Abstract
The Bacteroides conjugative transposon, CTnDOT, is an integrated conjugative element (ICE), found in many human colonic Bacteroides spp. strains. It has a complex regulatory system for both excision from the chromosome and transfer and mobilization into a new host. It was previously shown that a cloned DNA segment encoding the xis2c, xis2d, orf3, and exc genes was required for tetracycline dependent activation of the Ptra promoter. The Xis2c and Xis2d proteins are required for excision while the Exc protein stimulates excision. We report here that neither the Orf3 nor the Exc proteins are involved in activation of the Ptra promoter. Deletion analysis and electromobility shift assays showed that the Xis2c and Xis2d proteins bind to the Ptra promoter to activate the tra operon. Thus, the recombination directionality factors of CTnDOT excision also function as activator proteins of the Ptra promoter.
Keywords: Bacteroides, CTnDOT, Xis2c, Xis2d, Exc, gene regulation, transfer, RDFs, ICEs, conjugative transposon
INTRODUCTION
Bacteroides species compose approximately 40% of the normal microbiota of the human colon, but are capable of causing life threatening infections if they escape outside of the colon (Costello et al., 2009; Moore et al., 1978). Bacteroides spp. harbor many conjugative transposons (CTns), also called ICEs (integrative conjugative elements), which encode antibiotic resistance genes that can spread through horizontal gene transfer. One well-studied example of an ICE is CTnDOT, a 65-kb element containing a tetracycline resistance gene, tetQ, and an erythromycin resistance gene, ermF (2, 12, 13).
CTnDOT is stably maintained in the chromosome until the cells are exposed to tetracycline, which induces the production of proteins that promote excision and transfer. Upon exposure to tetracycline, translational attenuation increases production of proteins encoded by the three gene tetQ operon, tetQ, rteA and rteB (Wang et al., 2005; Wang et al., 2004). RteA and RteB proteins compose a two component regulatory system (Cheng et al., 2000; Cheng et al., 2001; Stevens et al., 1990). The RteA protein is the sensor component and the RteB protein is the transcriptional regulator (Moon et al., 2005). The RteB protein activates expression of another regulatory gene rteC and RteC induces expression of the excision operon containing the xis2c, xis2c, orf3, and exc genes (Moon et al., 2005; Park and Salyers, 2011). Previous work has shown that the orf3 gene is not needed for excision (Cheng et al., 2001). Xis2c and Xis2d are small basic proteins similar to lambda Xis and other recombination directionality factors (RDFs) (Lewis and Hatfull, 2001). Exc is a DNA topoisomerase III enzyme capable of relaxing DNA in vitro (26). These proteins, along with the host factor, promote excision by binding to the attL and attR sites of CTnDOT to form excisive intasomes with the tyrosine recombinase IntDOT. After the intasome forms a synaptic complex, IntDOT catalyzes two rounds of strand exchanges to form the attDOT and attB sites. In an in vitro intramolecular excision reaction, Exc is not required for excision but enhances excision 3-5 fold (Keeton and Gardner, 2012). The Exc protein is required in vivo for excision (Cheng et al., 2001).
Following excision of CTnDOT from the chromosome, a closed circular intermediate is formed. The transfer of this circular intermediate requires multiple transfer and mobilization proteins. The transfer genes are located in the tra operon which encodes a series of proteins TraA – TraQ. Previous work has shown that tetracycline dependent activation of this operon requires the excision operon (Jeters et al., 2009; Whittle et al., 2002). When a large portion of CTnDOT containing the mobilization region and the tra operon was cloned into the vector pLYL72, it was self-transmissible at a frequency of 10–5 to 10–6 transconjugants per recipient regardless of whether tetracycline was present (Li et al., 1995). However, transfer of pLYL72 was regulated by tetracycline if it was a co-resident with an intact CTn (Li et al., 1995). The genes required for this regulation were later shown to be contained within the excision operon (Whittle et al., 2002). However, it was unclear which of the excision proteins Xis2c, Xis2d, Orf3, or Exc was responsible for activation of the tra operon. In other excision systems, there is usually only one RDF. However, CTnDOT encodes two RDFs, Xis2c and Xis2d which are both required for excision.
We report here that the deletion of either the orf3 or exc genes did not reduce activation of the Ptra promoter. Deletion of either the xis2c or xis2d genes decreased activation of the Ptra promoter by over 20 fold. Using EMSA analysis we show that the RDFs of CTnDOT, Xis2c and Xis2d bind the Ptra promoter and activate the expression of the tra operon. When the Xis2c and Xis2d proteins bind the Ptra promoter together, the resultant complex that forms on the Ptra promoter does not migrate into the gel.
