2-induced LH release in the evening (positive feedback) as well as inhibition of LH in the morning by E2 (negative feedback), we used this model to explore the corresponding changes in GnRH and GPR54 mRNAs as well as in ion channel expression in GnRH neurons.
In these studies we acutely dispersed, collected and analyzed individual GnRH neurons as well as pools of many neurons in order to assess the distribution and quantitative expression of transcripts. Initially, we found that the cellular mRNA expression of ion channels could not be precisely quantified in individual neurons or in pools of 2–4 neurons for most transcripts. However, pools of 5–10 neurons were sufficient for quantification of ion channel mRNA expression in GnRH neurons (see Fig. 2). Importantly, we have documented that with increasing number of cells, there is a linear increase in mRNA levels, which would suggest that cellular RNA is preserved during the pooling procedure.
According to most studies, GPR54 mRNA and the response to kisspeptin are present in the majority of GnRH neurons (Han et al., 2005; Irwig et al., 2004; Zhang et al., 2008), which was also confirmed in the current study. In addition, we found that the cellular mRNA expression of GPR54, although robust, was not altered by E2 treatment. Our findings may indicate that the cellular mRNA expression of GPR54 is not regulated by E2 in native GnRH neurons, which is different from findings reported in GT 1–7 GnRH neuronal cell lines (Tonsfeldt et al., 2011).
It is well known that a bolus of GnRH is released into the portal vessels, which induces a surge of LH release from the pituitary (Caraty et al., 1989; Chappell and Levine, 2000; Levine and Ramirez, 1982; Pau et al., 1993). In contrast, reported changes in GnRH transcripts during different physiological conditions including the afternoon of proestrus and lactation, have been more variable (Gore and Roberts, 1997; Sagrillo et al., 1996). The previously reported lack of effect of E2 on GnRH mRNA levels (Kelly et al., 1989; Marks et al., 1993) was confirmed in the current study using a sensitive qPCR assay. There was a tendency for a decrease in mRNA expression immediately prior to the surge, possibly due to increased translation to GnRH protein. However, overall there was no significant change in GnRH mRNA levels. These findings may indicate that the cellular expression of GnRH mRNA is not regulated by E2. Instead, E2 controls the expression and function of ion channels important for GnRH neuronal excitability and GnRH release (Chu et al., 2012; Kelly and Wagner, 2002; Rønnekleiv et al., 2011; Zhang et al., 2009).
Numerous studies have documented the importance of HCN channels for induction of neuronal burst firing activities, and the h (pacemaker) current is expressed in GnRH neurons (Arroyo et al., 2006; Chu et al., 2010; Erickson et al., 1993a; Lüthi and McCormick, 1998; Zhang et al., 2007). Our results that the HCN1 and also HCN2 subtypes were highly expressed in GnRH neurons in adult female mice are consistent with the electrophysiological data in adult males (Chu et al., 2010). Therefore, in contrast to results in immature GnRH neurons in which HCN3 is the main HCN isoform (Constantin and Wray, 2008), in adult mice HCN1 and HCN2 are the main subtypes. Of the 4 different isoforms of HCN channels, HCN1 has the fastest kinetics and HCN4 the slowest. In addition, HCN2 and 4 are sensitive to cAMP, whereas HCN1 channels are less sensitive and HCN3 channels are insensitive to cyclic nucleotides (Biel et al., 2009). It is, therefore, interesting that the mRNA expression of HCN1 but not HCN2 was increased in E2-treated females (current findings), suggesting that HCN 1, but not HCN2, channel synthesis is regulated by E2. The effects of E2 on the h-current (Ih) have not been measured in females. But based on the increased HCN1 mRNA expression by E2, we would predict an E2-induced increase in Ih amplitude. A different regulation occurs in male GnRH neurons, where the Ih is increased in castrated as compared to intact animals, and E2-treatment reverses the effects of castration (Chu et al., 2010). The different steroid effects in female versus male mouse GnRH neurons could be due to a sex difference since males do not exhibit a GnRH or LH surge, and E2 is primarily inhibitory to male GnRH neurons (Pielecka-Fortuna and Moenter, 2006).
