Abstract
Significant circadian variations exist in the frequency of cardiac arrhythmia, but few studies have examined the relation between cardiac ion channels genes and biological clocks. We investigated this relation using suprachiasmatic nuclei lesion (SCNX) and pharmacological autonomic nervous system block (ANSB) mice. Significant 24-h variations were observed in the expression of clock genes Per2, Bmal1, and Dbp and ion channel genes KCNA5, KCND2, KCHIP2, and KCNK3 in the control mice hearts. In the SCNX mice, all genes examined lost circadian rhythm. In the ANSB mice, the expressions of the three clock genes were dampened significantly but still had circadian rhythm, whereas the four ion channel gene expressions lost rhythm. Heart rate also lost circadian rhythm in both the SCNX and ANSB mice. These results suggest that some ion channel gene expressions might be regulated by the central clock in the SCN through the ANS but not the peripheral clock in the heart.
Keywords: arrhythmia, suprachiasmatic nuclei, autonomic nervous system, circadian rhythm
1. Introduction
In mammals, the master circadian pacemaker resides in the suprachiasmatic nuclei (SCN) of the hypothalamus, and slave oscillators exist in peripheral organs. Output signals from the master pacemaker drive and synchronize the circadian oscillation of various behavioural and physiological processes through autonomic and/or neurohumoural activation. When the SCN is destroyed, the temporal programs are disturbed (LeSauter and Silver 1998).
Apparent circadian variations exist in cardiac arrhythmia, and they have been posited to be associated with the circadian expression of ion channels or physiological responses to autonomic nervous function, evidenced by the multisynaptic autonomic connection from SCN neurons to the heart (Inoue and Zipes 1987; Scheer et al. 2001). Yamashita et al. (2003) demonstrated that two K+ channel genes (KCNA 5 and KCND2) in rat heart show significant circadian rhythm in their mRNA levels, and these circadian rhythms were modulated by a pharmacological block of the autonomic nervous system (ANS). However, there is no evidence of regulation of rat cardiac gene expression by the biological clock, and it is also unclear whether such a circadian expression of cardiac genes is a universal phenomenon. The goals of the present study were (1) to determine whether other cardiac ion channel genes, in addition to KCNA5 and KCND2, also exhibit circadian oscillation in mouse heart, and (2) to evaluate the effect of an SCN lesion and ANS blockade on the expressions of the cardiac ion channels and clock genes, and electrocardiographic parameters.
2. Methods
Experimental animal care was conducted with permission from the Experimental Animal Welfare Committee of the Fujita Health University School of Medicine.
2.1 Animals
Ninety 4–6-week-old ddy mice (Chubu Kagaku Shizai Co., Ltd., Naogya) were housed individually and maintained under a 12 h/12 h light/dark cycle (light on: 8 am, CT0; light off: 8 pm, CT12) for 2 weeks and allowed access to food and water ad libitum. The animals were sacrificed at 6-h intervals in a single day. The right and left atrium and apex of the left ventricle were quickly removed and frozen in the liquid nitrogen at — 80°C. The samples for mRNA isolation were kept in RNA stabilization solution (RNAlater, Life Technologies, Carlsbad, CA, USA).
2.2 Telemetry transmitter implantation
Implantation of the telemetry transmitter in mice (n = 18) was performed with an intraperitoneal injection of ketamine/xylazine (125 mg/kg, 50 mg/kg). The transmitter was fixed on the back of the mouse with its two electrodes placed on the bilateral chest subcutaneously. The mouse was then put in an individual cage and placed on the transmitter's receiver board and allowed to recover. Each mouse's body temperature and electrocardiogram (ECG) were measured every 6 h. Heart rate was determined as the average of consecutive 20 beats of initiation of every 6 h. The QT interval was defined as the time between the QRS onset and the point at which the isoelectric line intersected a line drawn tangentially to the maximal downslope of the T wave. Data were collected by a DSI Dataquest A.R.T.2.3 system at the sampling rate of 1 kHz (Data Sciences International, New Brighton, MN, USA).