MATERIAL AND METHODS
Plasmids, Bacterial strains, and Growth Conditions
Plasmids and the Bacteroides strains used in this study are listed in Table 1 or are described in the text. All the Escherichia coli strains were grown in Luria Broth (LB) or on LB agar at 37 C and the Bacteroides strains were grown in either Trypticase Yeast Extract Glucose (TYG) medium or Supplemented Brain Heart Infusion (BHIS) medium at 37 C under anaerobic condition (Holdeman, 1975). Antibiotics were supplied by Sigma and used at the following concentrations: ampicillin (amp) 100 g/ml; chloramphenicol (cam) 15 g/ml; erythromycin (erm) 10 g/ml; gentamicin (gen) 200 g/ml; rifampicin (rif) 10 g/ml; tetracycline (tet) 1 g/ml.
Table 1.
Strains and vectors used in this study.
| Strain or plasmid | aRelevant phenotype | Description |
|---|---|---|
| B. thetaiotaomicron 5482 strains | ||
| BT4001 | (Rif) | Spontaneous rifampicin resistant strain of B. thetaiotaomicron 5482 |
| BT4001ΩQABC | (Rif Tc) | tetQrteArteBrteC regulatory genes of CTnDOT site specifically integrated into the BT4001 chromosome via NBU1 integrase (28). |
| BT4004 | (Rif Tc) | BT4001 containing a single integrated copy of CTnERL which provides the regulatory genes and the excision operon. |
| Plasmids and vectors | ||
| pC-COW | ApCmTc (Cm) | A pVAL1 (25) derivative which contains a chloramphenicol resistance marker and pB8-51 which makes this vector compatible in Bacteroides hosts with pBI143 based vectors such as pAFD1 (11). |
| pGW40.5 | ApCm(Cm) | pC-COW with the Ptra::uidA fusion cloned into the NruI site (14). |
| Clones of excision operon: | ||
| pAFD1 | Ap (Em) | The pBI143 based shuttle vector pFD160 (22) containing ermF cloned into the AatII site. Compatible in Bacteroides hosts with pB8-51 based vectors such as pGW40.5. |
| pHopp1 | Ap (Em) | Contains entire excision operon: xis2c, xis2d, orf3 and exc cloned on pAFD1. |
| pGRW53 | Ap (Em) | 3.7kb PstI-SstI from pGW46 (28) which contains an in-frame deletion of orf3 cloned into pAFD1. |
| pGRW53Δ2c | Ap (Em) | pGRW53 with an in-frame deletion of the 118aa xis2c (aa4-aa91). |
| pGRW53Δ2d | Ap (Em) | Deletion of xis2d from aa3 to end of deletion in orf3 on pGRW53. |
| pGRW53ΔC'exc | Ap (Em) | Deletion of aa406-aa666 leaving 405 N-terminal aa’ and, 30 C-terminal aa's of exc and all of rteR. |
| pGRW53Δexc | Ap (Em) | Deletion of 2010 nucleotides of the 5’ end of exc leaving 78 nucleotides at the 3’ prime. |
| Ptra mutations clones on the pMJF2 GUS fusion vector. | ||
| pJPARK100 | Ap (Em) | 0.5kb region upstream of traA, -bp138 to +bp352, relative to the transcriptional start site of tra operon, cloned in-frame to uidA gene on pMJF2 (10). Wild type region for GUS activity and gel shift assays. (this study) |
| pJPARK104 | Ap (Em) | pJPARK100 with mutations of -bp82 to -bp78 sequence upstream of Ptra start site. (this study). |
| pJPARK108 | Ap (Em) | pJPARK100 with mutations of –bp64 to –bp58 sequence upstream of Ptra start site. (this study). |
| pJPARK122 | Ap (Em) | pJPARK100 with mutations of –bp72 to –bp66 sequence upstream of Ptra start site. (this study). |
| pJPARK123 | Ap (Em) | pJPARK100 with mutations of –bp49 to –bp45 sequence upstream of Ptra start site. (this study). |
| pJPARK121 | Ap (Em) | pJPARK100 with mutations of –bp40 to –bp35 sequence upstream of Ptra start site. (this study). |
Antibiotic resistance abbreviations: Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Tc, tetracycline; Rif, rifampicin. The resistances within the parenthesis are expressed in Bacteroides strains and those outside the parenthesis are expressed in E. coli strains.
Protein Purification and Reagents
The purification of the Xis2d proteins was described elsewhere (Keeton and Gardner, 2012). Briefly, the Xis2d protein was overexpressed in E. coli BL21 (DE3) ihfA-and purified by a Heparin-Agarose Column. The Xis2d protein is approximately 80% pure. The Exc protein was prepared as previously described and is >95% pure (Sutanto et al., 2002).