Consistent with published data in rat, we found that most of the HVA channel subtypes are expressed in mouse GnRH neurons with the R-type channel expression being the most prominent (Kato et al., 2003; Tanaka et al., 2010). Moreover CaV1.3 (L), CaV2.2 (N) and CaV2.3 (R) mRNAs, were significantly increased in E2-treated animals.. Interestingly, the HVA L - and N-type peak currents in GnRH neurons exhibit diurnal variation in E2-treated, but not in oil-treated mice, with reduced current amplitude in the morning and increased current amplitude in the evening (Sun et al., 2010). Although, we did not directly compare HVA channel mRNA expression in E2-treated animals during the morning and evening, we found no change in oil-treated animals. Therefore, our data that L- and R-type channel mRNAs expression was increased with E2-treatment relative to oil-treatment irrespective of time of day, would indicate that the augmented HVA channel mRNA expression is dependent on E2 levels and not time of day in our animal model. Our previous results comparing T-type ion channel mRNA expression and current density in GnRH neurons and KCNQ channel mRNAs to M-current in NPY neurons, have found a close correlation between alterations in mRNA expression and function (Roepke et al., 2011; Zhang et al., 2009). Numerous studies have illustrated that calcium entry through voltage-gated channels contributes to neuronal excitability and is important for GnRH neuronal excitability and GnRH release (Constantin et al., 2011; Constantin et al., 2010; Iremonger and Herbison, 2012; Kroll et al., 2011; Krsmanovic et al., 1992; Lee et al., 2010; Sun et al., 2010; Zhang et al., 2009; Zhang and Spergel, 2011). However, more studies are needed to explore the correlation between HVA channel expression and HVA currents in GnRH neurons under different experimental conditions.
Our quantitative analysis of TRPC channel subtypes in GnRH neurons revealed that TRPC 1 and 4 mRNAs were the most highly expressed in mouse GnRH neurons, whereas TRPC 5 transcripts were expressed in significantly lower quantities. Interestingly, arcuate POMC cells express high levels of TRPC5 but low levels of TRPC4 transcripts (Qiu et al., 2010), suggesting a neuron-specific expression of TRPC channel subtypes. It is well known that the current-voltage relationship and mechanism of regulation of TRPC channels are dependent on the channel subunit composition (Clapham, 2003). The high expression of TRPC4 may help explain why kisspeptin activation of TRPC channels, underlying its robust depolarization, depends on phosphatidylinositol 4,5-biphosphate (PIP2) depletion and cSrc tyrosine kinase activation in GnRH neurons (Zhang et al; submitted; Odell et al., 2012; Otsuguro et al., 2008). Moreover, the mRNA expression of TRPC4 channels was increased in E2-treated as compared to oil-treated mice. Interestingly, kisspeptin actions in GnRH neurons do not appear to be influenced by the estrous stage of the animals or E2-treatment (Pielecka-Fortuna et al., 2008; Zhang et al., 2008). Rather, kisspeptin increases GABA-A and glutamate inputs to GnRH neurons in an E2-dependent manner (Pielecka-Fortuna and Moenter, 2010). However, to fully elucidate a potential E2 modulation of kisspeptin activation of TRPC current in GnRH neurons a dose response analysis is necessary, which has not yet been performed. Therefore, additional experiments are necessary to explore the functional significance of E2 modulation of TRPC4 mRNA expression in GnRH neurons.
Neuronal excitability and frequency of firing are also sculpted by the AHP that follows an action potential, and SK channels underlie a component of the medium AHP (mAHP) K+ currents in CA1 pyramidal cells and other neurons (Bond et al., 2004). SK channel subunits are expressed in GnRH neurons (Bosch et al., 2002; Kato et al., 2006), and SK channel activation exerts a powerful influence on firing properties of GnRH neurons (Kato et al., 2006; Lee et al., 2010; Liu and Herbison, 2008). Currently we have found that SK3 is the main subtype in GnRH neurons, and its mRNA expression is reduced in E2 treated mice. The mAHP current is inhibited by bath applied E2 in GnRH neurons (Chu et al., 2009). Therefore, these findings would indicate that E2 reduces the synthesis of SK channels and the expression of the AHP current in GnRH neurons. Since the apamine-sensitive AHP (SK) current regulates both intraburst and interburst neuronal firing (Lee et al., 2010), the E2-induced reduction of expression and current amplitude would play a significant role in modulating GnRH neuronal excitability and firing pattern.