2.3. SCNX and ANS block
Bilateral thermal lesions of the SCN were performed stereotaxically (Narishige Co., Tokyo) under ketamine/hydrazine anaesthesia. A stainless steel electrode (0.35 mm diameter) was inserted in the SCN (0.2 mm posterior and 0.0 mm lateral to the bregma, 6.2 mm below the skull surface) using a lesion-making device (Ugo Basile Biological Research Apparatus; Ugo Basile, Comerio VA, Italy). A 2-mA current was passed for 4 s. The sham operation was performed by inserting the lesion needle into the brain without passing a current. The body temperature, heart rate and body weight of the mice were monitored for 3 weeks after the lesion date. The mice that lost circadian rhythm in both heart rate and body temperature were used (Figure 1). A mouse was excluded from the study when its body weight increased more than 10 g or when it died less than 14 days before experiment. The autonomic nervous system block (ANSB) was performed by intraperitoneal injection of atropine (0.5 mg/kg) and propranolol (1 mg/ kg) every 6 h for 2 weeks. Mice of this group did not receive an SCN lesion. On the 15th day, mice were sacrificed at 6-h intervals for sampling (24 mice in each group).
Figure 1.
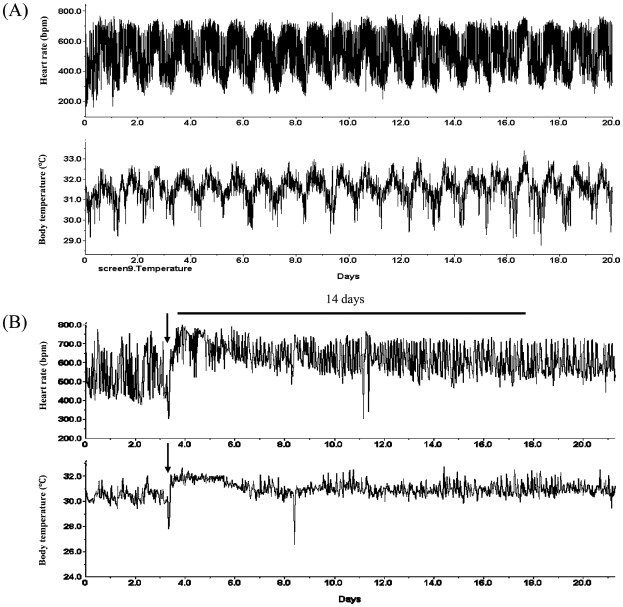
Confirmation of the successful suprachiasmatic nucleus (SCN) lesion. (A) Sham-operated mice: Substantial circadian rhythms in the heart rate and body temperature were observed. (B) SCN-lesioned mice: The arrow indicates the date that the SCN was lesioned. The SCN lesion was considered successful if the loss of circadian rhythms of heart rate and body temperature persisted for more than 14 days.
2.4. RNA preparation and real-time quantitative RT-PCR
The heart was removed from sham-operated, SCN-lesion and ANSB mice under ketamine/hydrazine anaesthesia. Total RNA was extracted from the samples by using R Neasy® Fibrous Tissue Mini kit (Qiagen, Hiden, Germany) and then reverse-transcribed and amplified by using real-time Taq Man® technology and analysed on an ABI PRISM7900 (Applied Biosystems, Foster, CA, USA). The primer pairs were designed from the mouse sequences available in GenBank, including three clock genes (Emal1, Per2, and DBP) and four K+ channel genes: KCNA5 (ultrarapid delayed rectifier K+ current), KCND2 (α subunit of transient outward current), KCHIP2 (β subunit of transient outward current), and KCNK3 (cardiac two-pore background K+ channel). PCR was executed under the following conditions: cDNA synthesis at 25°C for 10 min and then 37°C for 120 min, PCR amplification for 40 cycles at in a two-step method with denaturation at 95°C for 15 s, and annealing and extension at 60°C for 1 min. The target clock gene cDNA was co-amplified with GAPDH cDNA in a single PCR tube. The PCR data are reported as the number of transcripts per number of GAPDH. The sequences of PCR primers and sequence-specific probes are presented in Table 1.
Table 1.
Sequences of PCR primers and sequence-specific probes.
| Target sequence | Accession no. | Assay ID of Primers and TaqMan® Probe | Amplicon length (bp) |
|---|---|---|---|
| Per2 | NM-011066 | Mm00478113-m1 | 73 |
| Bmail1 | NM-007489 | Mm00500226-m1 | 87 |
| Dbp | NM-016974 | Mm00497539-m1 | 57 |
| KCNA5 | NM-145983 | Mm00524346-s1 | 61 |
| KCND2 | NM-019697 | Mm00498065-m1 | 88 |
| KCHIP2 | NM-030716 | Mm00518914-m1 | 74 |
| KCNK3 | NM-010608 | Mm00807036-m1 | 128 |
2.5. DNA microarray
Total RNA was isolated using an RNeasy kit (Qiagen, Valencia, CA, USA) and assessed for integrity using an RNA 6000 Nano Assay (Agilent Technologies, La Jolla, CA, USA). Sample amplification labelling and hybridization essentially followed the protocol recommended by the manufacturer. Briefly, high-quality RNA samples acquired from non-treated human umbilical vein endothelial cells (HUVEC) and Bmal/Clock-treated HUVEC were labelled with cyanine 3 (Cy3) and cyanine 5 (Cy5) using an Agilent Low-input RNA Fluorescent Linear Amplification kit. Hybridizations were performed on Agilent Human 1A (V2) Oligo Microarray slides, and a dye-swap experimental design was applied. Arrays were scanned on an Agilent Microarray Scanner (model GMS417 Array scanner, Affimetrix, Santa Clara, CA). The raw images were processed using Agilent's Feature Extraction software, and their analysis was conducted using Agilent Genespring software. The genes that were upregulated by fivefold or more in Bmal/Clock-treated HUVEC samples but not in nontreated HUVEC samples were selected.