The Xis2c protein was overexpressed in E. coli Rosetta (DE3) pLysS ihfA. Cells were grown to an optical density of 0.6 at A600 nm at 30°C and induced with 1mM IPTG. After induction, cells were grown at 25°C for 20 minutes. Then, rifampicin was added to a concentration of 200 ug/ml and the cells were shaken for 2 hours and pelleted by centrifugation. A 500ml cell pellet was resuspended in 5 ml of low salt sodium phosphate buffer (50 mM sodium phosphate, pH 7.0, 600 mM NaCl, 1 mM EDTA pH 7.0, 5% glycerol, 1 mM dithiothreitol (DTT), a Roche Complete EDTA-free protease inhibitor tablet, and lysozyme at 1mg/ml. Cells were sonicated and centrifuged at 10000 rpm for 30 minutes. The supernatant was loaded onto a HiTrap SP HP column (GE Life Sciences) and washed with 5 column volumes of low salt sodium phosphate buffer. A salt linear gradient ranging from 600mM to 2M NaCl was used for elution and Xis2d eluted from the column at approximately 1.3M NaCl. Active fractions were immediately dialyzed in Xis2c storage buffer (50 mM sodium phosphate, pH 7.0, 0.25M NaCl, 1 mM EDTA pH 7.0, 40% glycerol, 1 mM DTT) 2 times for 2 hours and overnight. Activity was verified by EMSA analysis. The supernatant was aliquoted, frozen in a dry ice in ethanol bath, and stored at -80°C. The identity of Xis2c was confirmed by mass spectrometry performed at the University of Illinois Mass Spectrometry lab. The protein is approximately 90% pure. An Agilent Technologies’ QuikChange XL site-directed mutagenesis kit was used for site-directed mutagenesis. PCR reactions were performed with KOD Hot Start DNA Polymerase from Novagen. The 100 bp Tri-Dye ladder is from NEB. Sequencing of the plasmids was done by either the UIUC Core Sequencing Facility or ACGT, Inc. Oligonucleotides were synthesized by Integrated DNA Technologies, Inc.
Construction of Deletions of the Excision Operon
The plasmid pHopp1 was constructed from the plasmid pAFD1 and encodes the entire excision operon PE-xis2c-xis2d-orf3-exc (Smith, 1985). A series of plasmids was created containing different deletions in the excision operon to test the effect on the activation of the Ptra::uidA fusion vector, pGW40.5, which is a translational fusion of the traA gene and the E. coli β-glucuronidase (Gus) reporter gene. The Ptra promoter was cloned upstream of the ATG start codon of the uidA gene (Gardner et al., 1996; Jeters et al., 2009; Valentine, 1988). The plasmid pGRW53 encodes the entire excision operon PE-xis2c-xis2d-exc but contains a deletion of the orf3 gene. The plasmid pGRW53Δ2c contains an in-frame deletion of amino acids 4-91 of the xis2c gene. To make an in-frame deletion of the xis2d gene, a construct was made which deleted the xis2d gene from amino acid 3 to the end of the orf3 gene to generate pGRW53Δ2d. An in-frame deletion of the exc gene was created leaving only 78 bp of the C-terminal amino acids to form pGRW53Δexc.
Construction of Site-directed Mutants in the Ptra Promoter
The Ptra promoter region is 490 bp containing 164 bp upstream of the transcription start site (Jeters et al., 2009). To test the effects of various mutations in the region upstream of the -33 region of the Ptra promoter, a series of 5-7 bp site-directed substitutions were constructed in the Ptra promoter from -82 to -35 of the E. coli- Bacteroides shuttle vector, pMJF2, to create an in-frame Ptra::uidA translational fusion.
Transfer of Shuttle Vectors
Vectors were transferred from E. coli DH5αMCR to Bacteroides recipients by HB101 RP4 donors during a triparental matings using nitrocellulose filters on BHIS agar plates incubated aerobically as previously described (Wang et al., 2004). Depending on the experiment being performed, the Bacteroides thetaiotaomicron recipients contained either CTnERL which is very closely related to CTnDOT or BT4001 QABC, which contains the tetQ-rteA-rteB-rteC genes integrated in the chromosome with the plasmid pGW40.5 (Jeters et al., 2009).
Gus Assays
The Gus assays were performed in one of two ways. For testing the effects of the plasmids containing deletions of the excision operon, Gus assays was performed as described in Feldhaus et al (Feldhaus et al., 1991). One unit (U) was defined as 0.01A415 (absorbance at 415nm) per minute at 37 C and is expressed as U/mg protein. Protein concentrations were determined by the method of Lowry et al (Lowry et al., 1951). When we tested the effects of the site directed mutants of the Ptra::uidA mutations in the pMJF2 vectors, Gus assays were performed on chloroform permeabilized cells adapted from Jefferson et al, 1986 (Jefferson et al., 1986). Bacteroides cultures were grown to an O.D. of 0.6 at A650 and 1ml of culture was resuspended in 1ml Gus buffer (50mM phosphate buffer pH 7.0 with 14 mM β-mercaptoethanol) pre-warmed to 37°C. The cells were permeabilized by adding 20ul of 0.1% SDS and 40ul of chloroform and vortexed for 10 seconds. All samples were incubated for 5 minutes at 37°C followed by adding para-4-nitrophenylβ –D-glucuronide to a final concentration of 1mM and vortexed for 5 seconds. After the reaction turned yellow, it was stopped with 2.5M 2-amino-2methyl-1,3-propanediol solution and the time (T) was recorded. Samples were centrifuged for 5 minutes and the OD at A415 was determined. Gus activity was calculated as 1000x A415/Time stopped x A650.