For most, if not all, of the channel subtypes regulated by E2, the mRNA levels were increased (or decreased) in the morning during E2-inhibition of plasma LH levels (negative feedback?) and remained unchanged during the evening similar to that reported previously for T-type CaV3.3 calcium channels (Zhang et al., 2009). These findings would indicate that prolonged (>24 h) of surge levels of E2 lead to alteration in mRNA expression of a number of ion channel subunits in GnRH neurons already in the morning of induced proestrus.. We hypothesize that surge levels of E2 alters the number of channels and thereby alters the excitability of GnRH neurons as we have shown for T-type calcium channels in GnRH neurons irrespective of time of day (Zhang et al., 2009). The precise mechanisms by which E2 maintains inhibition of GnRH neuronal activity and secretion until the afternoon (evening) of proestrus is currently not well understood. However, an E2-dependent circadian signal to GnRH neurons as well presynaptic neurons is clearly involved (Christian et al., 2005; Christian and Moenter, 2007; Robertson et al., 2009).
In summary, our working hypothesis is that rising levels of E2 increase mRNA expression of multiple ion channels in GnRH neurons, including HCN channels, T-type and HVA calcium channels and TRPC channels, which would increase the overall excitability of GnRH neurons (Zhang et al., 2009, and current findings). At a hyperpolarized membrane potential due to the ATP-sensitive potassium (K-ATP) and G protein-activated inwardly rectifying K+ (GIRK) channel activity (Lagrange et al., 1995; Zhang et al., 2010; Zhang et al., 2007), the hyperpolarization-activated pacemaker (h) current is triggered that drives the membrane potential towards a more depolarized level. This depolarization activates T-type channels, which were deinactivated during the preceding hyperpolarization. Rising E2 levels also increase kisspeptin excitatory drive to GnRH neurons, which inhibits K+ channel activity and activates TRPC channel inputs to depolarize GnRH (Liu et al., 2008; Pielecka-Fortuna et al., 2008; Zhang et al., 2008). E2 also decreases the expression (current findings) and function of SK channels, which underlie the medium afterhyperpolarization in GnRH neurons (Chu et al., 2009). Collectively, these actions of surging E2 levels in conjunction with the neural clock inputs, lead to an alteration in ion channel mRNA expression and GnRH neuronal burst firing sufficient for GnRH release, the LH surge and ovulation.
Highlights.
Single-cell quantitative PCR of channel transcripts in GnRH neurons was validated and used
E2 increased or decreased the mRNA expression of ion channels in GnRH neurons
Altered channel mRNA expression leads to changes in the excitability of GnRH neurons
Acknowledgements
Research reported in this publication was supported by National Institutes of Health Grant NS 43330. KJT was supported by the Reproductive Biology Training Grant NIH T32HD007133 and Steinberg Funds from OHSU Department of Physiology and Pharmacology. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health.
Abbreviations
- AHP
afterhyperpolarization
- E2
17β-estradiol
- GnRH
gonadotropin-releasing hormone
- GPR54
G protein-coupled receptor 54
- HCN
hyperpolarization-activated cyclic nucleotide-gated
- HVA
high voltage activated
- LH
luteinizing hormone
- POA
preoptic area
- qPCR
quantitative real-time PCR
- RT
reverse transcription
- sc-PCR
single cell PCR
- SK
small-conductance calcium-activated potassium channels
- TRPC
canonical transient receptor potential
- ZT
zeitgeber time
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Martha A. Bosch, Email: boschm@ohsu.edu.
Karen J. Tonsfeldt, Email: tonsfelk@ohsu.edu.
Oline K. Rønnekleiv, Email: ronnekle@ohsu.edu.