2.6. Statistical analysis
The 24-h variations in the mean m RNA levels of the studied genes were double-plotted and analysed using an algorithm for least-squares cosine-curve fitting. The properties of the circadian rhythms were quantified by the following four parameters:
-
(1)
significance of rhythm (p-value for the hypothesis of zero-rhythm amplitude),
-
(2)
midline-estimating statistic of rhythm (MESOR, a rhythm-adjusted 24-h time-series mean),
-
(3)
amplitude (one-half of the peak-to-trough amplitude by fitting with a single cosine-curve approximation), and
-
(4)
acrophase (crest time of the peak fitted approximation to rhythm referenced to local midnight).
A circadian rhythm was considered to exist when the null hypothesis regarding the significance of rhythm was <0.05. The gene expressions of 24-h components (MESOR, amplitude, and acrophase) among the three treatments (sham-operated, SCNX, and ANSB) were analysed by one-way analysis of variance (ANOVA). After a demonstration of significant differences among treatments by ANOVA, post hoc comparisons between treatment pairs were made by the Bonferroni multiple comparison procedure. Quantitative data are expressed as means ± standard deviation (SD) values. A two-tailed p-value of <0.05 was considered significant.
3. Results
3.1. Screening for candidate cardiac gene by DNA microarray
The DNA microarray showed that KCNA5, KCND2, KCNK3, KCNJ12 and ATP2A2, SLC9A3R2 were augmented by Bmal1/Clock induction (data not shown). Among those six genes, KCNA5, KCND2, KCHIP2 and KCNK3 exhibited significant circadian variation and were examined further.
3.2. Clock gene expression in the mouse heart
The sham-operated mice showed significant circadian rhythm in the three clock genes of the atrium (Figure 2A, Table 2). The SCNX mice lost circadian variations of the expression in all clock genes. In the ANSB mice, the circadian rhythm of all clock gene expressions was preserved, and Dbp expression was significantly reduced. Also, the expression of the Dbp gene in both the SCNX and ANSB mice was significantly lower than that in the sham-operated mice. Neither SCNX nor ANSB influenced the expression level of Per2.
Figure 2.
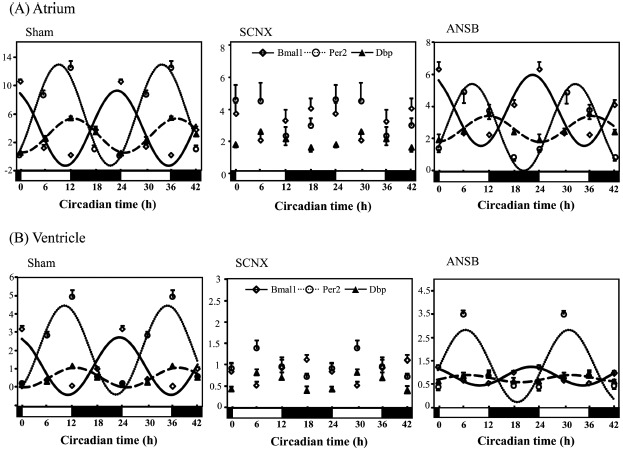
Influence of SCNX and ANSB on the mRNA level of clock genes in the mouse heart (for each group, n = 3–6). Clock gene expressions in the atrium (A) and ventricle (B). All data were normalized against GAPDH and double-plotted. The solid line or dotted line indicates circadian rhythm fitted by least-squares cosine-curve fitting. Open diamonds, Email expression; open circles and closed triangles, Per2; closed squares, Dbp. The open and closed bars under the x-axis represent the light and dark periods, respectively, in one day. The expression of all of the clock genes lost circadian rhythm in the SCNX group and was dampened significantly in the ANSB group. ANSB, autonomic nervous system block; SCNX, suprachiasmatic nucleus lesioned.