Electrophoretic mobility shift assays (EMSAs)
Electrophoretic mobility shift assays (EMSAs) were performed using a 182 bp fragment of the Ptra promoter containing the sequence from -118 to +65 using the primers GCCCTTAAAGCGGTATCGGG and GCCCGGATTGCTGTCCGAACATCC. A 182 bp non-specific DNA was generated from phage P22 DNA using the primers GGTAGGCATACGGAAGTTAAAGTGCGG and TCTCTAGGCCACCGCAAATTGAACTGCGG. Finally, to test the effects of these mutations, a 138 bp fragment was constructed using the primers GGTATCGGACAGATGCTTGACCC and GGTGTGCTTTCAGTCCTTCATCC.
EMSAs were performed in buffer consisting of 50 mM Tris-HCl (pH 8), 1 mM EDTA, 50 mM NaCl, 10% glycerol, and 0.18 mg/ml of heparin. Substrates were incubated with various concentrations of protein for 20 minutes and then loaded onto a prerun 6% DNA retardation gel from Invitrogen. Gels were stained with Sybergreen for 40 minutes and the DNA fragments were visualized using a BioRad transilluminator.
RESULTS
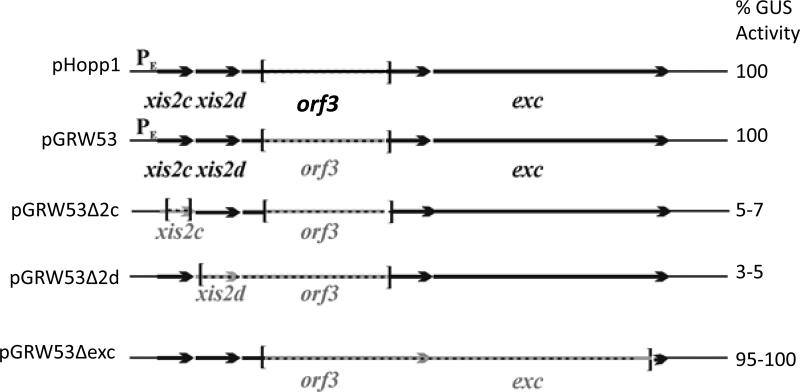
The tetracycline regulated excision and transfer of CTnDOT has been shown previously to require genes encoded within the excision operon. There are 4 proteins encoded in the operon, Xis2c, Xis2d, Orf3, and Exc (Jeters et al., 2009; Li et al., 1995; Whittle et al., 2002). It was unclear which one or a combination of these 4 proteins was responsible for activation. A series of plasmids was constructed containing in-frame deletions of the xis2c, xis2d, orf3, and exc genes and tested for the ability to activate expression of the tra operon (Figure 1). These plasmids were mated into BT4001ΩQABC, which contains the tetQ-rteA-rteB-rteC regulatory genes of CTnDOT site specifically integrated into the chromosome (Whittle et al., 2002). This strain also contains the plasmid pGW40.5 which contains the 490 bp promoter region Ptra promoter fused to the uidA gene, which is a translational fusion of the traA gene and the Gus reporter gene. Cells were grown with tetracycline to induce expression of the excision genes. The plasmid pHopp1 contains the entire excision operon and Gus assays produced wild type levels of activation. The first construct, pGRW53, which contains an in frame deletion of the orf3 gene, yielded the same level of Gus activity as the pHopp1. This shows that Orf3 is not required for activation of the Ptra promoter when Xis2c and Xis2d are present. Since Orf3 was not required for the excision reaction and is not required for activation of the Ptra promoter, we used constructs that contained deletions of the orf3 gene.
Figure 1.
In-frame deletions were made in the Exc operon contained on pGRW53 and the resultant constructs were tested for their abilities to induce activity of the Ptra ::uidA fusion pGW40.5 in the Bacteroides host, BT4001ΩQABC. The GUS activity was measured when the strains were grown with and without tetracycline. The range of the tetracycline induced GUS activities for each isolate relative to the pHopp1 is seen on the right of the schematic for each construct tested. The schematics depict the genes on pGRW53 and then the in-frame deletions of the gene being tested. Plasmids containing deletions in the orf3 gene had no phenotype in these experiments.