References
- Arroyo A, Kim B, Rasmusson RL, Yeh J. Hyperpolarization-activated cation channels are expressed in rat hypothalamic gonadotropin-releasing hormone (GnRH) neurons and immortalized GnRH neurons. J. Soc. Gyn. Invest. 2006;13:442–450. doi: 10.1016/j.jsgi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Biel M, Wahl-Schott C, Michalakis S, Zong X. Hyperpolarization-activated cation channels: From genes to function. Physiol. Rev. 2009;89:847–885. doi: 10.1152/physrev.00029.2008. [DOI] [PubMed] [Google Scholar]
- Bond CT, Herson PS, Strassmaier T, Hammond R, Stackman R, Maylie J, Adelman JP. Small conductance Ca2+-activated K+ channel knockout mice reveal the identity of calcium-dependent afterhyperpolarization currents. J. Neurosci. 2004;24:5301–5306. doi: 10.1523/JNEUROSCI.0182-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch MA, Kelly MJ, Rønnekleiv OK. Distribution, neuronal co-localization and 17β-E2 modulation of small conductance calcium-activated K+ channel (SK3) mRNA in the guinea pig brain. Endocrinology. 2002;143:1097–1107. doi: 10.1210/endo.143.3.8708. [DOI] [PubMed] [Google Scholar]
- Bronson FH, Vom Saal FS. Control of the preovulatory release of luteinizing hormone by steroids in the mouse. Endocrinology. 1979;104:1247–1255. doi: 10.1210/endo-104-5-1247. [DOI] [PubMed] [Google Scholar]
- Bronson FH, Whitten WK. Oestrus-accelerating pheromone of mice: assay, androgen-dependency and presence in bladder urine. J. Reprod. Ferti.l. 1968;15:131–134. doi: 10.1530/jrf.0.0150131. [DOI] [PubMed] [Google Scholar]
- Caraty A, Locatelli A, Martin GB. Biphasic response in the secretion of gonadotrophin-releasing hormone in ovariectomized ewes injected with oestradiol. J. Endocrinol. 1989;123:375–382. doi: 10.1677/joe.0.1230375. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Striessnig J, Snutch TP, Perez-Reyes E. International Union of Pharmacology.XXXIX. Compendium of voltage-gated ion channels: calcium channels. Pharmaco.l Rev. 2003;55:575–578. doi: 10.1124/pr.55.4.8. [DOI] [PubMed] [Google Scholar]
- Chappell PE, Goodall CP, Tonsfeldt KJ, White RS, Bredeweg E, Latham KL. Modulation of Gonadotropin-releasing hormone (GnRH) secretion by an endogenous circadian clock. J. Neuroendocrinol. 2009;21:339–345. doi: 10.1111/j.1365-2826.2009.01845.x. [DOI] [PubMed] [Google Scholar]
- Chappell PE, Levine JE. Stimulation of gonadotropin-releasing hormone surges by estrogen. I. Role of hypothalamic progesterone receptors. Endocrinology. 2000;141:1477–1485. doi: 10.1210/endo.141.4.7428. [DOI] [PubMed] [Google Scholar]
- Chemin J, Monteil A, Perez-Reyes E, Bourinet E, Nargeot J, Lory P. Specific contribution of human T-type calcium channel isotypes (alpha(1G), alpha(1H) and alpha(1I)) to neuronal excitability. J. Physiol. 2002;540:3–14. doi: 10.1113/jphysiol.2001.013269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian CA, Mobley JL, Moenter SM. Diurnal and estradiol-dependent changes in gonadotropin-releasing hormone neuron firing activity. Proc. Natl. Acad. Sci. USA. 2005;102:15682–15687. doi: 10.1073/pnas.0504270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian CA, Moenter SM. Estradiol induces diurnal shifts in GABA transmission to gonadotropin-releasing hormone neurons to provide a neural signal for ovulation. J. Neurosci. 2007;27:1913–1921. doi: 10.1523/JNEUROSCI.4738-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian CA, Moenter SM. Vasoactive intestinal polypeptide can excite gonadotropin-releasing hormone neurons in a manner dependent on estradiol and gated by time of day. Endocrinology. 2008;149:3130–3136. doi: 10.1210/en.2007-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z, Andrade J, Shupnik MA, Moenter SM. Differential regulation of gonadotropin-releasing hormone neuron activity and membrane properties by acutely applied estradiol: dependence on dose and estrogen receptor subtype. J. Neurosci. 