Table 2.
Clock gene expressions in the mouse heart.
| Gene | Treatment | Site | MESOR | Amplitude | Acrophase (h) |
|---|---|---|---|---|---|
| Bmal1 | Sham | Atrium | 4.0 ± 0.1§* | 5.4 ± 0.1§*# | 23.2 ± 0.3 |
| Ventricle | 1.2 ± 0.1§*# | 1.7 ± 0.1§*# | 23.2 ± 0.2 | ||
| SCNX | Atrium | 2.9 ± 0.3§ | 1.5 ± 0.2§ | N/A | |
| Ventricle | 0.7 ± 0.1§ | 0.4 ± 0.1§ | N/A | ||
| ANSB | Atrium | 3.8 ± 0.2§ | 2.2 ± 0.3§ | 22.5 ± 0.1 | |
| Ventricle | 0.9 ± 0.1§ | 0.4 ± 0.1§ | 22.1 ± 0.4 | ||
| Per2 | Sham | Atrium | 2.8 ± 0.3 | 2.5 ± 0.1§*# | 12.8 ± 0.4 |
| Ventricle | 0.6 ± 0.1 | 0.5 ± 0.1§*# | 12.6 ± 0.3# | ||
| SCNX | Atrium | 2.0 ± 0.2 | 1.3 ± 0.2§ | N/A | |
| Ventricle | 0.6 ± 0.1 | 0.3 ± 0.1§ | N/A | ||
| ANSB | Atrium | 2.6 ± 0.4 | 0.9 ± 0.4§ | 11.5 ± 1.2 | |
| Ventricle | 0.7 ± 0.1 | 0.04 ± 0.01§ | 7.2 ± 2.3 | ||
| Dbp | Sham | Atrium | 5.8 ± 0.3§*# | 7.2 ± 0.6§*# | 9.9 ± 0.2 |
| Ventricle | 1.9 ± 0.1§*# | 2.2 ± 0.2§*# | 9.9 ± 0.3# | ||
| SCNX | Atrium | 3.6 ± 0.4§ | 2.1 ± 0.4§ | N/A | |
| Ventricle | 1.0 ± 0.1§ | 0.5 ± 0.1§γ | N/A | ||
| ANSB | Atrium | 2.7 ± 0.2§ | 2.4 ± 0.4§ | 8.0 ± 0.4 | |
| Ventricle | 1.3 ± 0.1§ | 1.5 ± 0.1§ | 6.6 ± 0.1 |
Note: Data are mean ± SEM for 3–6 mice per group. MESOR, midline estimating statistic of rhythm; Sham, sham-operated; SCNX, suprachiasmatic nucleus lesioned; ANSB, autonomic nervous system block; N/A, not applicable.
One-way ANOVA p < 0.05, Bonferroni/Dunn, p < 0.05.
Sham vs. lesion;
Sham vs. ANSB;
Lesion vs. ANSB.
In the ventricle, the sham-operated mice showed significant circadian rhythm in the three clock genes (Figure 2B, Table 2). The SCNX mice lost circadian variation of the expression of all three clock genes. In the ANSB mice, the circadian rhythm of the three clock gene expressions was preserved, and the Dbp expression was significantly phase-advanced. The MESOR values for Bmal1 and Dbp were significantly different among the three treatment groups, whereas the expression of Per2 was not influenced by the SCNX or ANSB. The MESOR values for the three clock genes in the atria of the sham-operated mice were significantly larger than those in the ventricles (Bmal 1, p < 0.05; Per2, p < 0.05; and Dbp, p < 0.05).
3.3. Effects of SCNX and ANSB on ion channel gene expression
The sham-operated mice exhibited substantial circadian rhythms in all four cardiac ion channel genes (KCNA5, KCND2, KCHIP2, and KCNK3) in both the atrium and ventricle (Figure 3). The SCNX eliminated the circadian rhythm of all of these cardiac genes in the atrium (Figure 3A, Table 3). Of note, SCNX increased the MESOR in KCND2 and KCNK3 expressions. ANSB, similarly, abolished circadian rhythms of all four ion channel genes, and the MESOR values of all cardiac gene expressions were increased significantly in the atrium.
Figure 3.