When plasmids containing deletions of the xis2c or xis2d genes were included with tetracycline, there was a 20 fold decrease in activation of the Ptra operon returning levels to background (Figure 1). However, plasmids containing deletions of the exc gene activated expression of the promoter at the same level as the plasmid pHopp1. This was surprising because it was previously shown that the exc gene is required for the entire process of transfer and mobilization (Whittle et al., 2002). However, this may suggest that the function of the Exc protein occurs at a different point during transfer and mobilization and not during activation of the promoter.
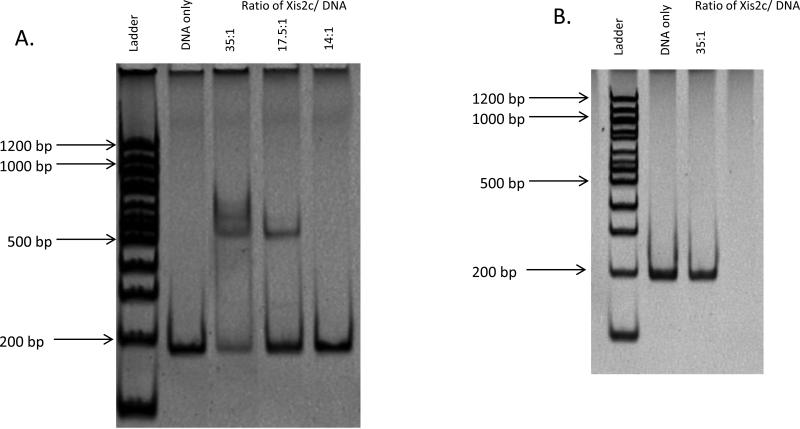
There are many mechanisms by which promoter activation may occur (Barnard et al., 2004; Beck et al., 2007). Of the many examples, there are at first two distinct overall patterns, activators that directly bind the promoter and those that can activate without binding DNA. To determine if the Xis2c and Xis2d proteins could bind the upstream promoter sequence, we performed EMSAs. The Xis2d protein was partially purified (greater than 80% pure) and was tested for the ability to shift the entire Ptra promoter region of 490bp. The Xis2d protein bound and shifted this fragment of DNA (data not shown). Binding was further narrowed down to a 182 bp fragment containing DNA from positions -118 to +65 of the tra promoter (Figure 2A). At low concentrations of Xis2d protein a single shift is observed. At higher concentrations of protein, multiple bands appear that may represent two or more binding sites. In other systems, such as phage lambda, the Xis protein binds 2 adjacent direct repeats to form a filament with a 3rd non-specific Xis protein binding between the two sites (Abbani et al., 2007). A similar binding pattern seen with the Xis2d protein on the Ptra promoter could explain the multiple bands made with the 182 bp fragment of the Ptra promoter region. An EMSA analysis of Xis2d using non-specific DNA from the bacteriophage P22 did not produce a shift suggesting that the shifts are specific (Figure 2B).
Figure 2.
A. EMSA of the Xis2d protein on the -118 to +65 region of the Ptra promoter. Lane 1 is the 100 bp Tri-Dye Ladder. In Lane 2, contains the 182 bp fragment of the Ptra promoter region. Lane 3 is an empty vector of the Xis2d protein. Lanes 4-8 contain shifts using different concentrations of the Xis2d protein. The ratio of Xis2d to DNA is shown above each lane. B. EMSA of the Xis2d protein on non-specific DNA. Lane 1 is the 100 bp Tri-Dye Ladder. Lane 2 is a DNA fragment of the same length as the Ptra promoter. Lane 3 contains Xis2d and non- specific DNA.
EMSAs using the partially purified Xis2c protein (greater than 90% pure) are shown in Figure 3. The shift results in two bands at higher concentrations which resolve to a single shift when the concentration of the Xis2c protein is decreased (Figure 3A). An EMSA analysis using non-specific DNA from P22 did not produce the same banding pattern at higher concentrations suggesting that the shifts are specific (Figure 3B). When the Xis2c and Xis2d proteins were incubated together on the Ptra promoter, a complex formed that prevented migration into the gel.
Figure 3.
EMSA analysis of the binding of the Xis2c protein to the -118 to +65 region of the Ptra promoter . The Xis2c protein is partially purified. Lane 1 contains the 100bp ladder from NEB. Lane 2 contains the DNA fragment of the Ptra promoter. Lane 3-5 contain decreasing concentrations of the Xis2c protein. At higher concentrations, two bands are detected (Lane 3). As the concentration of Xis2c is decreased, a single shift is detected (Lane 4). The shift of the DNA is lost at lower concentrations (Lane 5). B. EMSA of the Xis2c protein on non-specific DNA. Lane 1 is the 100 bp Tri-Dye Ladder. Lane 2 is a DNA fragment of the same length as the Ptra promoter. Lane 3 contains Xis2c and non- specific DNA.