2009;29:5616–5627. doi: 10.1523/JNEUROSCI.0352-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z, Takagi H, Moenter SM. Hyperpolarization-activated currents in gonadotropin-releasing hormones (GnRH) neurons contribute to intrinsic excitability and are regulated by gonadal steroid feedback. J. Neurosci. 2010;30:13373–13383. doi: 10.1523/JNEUROSCI.1687-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z, Tomaiuolo M, Bertram R, Moenter SM. Two types of burst firing in gonadotropin-releasing hormone neurones. J. Neuroendocrinol. 2012;24:1065–1077. doi: 10.1111/j.1365-2826.2012.02313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- Clarkson J, d'Anglemont de Tassigny X, Moreno AS, Colledge WH, Herbison AE. Kisspeptin-GPR54 signaling is essential for preovulatory gonadotropin-releasing hormone neuron activation and the luteinizing hormone surge. J. Neurosci. 2008;28:8691–8697. doi: 10.1523/JNEUROSCI.1775-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin S, Jasoni C, Romano N, Lee K, Herbison AE. Understanding calcium homeostasis in postnatal gonadotropin-releasing hormone neurons using cell-specific Pericam transgenics. Cell Calcium. 2011;51:267–276. doi: 10.1016/j.ceca.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Constantin S, Jasoni CL, Wadas B, Herbison AE. γ-Aminobutyric acid and glutamate differentially regulate intracellular calcium concentrations in mouse gonadotropin-releasing hormone neurons. Endocrinology. 2010;151:262–270. doi: 10.1210/en.2009-0817. [DOI] [PubMed] [Google Scholar]
- Constantin S, Wray S. GnRH-1 neuronal activity in independent of HCN channels but is sensitive to PKA-dependent phosphorylation. Endocrinology. 2008:1–24. doi: 10.1210/en.2007-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal SJ, Estep JS, Valentin-Bon IE, Jerse AE. Standardization of the Whitten Effect to induce susceptibility to Neisseria gonorrhoeae in female mice. Contemp. Top Lab Anim. Sci. 2001;40:13–17. [PubMed] [Google Scholar]
- Erickson KR, Rønnekleiv OK, Kelly MJ. Electrophysiology of guinea-pig supraoptic neurones: Role of a hyperpolarization-activated cation current in phasic firing. J. Physiol. (Lond.) 1993a;460:407–425. doi: 10.1113/jphysiol.1993.sp019478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson KR, Rønnekleiv OK, Kelly MJ. Role of a T-type calcium current in supporting a depolarizing potential, damped oscillations, and phasic firing in vasopressinergic guinea pig supraoptic neurons. Neuroendo. 1993b;57:789–800. doi: 10.1159/000126438. [DOI] [PubMed] [Google Scholar]
- Gee DM, Flurkey K, Mobbs CV, Sinha YN, Finch CE. The regulation of luteinizing hormone and prolactin in C57BL/6J mice: effects of estradiol implant size, duration of ovariectomy, and aging. Endocrinology. 1984;114:685–693. doi: 10.1210/endo-114-3-685. [DOI] [PubMed] [Google Scholar]
- Gore AC, Roberts JL. Regulation of gonadotropin-releasing hormone gene expression in vivo and in vitro. Front. Neuroendocrino.l. 1997;18:209–245. doi: 10.1006/frne.1996.0149. [DOI] [PubMed] [Google Scholar]
- Han S-K, Gottsch ML, Lee KJ, Popa SM, Smith JT, Jakawich SK, Clifton DK, Steiner RA, Herbison AE. Activation of gonadotropin-releasing hormone neurons by kisspeptin as a neuroendocrine switch for the onset of puberty. J. Neurosci. 2005;25:11349–11356. doi: 10.1523/JNEUROSCI.3328-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iremonger KJ, Herbison AE. Initiation and propagation of action potentials in gonadotropin-releasing hormone neuron dendrites. J. Neurosci. 2012;32:151–158. doi: 10.1523/JNEUROSCI.3739-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwig MS, Fraley GS, Smith JT, Acohido BV, Popa SM, Cunningham MJ, Gottsch ML, Clifton DK, Steiner RA. Kisspeptin activation of gonadotropin releasing hormone neurons and regulation of KiSS-1 mRNA in the male rat. Neuronendocrinol. 2004;80:264–272. doi: 10.1159/000083140. [DOI] [PubMed] [Google Scholar]