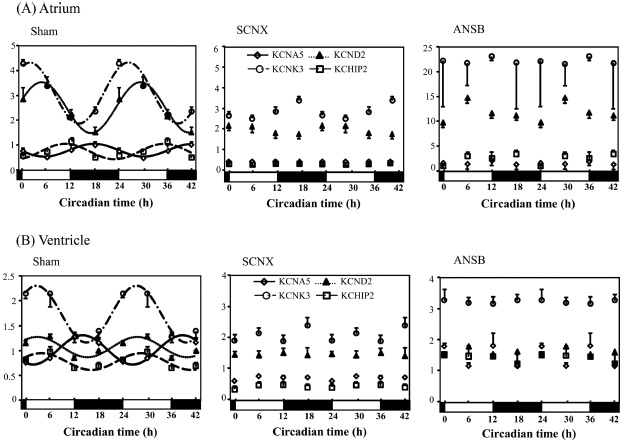
Cardiac ion channel genes expressions were similarly altered in the SCNX and ANSB mice (for each group, n = 3–6). Cardiac ion channel genes expressions in the atrium (A) and ventricle (B). All data were normalized against GAPDH and double-plotted. The solid line or dotted line indicates circadian rhythm fitted by least-squares cosine-curve fitting. Open diamonds, KCNA5; closed triangles, KCND2; open squares, KCHIP2; open circles, KCNK3. The open and closed bars under the x-axis represent the light and dark periods, respectively, in one day. All four ion channel gene expressions lost circadian rhythm in the SCNX and ANSB mice.
Table 3.
Cardiac ion channel gene expressions in the mouse heart.
| Gene | Treatment | Site | MESOR | Amplitude | Acrophase (h) |
|---|---|---|---|---|---|
| KCNA5 | Sham | Atrium | 0.8 ± 0.1§*# | 0.4 ± 0.1§* | 15.6 ± 0.6 |
| Ventricle | 1.0 ± 0.1§*# | 0.3 ± 0.1 | 13.2 ± 0.2 | ||
| SCNX | Atrium | 0.5 ± 0.1§γ | 0.1 ± 0.1§ | N/A | |
| Ventricle | 0.7 ± 0.1§γ | 0.1 ± 0.1 | N/A | ||
| ANSB | Atrium | 1.5 ± 0.2§ | 0.3 ± 0.3§ | N/A | |
| Ventricle | 1.5 ± 0.1§ | 0.2 ± 0.1 | N/A | ||
| KCND2 | Sham | Atrium | 2.8 ± 0.1§* | 1.2 ± 0.2§* | 5.4 ± 0.4 |
| Ventricle | 1.1 ± 0.1§ | 0.2 ± 0.1 | 3.0 ± 0.3 | ||
| SCNX | Atrium | 1.8 ± 0.1§γ | 0.6 ± 0.1§γ | N/A | |
| Ventricle | 1.5 ± 0.1§ | 0.2 ± 0.1 | N/A | ||
| ANSB | Atrium | 11.7 ± 0.8§ | 2.9 ± 0.4§ | N/A | |
| Ventricle | 1.6 ± 0.1§ | 0.1 ± 0.1 | N/A | ||
| KCHIP2 | Sham | Atrium | 0.7 ± 0.1§# | 0.4 ± 0.1§ | 11.1 ± 0.9 |
| Ventricle | 0.8 ± 0.1§*# | 0.3 ± 0.2 | 12.2 ± 1.1 | ||
| SCNX | Atrium | 0.4 ± 0.1§γ | 0.1 ± 0.1§γ | N/A | |
| Ventricle | 0.6 ± 0.1§γ | 0.2 ± 0.1 | N/A | ||
| ANSB | Atrium | 2.4 ± 0.5§ | 0.9 ± 0.6§ | N/A | |
| Ventricle | 1.4 ± 0.1§ | 0.1 ± 0.1 | N/A | ||
| KCNK3 | Sham | Atrium | 3.1 ± 0.1§# | 1.2 ± 0.1§# | 1.7 ± 0.8 |
| Ventricle | 2.2 ± 0.1§# | 0.4 ± 0.1§ | 16.8 ± 2.5 | ||
| SCNX | Atrium | 3.0 ± 0.1§γ | 0.7 ± 0.1§γ | N/A | |
| ANSB | Ventricle | ||||
| Atrium | 2.0 ± 0.1§γ | ||||
| 22.0 ± 3.2§ | 0.5 ± 0.1§γ | ||||
| 10.5 ± 2.3§ | N/A | ||||
| N/A | |||||
| Ventricle | 3.2 ± 0.1§ | 0.1 ± 0.1§ | N/A |
Note: Data are mean ± SEM for 3–6 mice per group. Abbreviations are as in Table 1.
One-way ANOVA p < 0.05, Bonferroni/Dunn, p < 0.05.