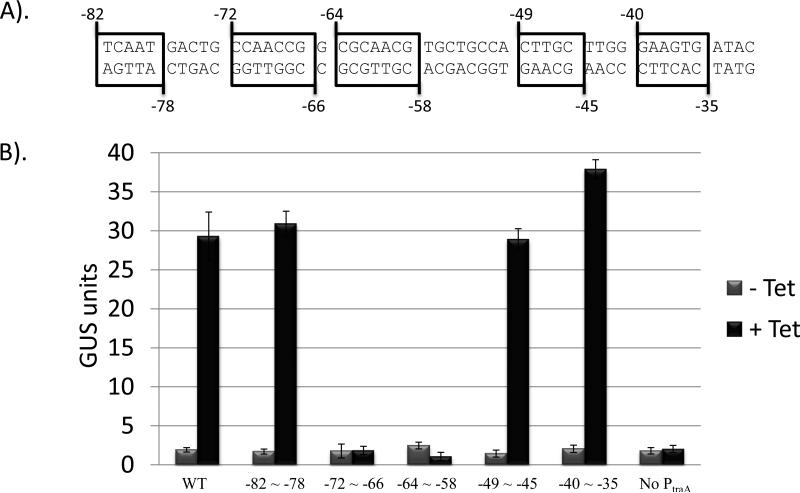
Since both Xis2c and Xis2d bind the Ptra promoter, it is possible that they may bind upstream of the Ptra promoter. The Ptra promoter is a 490 bp region containing 137 bp upstream of the transcription start site followed by 353 bp of DNA upstream of the traA gene (Jeters et al., 2009). To test the effects of various mutations in the region upstream of the -33 region of the Ptra promoter, a series of 5-7 bp site-directed substitutions were constructed in the Ptra promoter from basepairs -82 to -35 on the plasmid pJPARK100 (Figure 4). The plasmid, pJPARK100, contains the same translational fusion of the traA gene and the Gus reporter gene as in pGW40.5 but is in a different plasmid (Gardner et al., 1996; Jeters et al., 2009; Valentine, 1988). Plasmids were mated into BT4004, which contains a single integrated copy of CTnERL, a closely related transposon to CTnDOT which lacks the ermF gene. Tetracycline induces the regulatory cascade which activates expression of the CTnERL regulatory cascade which produces the excision genes. Activation of this promoter produces the wild-type Gus protein. A series of 5-7 bp substitutions were constructed in the plasmid pJPARK100 between the positions -82 to -35 of the Ptra promoter. Each plasmid contained 5-7 bp changed by to their complement and each plasmid constructs was tested for Gus activity (Figure 4). Mutations in the Ptra promoter in two plasmids extending from positions -72 to -58 resulted in loss of activation of the uidA gene when cells were induced with tetracycline. This region from -72 to -58 may include a possible binding site for Xis2d and an EMSA analysis using the Xis2d protein was conducted on each 138 bp fragment (Figure 5). The Xis2d protein was incubated with each fragment containing the mutations described above and none of the fragments shifted as well as the wild-type DNA fragment (Figure 5: Compare Lane 4 to Lanes 5-9). In addition, some of the shifts did not migrate to the same position as the wild-type fragment (Figure 5: Compare Lane 4 to Lane 6). Because Xis2d cannot shift non-specific DNA, the binding is specific suggesting that these shifts may represent multiple binding sites for the Xis2d protein.
Figure 4.
Tetracycline induced GUS activity of constructs of the Ptra ::uidA fusion containing mutations upstream of the Ptra promoter. A. A series of site directed substitutions were constructed in the Ptra promoter by changing the bases to their complement and the level of activity was determined in strains containing ΩCTnERL, BT4004. B. Induction of the uidA operon was tested with and without tetracycline. The GUS activity is expressed in modified Miller units.
Figure 5.
A. Systematic mutations were constructed in the region from -82 to -31 of the DNA fragment of the Ptra promoter. B. EMSA analysis of these fragments was preformed. Lane 1 contains the 100bp ladder from NEB. Lane 2 contains the DNA fragment of the Ptra promoter. Lane 3 contains the DNA fragment of the Ptra promoter with a protein preparation lacking Xis2d. Lane 5 contains the DNA fragment of the Ptra promoter shifted by the Xis2d protein. Lanes 5-9 contain the DNA fragment of the Ptra promoter with one of the mutations shown in A.