- Kato M, Tanaka N, Usui S, Sakuma Y. SK channel blocker apamin inhibits slow afterhyperpolarization currents in rat gonadotropin-releasing hormone neurones. J. Physiol. 2006;574.2:431–442. doi: 10.1113/jphysiol.2006.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Ui-Tei K, Watanabe M, Sakuma Y. Characterization of voltage-gated calcium currents in gonadotropin-releasing hormone neurons tagged with green fluorescent protein in rats. Endocrinology. 2003;144:5118–5125. doi: 10.1210/en.2003-0213. [DOI] [PubMed] [Google Scholar]
- Kelly MJ, Garrett J, Bosch MA, Roselli CE, Douglass J, Adelman JP, Rønnekleiv OK. Effects of ovariectomy on GnRH mRNA, proGnRH and GnRH levels in the preoptic hypothalamus of the female rat. Neuroendocrinol. 1989;49:88–97. doi: 10.1159/000125095. [DOI] [PubMed] [Google Scholar]
- Kelly MJ, Rønnekleiv OK. Electrophysiological analysis of neuroendocrine neuronal activity in hypothalamic slices. In: Levine JE, editor. Methods in Neurosciences: Pulsatility in Neuroendocrine Systems. San Diego: Academic Press, Inc.; 1994. pp. 47–67. [Google Scholar]
- Kelly MJ, Wagner EJ. GnRH neurons and episodic bursting activity. Trends Endocrinol. Metab. 2002;13:409–410. doi: 10.1016/s1043-2760(02)00698-7. [DOI] [PubMed] [Google Scholar]
- Kim D, Song I, Keum S, Lee T, Jeong MJ, Kim SS, McEnery MW, Shin HS. Lack of the burst firing of thalamocortical relay neurons and resistance to absence seizures in mice lacking alpha(1G) T-type Ca2+ channels. Neuron. 2001;31:35–45. doi: 10.1016/s0896-6273(01)00343-9. [DOI] [PubMed] [Google Scholar]
- Kroll H, Bolsover S, Hsu J, Kim S-H, Bouloux P-M. Kisspeptin-evoked calcium signals in isolated primary rat gonadotropin-releasing hormone neurones. Neuroendocrinol. 2011;93:114–120. doi: 10.1159/000321678. [DOI] [PubMed] [Google Scholar]
- Krsmanovic LZ, Stojikovic SS, Merelli F, Dufour SM, Virmani MA, Catt KJ. Calcium signaling and episodic secretion of gonadotropin-releasing hormone in hypothalamic neurons. Proc. Natl. Acad. Sci. USA. 1992;89:8462–8466. doi: 10.1073/pnas.89.18.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange AH, Rønnekleiv OK, Kelly MJ. Estradiol-17β and μ-opioid peptides rapidly hyperpolarize GnRH neurons: A cellular mechanism of negative feedback? Endocrinology. 1995;136:2341–2344. doi: 10.1210/endo.136.5.7720682. [DOI] [PubMed] [Google Scholar]
- Lee K, Duan W, Sneyd J, Herbison AE. Two slow calcium-activated afterhyperpolarization currents control burst firing dynamics in gonadotropin-releasing hormone neurons. J. Neurosci. 2010;30:6214–6224. doi: 10.1523/JNEUROSCI.6156-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legan SJ, Coon GA, Karsch FJ. Role of estrogen as initiator of daily LH surges in the ovariectomized rat. Endocrinology. 1975;96:50–56. doi: 10.1210/endo-96-1-50. [DOI] [PubMed] [Google Scholar]
- Levine JE, Ramirez VD. Luteinizing hormone-releasing hormone release during the rat estrous cycle and after ovariectomy, as estimated with push-pull cannulae. Endocrinology. 1982;111:1439–1448. doi: 10.1210/endo-111-5-1439. [DOI] [PubMed] [Google Scholar]
- Liu X, Herbison AE. Small-conductance calcium-activated potassium channels control excitability and firing dynamics in gonadotropin-releasing hormone (GnRH) neurons. Endocrinology. 2008;149:3598–3604. doi: 10.1210/en.2007-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lee K, Herbison AE. Kisspeptin excites gonadotropin-releasing hormone (GnRH) neurons through a phosphilipase C/calcium-dependent pathway regulating multiple ion channels. Endocrinology. 2008;149:4605–4614. doi: 10.1210/en.2008-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta-delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lüthi A, McCormick DA. H-current: properties of a neuronal and network pacemaker. Neuron. 1998;21:9–12. doi: 10.1016/s0896-6273(00)80509-7. [DOI] [PubMed] [Google Scholar]
- Marks DL, Smith SM, Clifton DK, Steiner RA. Regulation of gonadotropin-releasing hormone (GnRH) and galanin gene expression in GnRH neurons during lactation in the rat. Endocrinology. 1993;133:1450–1458. doi: 10.1210/endo.133.3.7689958. [DOI] [PubMed] [Google Scholar]
- Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr. Rev. 2009;30:713–743. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell AF, Scott JL, Van Helden DF. Epidermal growth factor induces tyrosine phosphorylation, membrane insertion, and activation of transient receptor potential channel 4. J. Biol. Chem. 2012;280:37974–37987. doi: 10.1074/jbc.M503646200. [DOI] [PubMed] [Google Scholar]
- Otsuguro K-I, Tang J, Tang Y, Xiao R, Freichel M, Tsvilovskyy V, Ito S, Flockerzi V, Zhu MX, Zholos AV. Isoform-specific inhibition of TRPC4 channel by phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 2008;283:10026–10036. doi: 10.1074/jbc.M707306200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pau K-YF, Berria M, Hess DL, Spies HG. Preovulatory gonadotropin-releasing hormone surge in ovarian-intact Rhesus Macaques. Endocrinology. 1993;133:1650–1656. doi: 10.1210/endo.133.4.8404606. [DOI] [PubMed] [Google Scholar]
- Pau K-YF, Orstead KM, Hess DL, Spies HG. Feedback effects of ovarian steroids on the hypothalamic-hypophyseal axis in the rabbit. Biol. Repro. 1986;35:1009–1023. doi: 10.1095/biolreprod35.4.1009. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acid Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielecka J, Moenter SM. Effect of steroid milieu on gonadotropin-releasing hormone-1 neuron firing pattern and luteinizing hormone levels in male mice. Biol. Reprod. 2006;74:931–937. doi: 10.1095/biolreprod.105.049619. [DOI] [PubMed] [Google Scholar]
- Pielecka-Fortuna J, Chu Z, Moenter SM. Kisspeptin acts directly and indirectly to increase gonadotropin-releasing hormone neuron activity and its effects are modulated by estradiol. Endocrinology. 2008;149:1979–1986. doi: 10.1210/en.2007-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielecka-Fortuna J, Moenter SM. Kisspeptin increases γ-aminobutyric acidergic and glutamatergic transmission directly to gonadotropin-releasing hormone neurons in an estradiol-dependent manner. Neuroendo. 2010;151:291–300. doi: 10.1210/en.2009-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Fang Y, Bosch MA, Rønnekleiv OK, Kelly MJ. Guinea pig kisspeptin neurons are depolarized by leptin via activation of TRPC channels. Endocrinology. 2011;152:1503–1514. doi: 10.1210/en.2010-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Fang Y, Rønnekleiv OK, Kelly MJ. Leptin exceites proopiomelanocortin neurons via activation of TRPC channels. J. Neurosci. 2010;30:1560–1565. doi: 10.1523/JNEUROSCI.4816-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JL, Clifton DK, de la Iglesia HO, Steiner RA, Kauffman AS. Circadian regulation of Kiss1 neurons: implications for timing the preovulatory gonadotropin-releasing hormone/luteninizing hormone surge. Neuroendocrinol. 2009;150:3664–3671. doi: 10.1210/en.2009-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepke TA, Qiu J, Smith AW, Rønnekleiv OK, Kelly MJ. Fasting and 17β-estradiol differentially modulate the M-current in neuropeptide Y neurons. J. Neurosci. 2011;17:11825–11835. doi: 10.1523/JNEUROSCI.1395-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rønnekleiv OK, Bosch MA, Zhang C. 17β-Oestradiol regulation of gonadotrophin-releasing hormone neuronal excitability. J. Neuroendocrinol. 2011;24:122–130. doi: 10.1111/j.1365-2826.2011.02160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagrillo CA, Grattan DR, McCarthy MM, Selmanoff M. Hormonal and neurotransmitter regulation of GnRH gene expression and related reproductive behaviors. Behav. Genet. 1996;26:241–277. doi: 10.1007/BF02359383. [DOI] [PubMed] [Google Scholar]