Sham vs. lesion;
Sham vs. ANSB;
Lesion vs. ANSB.
In the ventricle, as in the atrium, both SCNX and ANSB abolished the circadian rhythm of all four ion channel genes (Figure 3B, Table 3). The MESOR values of all gene expressions except KCND2 were significantly increased in the ANSB mice, and the expression of KCNA5 was significantly reduced in the SCNX mice.
3.4. Effects of SCNX and ANSB on cardiac electrophysiology
The sham-operated mice exhibited substantial circadian rhythms in the heart rate and QT interval, whereas in the SCNX and ANSB mice, the circadian rhythms of the heart rate and QT interval were lost (Figure 4, Table 4). Interestingly, the MESOR values for heart rate in the SCNX mice (669 ± 21 bpm) were significantly higher than those in the ANSB mice (466 ± 30 bpm, p < 0.05). The MESOR values for the QT interval in the ANSB mice were, therefore, longer than those in the SCNX mice (51 ± 2 ms vs. 37 ± 1 ms, p < 0.05).
Figure 4.
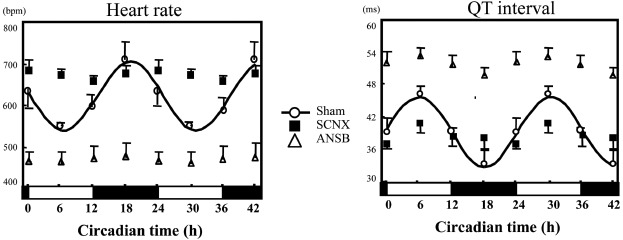
Heart rate and QT interval lost circadian rhythm in both SCNX and ANSB mice (for each group, n = 3–6). Heart rate (beats per min) and QT interval (ms) were recorded every 6 h and analysed by the least-squares cosine-curve fitting. The solid line indicates circadian rhythm fitted by least-squares cosine-curve fitting. Open circles, sham-operated mice; open triangles, ANSB mice; closed squares, SCNX mice. The open and closed bars under the x-axis represent the light and dark periods, respectively, in one day. The heart rate lost circadian rhythm in both SCNX and ANSB mice. The mean heart rate in the SCNX mice was significantly higher than that in the ANSB mice.
Table 4.
Heart rate and QT interval.
| Parameter | Treatment | MESOR | Amplitude | Acrophase (h) |
|---|---|---|---|---|
| Heart rate (bpm) | Sham | 622 ± 9§# | 109 ± 21§*# | 19 ± 1.4 |
| SCNX | 669 ± 21§ | 20 ± 4§ | N/A | |
| ANSB | 466 ± 30§γ; | 9 ± 6§ | N/A | |
| QT interval (ms) | Sham | 40 ±1§*# | 8 ± 2§*# | 5 ± 1.1 |
| SCNX | 37 ± 1§ | 3 ± 1§ | N/A | |
| ANSB | 51 ± 2§γ; | 3 ± 1§ | N/A |
Note: Data are mean ± SEM for 3–6 mice per group. Abbreviations are as in Table 1.
One-way ANOVA p < 0.05, Bonferroni/Dunn, p < 0.05.
Sham vs. lesion;
Sham vs. ANSB;
Lesion vs. ANSB; N/A, not applicable (aperiodic).
4. Discussion
4.1. Major findings
We examined the clock gene and cardiac ion channel gene transcripts in the hearts of sham-operated, SCNX, and ANSB mice. Our major findings are as follows:
-
(1)
Besides KCNA5 and KCND2 (Yamashita et al. 2003), the other two cardiac genes, the expressions of KCNK3 and KCHIP2 also exhibited substantial circadian rhythms in the sham-operated mice.
-
(2)
In the SCNX mice, all three clock genes and all four ion channel genes lost circadian rhythm.
-
(3)
ANSB eliminated the circadian oscillation of all of the ion channel gene expressions but not the clock genes’ expression.
-
(4)
The SCNX mice and the ANSB mice exhibited losses of circadian rhythm in the heart rate and QT interval.
4.2. Regulation of the cardiac gene expression
It is well known that the master pacemaker controlling circadian rhythm is located in the SCN in mammals. In addition, accumulated evidence indicates that various peripheral tissues including cardiovascular cells (Ko and Takahashi 2006) also possess circadian oscillators, and that impairment of the function of such clock systems may be associated with some cardiovascular diseases (Maemura et al. 2000; McNamara et al. 2001; Nonaka et al. 2001; Young et al. 2001a; Durgan et al. 2005; Curtis and Fitzgerald 2006; Young 2006; Viswambharan et al. 2007).