DISCUSSION
The Ptra promoter of CTnDOT is activated by the Xis2c and Xis2d proteins. These proteins have a dual role in excision and transfer. Presumably, following the addition of tetracycline to the cells, the excision operon is induced and the Xis2c and Xis2d proteins are produced. These proteins are required for excision of CTnDOT from the chromosome by binding the attL and attR sites and promoting formation of the excisive intasome (Keeton and Gardner, 2012). As demonstrated in this work, the Xis2c and Xis2d proteins also bind the Ptra promoter to induce expression of the tra genes which enable transfer of CTnDOT to the recipient. Surprisingly, the Exc protein is not involved in the activation step of the promoter of the tra operon. It was previously shown that the plasmid pLYL72 required the Exc protein for the entire process of transfer and mobilization (Whittle et al., 2002). However, the Exc function appears to be downstream of the activation of the Ptra promoter.
The mechanism of activation of the Ptra promoter is unknown. The Xis2c and Xis2d proteins have been shown to bind the upstream region from -118 to +65 by EMSA analysis. Each protein can bind independently to the DNA fragment and produce a shift containing multiple bands. This suggests either multiple binding sites or the ability to form a filament on the DNA similar to Lambda Xis (Abbani et al., 2007). The multiple shifts are specific for the DNA as non-specific DNA fragments do not produce the same shifting pattern. When Xis2c and Xis2d are added together, the resultant shift does not migrate into the gel preventing analysis of this complex.
To further narrow the location of the binding sites, multiple basepair substitutions were generated in the region from -72 to – 58 of Ptra promoter region. Basepair substitutions extending from positions -72 to -58 resulted in the loss of activity of the Ptra promoter suggesting this is a possible binding site of one of the proteins.
The most straightforward way activation may occur is by either Class I or Class II activation mechanisms. In Class 1 systems, the activator binds upstream of the promoter and makes a direct interaction with the RNAP α subunit which enables RNA polymerase to begin transcription (Busby S, 1994). In Class II systems, the activator binds the promoter and interacts with domain 4 of the RNAP σ subunit to initiate transcription (Dove et al., 2003). In both cases, the proteins directly bind the DNA to interact with the promoter. An example of this is the E. coli proP P2 promoter which is activated by Fis and CRP. CRP acts as a Class I activator while Fis acts as a Class II activator (McLeod et al., 2002).
An example of an RDF acting as a transcriptional activator is found in the phage P2 system. The Cox protein of the phage P2 is also a RDF that activates a promoter and is involved in excision. The Cox protein activates the PLL promoter on the P4 phage by binding upstream, but the mechanism of activation is unknown (Eriksson and Haggård-Ljungquist, 2000). It has two required binding regions; one with a higher affinity for the protein and a lower affinity site located upstream of the -35 region indicating a possible Class I mechanism of activation (Saha, 1989). Another example of a system where two transcriptional activators are involved in excision is in the ColEI plasmid system. In the case of the ColE1 plasmid, two transcriptional factors ArgR and PepA bind to the 180bp adjacent to the cer site where XerC and XerD catalyze recombination (Alen et al., 1997; Colloms et al., 1996). These proteins also function as transcriptional activators.
The primary amino acid sequence and the predicted secondary structures of Xis2c and Xis2d are similar to each other but share no homology with any other RDFs including the Cox protein. Since the amino acids in the Cox protein involved in activation or interaction with RNA polymerase are unknown, we cannot predict if Xis2c or Xis2d share similar residues although the proteins share similar functions.
Another possible model is that these proteins bind the DNA and promote the binding of another protein, such as a transcription factor, to the promoter enabling activation. Further mutational analysis of the Ptra promoter may identify binding sites that are not utilized by Xis2c and Xis2d which would show that other proteins may be involved in this activation process. The next step will be to determine the exact binding site of each protein and the mechanism by which the promoter is activated.
Highlights.