- Spergel DJ. Calcium and small-conductance calcium-activated potassium channels in gonadotropin-releasing hormone neurons before, during, and after puberty. Endocrinology. 2007;148:2383–2390. doi: 10.1210/en.2006-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Chu Z, Moenter SM. Diurnal in vivo and rapid in vitro effects of estadiol on voltage-gated calcium channels in gonadotropin-releasing hormone neurons. J. Neurosci. 2010;30:3912–3923. doi: 10.1523/JNEUROSCI.6256-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter KJ, Song WJ, Sampson TL, Wuarin J-P, Saunders JT, Dudek FE, Moenter SM. Genetic targeting of green fluorscent protein to gonadotropin-releasing hormone neurons: Characterization of whole-cell electrophysiological properties and morphology. Endocrinology. 2000;141:412–419. doi: 10.1210/endo.141.1.7279. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Ishii H, Yin C, Koyama M, Sakuma Y, Kato M. Voltagegated Ca2+ channel mRNAs and T-type Ca2+ currents in rat gonadotropin-releasing hormone neurons. J. Physio.l Sci. 2010;60:195–204. doi: 10.1007/s12576-010-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonsfeldt KJ, Goodall CP, Latham KL, Chappell PE. Oestrogen induces rhythmic expression of the Kisspeptin-1 receptor GPR54 in hyopthalamic gonadotrophin-releasin hormone-secreting GT1–7 cells. J. Neuroendocrinol. 2011;23:823–830. doi: 10.1111/j.1365-2826.2011.02188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida B, Deli L, Hrabovszky E, Kalamatianos T, Caraty A, Coen CW, Liposits Z, Kalló I. Evidence for suprachiasmatic vasopressin neurons innervating kisspeptin neurons in the rostral periventricular area of the mouse brain: regulation by oestrogen. J. Neuroendocrinol. 2010;22:1032–1039. doi: 10.1111/j.1365-2826.2010.02045.x. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Rønnekleiv OK, Kelly MJ. The noradrenergic inhibition of an apamine-sensitive small conductance Ca2+ -activated K+ channel in hypothalamic γ-aminobutyric acid neurons: Pharmacology, estrogen sensitivity and relevance to the control of the reproductive axis. J. Pharmacol. Exp. Ther. 2001;299:21–30. [PubMed] [Google Scholar]
- Ward DR, Dear FM, Ward IA, Anderson SI, Spergel DJ, Smith PA, Ebling FJP. Innervation of gonadotropin-releasing hormone neurons by peptidergic neurons conveying circadian or energy balance information in the mouse. PLos One. 2009;4:1–8. doi: 10.1371/journal.pone.0005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermantel TM, Campbell RE, Porteous R, Bock D, Gröne H-J, Todman MG, Korach KS, Greiner E, Perez CA, Schultz G, Herbison AE. Definition of estrogen receptor pathway critical for estrogen positive feedback to gonadotropin-releasing hormone neurons and fertility. Neuron. 2006;52:271–280. doi: 10.1016/j.neuron.2006.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Bosch MA, Levine JE, Rønnekleiv OK, Kelly MJ. Gonadotropin-releasing hormone neurons express KATP channels that are regulated by estrogen and responsive to glucose and metabolic inhibition. J. Neurosci. 2007;27:10153–10164. doi: 10.1523/JNEUROSCI.1657-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Bosch MA, Rick EA, Kelly MJ, Rønnekleiv OK. 17β-estradiol regulation of T-type calcium channels in gonadotropin-releasing hormone neurons. J. Neurosci. 2009;29:10552–10562. doi: 10.1523/JNEUROSCI.2962-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Kelly MJ, Rønnekleiv OK. 17β-estradiol rapidly increases adenosine 5'-triphosphate-sensitive potassium channel activity in gonadotropin-releasing hormone neurons via a protein kinase signaling pathway. Endocrinology. 2010;151:4477–4484. doi: 10.1210/en.2010-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Roepke TA, Kelly MJ, Rønnekleiv OK. Kisspeptin depolarizes gonadotropin-releasing hormone neurons through activation of TRPC-like cationic channels. J Neurosci. 2008;28:4423–4434. doi: 10.1523/JNEUROSCI.5352-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-B, Spergel DJ. Kisspeptin inhibits high-voltage activated Ca2+ channels in GnRH neurons via multiple Ca2+ influx and release pathways. Neuroendocrinol. 2012;96:68–80. doi: 10.1159/000335985. [DOI] [PubMed] [Google Scholar]