The results of the present study suggest that circadian expressions of cardiac genes might be related to the biological clock in the SCN. In the SCNX mice, the expressions of the clock genes and cardiac ion channel genes examined were arrhythmic. The MESOR and amplitude values in most of the cardiac genes were reduced significantly, which suggests that the ion channel gene expressions were “flattened” in the absence of a central clock. However, whether the central clock in the SCN directly influenced the cardiac ion channels or did so through peripheral clock genes in the heart remains unknown.
Evidence has accumulated supporting both neural and humoural control of peripheral rhythms, and this indicates that the SCN regulates the expression of circadian oscillations in various peripheral organs by diverse pathways. By using a model system, SCNX mice parabiotic-linked to intact partners, Guo et al. (2005) found that circadian rhythms of clock gene expressions were recovered in the liver and kidney of the SCNX mice, indicating that blood-borne cues from intact mice were enough to entrain the peripheral clock in these organs. On the other hand, clock gene expressions remained arrhythmic in the heart, muscle and spleen in the SCNX mice, and Guo et al. theorized that the heart might rely more heavily on neural signals.
This study suggested that the ANS is an important pathway for the SCN to transmit time cues to the heart. As expected in the present study, ANSB also resulted in arrhythmic ion channel gene expressions in the heart. On the other hand, unexpectedly, the three clock genes’ expressions were still rhythmic in ANSB mouse hearts. The present results clearly demonstrated that removal of the sympathetic input led to a disrupted circadian expression in ion channels but was not enough to eliminate the daily rhythmicity of clock genes within a tissue. In light of the combined data from the SCNX and ANSB mice, it is reasonable to speculate that a clock system in the SCN but not in the heart might be associated with the circadian cardiac ion channel gene expression. More evidence must be gathered to test this hypothesis.
4.3. Changes in cardiac electrophysiology in accord with those of cardiac gene expression
We examined ECG parameters in the SCNX and ANSB mice. Our results demonstrated that the heart rate and QT interval had substantial circadian rhythms in the sham-operated mice, whereas the SCNX and ANSB abolished such rhythms, similar to the changes in the ion channel gene expressions.
Diurnal variations in several cardiovascular parameters (e.g., blood pressure, heart rate) have been attributed primarily to circadian variations in environmental stimuli (Turton and Deegan 1974; Muller et al. 1989). The biological clock system provides a means by which the heart can predict diurnal variations in the environment to produce optimized responses, enabling the cardiovascular system to prepare for a given stimulus, thereby optimizing the appropriate response. Attenuation of this molecular mechanism would therefore impair such an ability of the heart.
Young et al. (2001b) reported that pressure overload-induced hypertrophy altered the circadian clock in the rat heart, resulting in attenuation in the clock output genes. They also demonstrated that the phases of the clock genes isolated from streptozotocin-induced diabetic rats heart were altered compared to those observed for control hearts (Young et al. 2002). These two studies indicate that the biological clock system might be damaged under pathological conditions. Knutsson et al. (1986) showed that nightshift workers had an increased incidence of heart disease when compared with dayshift co-workers. Although there is no confirmed casual relation, the impairment in the circadian clock may cause the heart to fail to anticipate the changes during a day. Our study using an SCNX and ANSB animal model provides some indirect evidence supporting this hypothesis.
4.4. Study limitations
There are some limitations in the present study. The corresponding proteins to clock and cardiac genes and current density of cardiac ion channels were not measured. We cannot determine whether the protein and electrophysiological properties are also influenced by other aspects of the study treatments such as feed restriction or light control. Light stimulation was shown to partly modulate the expression of the circadian variation in KCNA5 in the rat heart [4]. The mouse is a nocturnal animal, and its circadian rhythm may be completely different from that of human beings.
5. Conclusions
In the present study, we observed that the cardiac ion channel genes KCNA5, KCND2, KCHIP2, and KCNK3 showed circadian expressions, suggesting that the central clock in the SCN but not a peripheral clock in the heart might be involved in the regulation of these cardiac ion channels, and the ANS might mediate such regulation. Further studies on the pathways by which the biological clock transmits its circadian signals to the heart might open a new avenue to our understanding of cardiac arrhythmia and give birth to some new therapy methods.
Acknowledgment
The authors thank Professor Shigenobu Shibata (Waseda University, Japan) for the technical assistance with the lesion mice.