We identified the proteins responsible for activation of the transfer operon- Xis2c and Xis2d
Xis2c and Xis2d are involved in excision of CTnDOT
Xis2c and Xis2d also bind upstream of the pTra promoter and activate expression of the transfer operon
The other two proteins, Exc and Orf3, in the excision operon do not activate the pTra promoter
ACKNOWLEDGEMENTS
The work was supported by grant AI/GM 22383 and GA28717 from the National Institute of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abbani MA, et al. Structure of the cooperative Xis–DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly. Proceedings of the National Academy of Sciences. 2007;104:2109–2114. doi: 10.1073/pnas.0607820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard A, et al. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Current Opinion in Microbiology. 2004;7:102–108. doi: 10.1016/j.mib.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Beck LL, et al. Look, no hands! Unconventional transcriptional activators in bacteria. Trends in Microbiology. 2007;15:530–537. doi: 10.1016/j.tim.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Busby S, E. R. Promoter structure, promoter recognition, and transcription activation in procaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Cheng Q, et al. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J Bacteriol. 2000;182:4035–43. doi: 10.1128/jb.182.14.4035-4043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, et al. Identification of genes required for excision of CTnDOT, a Bacteroides conjugative transposon. Mol Microbiol. 2001;41:625–32. doi: 10.1046/j.1365-2958.2001.02519.x. [DOI] [PubMed] [Google Scholar]
- Costello EK, et al. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SL, et al. Region 4 of σ as a target for transcription regulation. Molecular Microbiology. 2003;48:863–874. doi: 10.1046/j.1365-2958.2003.03467.x. [DOI] [PubMed] [Google Scholar]
- Eriksson JM, Haggård-Ljungquist E. The Multifunctional Bacteriophage P2 Cox Protein Requires Oligomerization for Biological Activity. Journal of Bacteriology. 2000;182:6714–6723. doi: 10.1128/jb.182.23.6714-6723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhaus MJ, et al. Use of an Escherichia coli beta-glucuronidase gene as a reporter gene for investigation of Bacteroides promoters. J Bacteriol. 1991;173:4540–3. doi: 10.1128/jb.173.14.4540-4543.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, et al. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed beta-1,4-D-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Appl. Environ. Microbiol. 1996;62:196–202. doi: 10.1128/aem.62.1.196-202.1996. PMCID: PMC167786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdeman LV, Moore WEC. Anaerobe laboratory manual. Virginia Polytechnical Institute and State University; Blacksburg: 1975. [Google Scholar]
- Jefferson RA, et al. beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proceedings of the National Academy of Sciences. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeters RT, et al. Tetracycline-associated transcriptional regulation of transfer genes of the Bacteroides conjugative transposon CTnDOT. J Bacteriol. 2009;191:6374–82. doi: 10.1128/JB.00739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton CM, Gardner JF. Roles of Exc Protein and DNA Homology in the CTnDOT Excision Reaction. Journal of Bacteriology. 2012;194:3368–3376. doi: 10.1128/JB.00359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Hatfull GF. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 2001;29:2205–16. doi: 10.1093/nar/29.11.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LY, et al. Location and characteristics of the transfer region of a Bacteroides conjugative transposon and regulation of transfer genes. Journal of Bacteriology. 1995;177:4992–9. doi: 10.1128/jb.177.17.4992-4999.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, et al. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- McLeod SM, et al. The C-terminal domains of the RNA polymerase α subunits: contact site with fis and localization during co-activation with CRP at the Escherichia coli proP P2 promoter. Journal of Molecular Biology. 2002;316:517–529. doi: 10.1006/jmbi.2001.5391. [DOI] [PubMed] [Google Scholar]
- Moon K, et al. Regulation of excision genes of the Bacteroides conjugative transposon CTnDOT. J Bacteriol. 2005;187(16):5732–41. doi: 10.1128/JB.187.16.5732-5741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W, et al. Some current concepts in intestinal bacteriology. The American Journal of Clinical Nutrition. 1978;31:33S–42S. doi: 10.1093/ajcn/31.10.S33. [DOI] [PubMed] [Google Scholar]
- Park J, Salyers AA. Characterization of the Bacteroides CTnDOT Regulatory Protein RteC. Journal of Bacteriology. 2011;193:91–97. doi: 10.1128/JB.01015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Haggård-Ljungquist E, Nordstrom K. Activation of prophage P4 by the P2 Cox protein and the site of action of the Cox protein on the two phage genomes. Proc. Natl. Acad. Sci. 1989;86:3973–3977. doi: 10.1073/pnas.86.11.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ. Development and use of cloning sustems for Bacteroides fragilis: cloning of a plasmid-encoded clindamycin resistance determinant. J. Bacteriol. 1985;164:294–301. doi: 10.1128/jb.164.1.294-301.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens AM, et al. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J. Bacteriol. 1990;172:4271–4279. doi: 10.1128/jb.172.8.4271-4279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutanto Y, et al. Characterization of Exc, a novel protein required for the excision of Bacteroides conjugative transposon. Mol Microbiol. 2002;46(5):1239–46. doi: 10.1046/j.1365-2958.2002.03210.x. [DOI] [PubMed] [Google Scholar]
- Valentine PJ, Shoemaker NB, Salyers AA. Mobilization of Bacteroides plasmids by Bacteroides conjugal elements. J. Bacteriol. 1988;170:1319–1324. doi: 10.1128/jb.170.3.1319-1324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. Translational control of tetracycline resistance and conjugation in the Bacteroides conjugative transposon CTnDOT. J Bacteriol. 2005;187:2673–80. doi: 10.1128/JB.187.8.2673-2680.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT. J Bacteriol. 2004;186:2548–57. doi: 10.1128/JB.186.9.2548-2557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle G, et al. Characterization of genes involved in modulation of conjugal transfer of the Bacteroides conjugative transposon CTnDOT. J Bacteriol. 2002;184(14):3839–47. doi: 10.1128/JB.184.14.3839-3847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]