References
- Curtis AM, Fitzgerald GA. Central and peripheral clocks in cardiovascular and metabolic function. Ann Med. 2006;38:552–559. doi: 10.1080/07853890600995010. [DOI] [PubMed] [Google Scholar]
- Durgan DJ, Hotze MA, Tomlin TM, et al. The intrinsic circadian clock within the cardiomyocyte. Am J Physiol Heart Circ Physiol. 2005;289:H1530–H1541. doi: 10.1152/ajpheart.00406.2005. [DOI] [PubMed] [Google Scholar]
- Guo H, Brewer JM, Champhekar A, Harris RB, Bittman EL. Differential control of peripheral circadian rhythms by suprachiasmatic-dependent neural signals. Proc Natl Acad Sci USA. 2005;102:3111–3116. doi: 10.1073/pnas.0409734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Zipes DP. Results of sympathetic denervation in the canine heart: supersensitivity that may be arrhythmogenic. Circulation. 1987;75:877–887. doi: 10.1161/01.cir.75.4.877. [DOI] [PubMed] [Google Scholar]
- Knutsson A, Akerstedt T, Jonsson BG, Orth-Gomer K. Increased risk of ischaemic heart disease in shift workers. Lancet. 1986;2:89–92. doi: 10.1016/s0140-6736(86)91619-3. [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No. 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- LeSauter J, Silver R. Output signals of the SCN. Chronobiol Int. 1998;15:535–550. doi: 10.3109/07420529808998706. [DOI] [PubMed] [Google Scholar]
- Maemura K, de la Monte SM, Chin MT, et al. CLIF, a novel cycle-like factor, regulates the circadian oscillation of plasminogen activator inhibitor-1 gene expression. J Biol Chem. 2000;275:36847–36851. doi: 10.1074/jbc.C000629200. [DOI] [PubMed] [Google Scholar]
- McNamara P, Seo SP, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–889. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Muller JE, Tofler GH, Stone PH. Circadian variation and triggers of onset of acute cardiovascular disease. Circulation. 1989;79:733–743. doi: 10.1161/01.cir.79.4.733. [DOI] [PubMed] [Google Scholar]
- Nonaka H, Emoto N, Ikeda K, et al. Angiotensin II induces circadian gene expression of clock genes in cultured vascular smooth muscle cells. Circulation. 2001;104:1746–1748. doi: 10.1161/hc4001.098048. [DOI] [PubMed] [Google Scholar]
- Scheer FA, Ter Horst GJ, van Der Vliet J, Buijs RM. Physiological and anatomic evidence for regulation of the heart by suprachiasmatic nucleus in rats. Am J Physiol Heart Circ Physiol. 2001;280:H1391–H1399. doi: 10.1152/ajpheart.2001.280.3.H1391. [DOI] [PubMed] [Google Scholar]
- Turton MB, Deegan T. Circadian variations of plasma catecholamine, cortisol and immunoreactive insulin concentrations in supine subjects. Clin Chim Acta. 1974;55:389–397. doi: 10.1016/0009-8981(74)90014-x. [DOI] [PubMed] [Google Scholar]
- Viswambharan H, Carvas JM, Antic V, et al. Mutation of the circadian clock gene Per2 alters vascular endothelial function. Circulation. 2007;115:2188–2195. doi: 10.1161/CIRCULATIONAHA.106.653303. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Sekiguchi A, Iwasaki YK, et al. Circadian variation of cardiac K+ channel gene expression. Circulation. 2003;107:1917–1922. doi: 10.1161/01.CIR.0000058752.79734.F0. [DOI] [PubMed] [Google Scholar]
- Young ME. The circadian clock within the heart: potential influence on myocardial gene expression, metabolism, and function. Am J Physiol Heart Circ Physiol. 2006;290:H1–H16. doi: 10.1152/ajpheart.00582.2005. [DOI] [PubMed] [Google Scholar]
- Young ME, Razeghi P, Cedars AM, Guthrie PH, Taegtmeyer H. Intrinsic diurnal variations in cardiac metabolism and contractile function. Circ Res. 2001a;89:1199–1208. doi: 10.1161/hh2401.100741. [DOI] [PubMed] [Google Scholar]
- Young ME, Razeghi P, Taegtmeyer H. Clock genes in the heart: characterization and attenuation with hypertrophy. Circ Res. 2001b;88:1142–1150. doi: 10.1161/hh1101.091190. [DOI] [PubMed] [Google Scholar]
- Young ME, Wilson CR, Razeghi P, Guthrie PH, Taegtmeyer H. Alterations of the circadian clock in the heart by streptozotocin-induced diabetes. J Mol Cell Cardiol. 2002;34:223–231. doi: 10.1006/jmcc.2001.1504. [DOI] [PubMed] [Google Scholar]


