A 3-year program evaluates usefulness of a Cancer Genome Atlas. The expectation is that improved therapies will follow. Will this finally be enough to win the War on Cancer? What about the role of prevention?
Abstract
The latest salvo in the War on Cancer is a program to explore the feasibility of producing the Cancer Genome Atlas, with the expectation of improved therapies to follow. Though noble, the effort will be time-and labor-intensive, raising the question of whether a prolonged application of preventive measures would ultimately prove to be more effective than resource-consuming molecule-to-molecule combat.
In December, another salvo was fired in the decades-old War on Cancer. Officially declared by President Nixon in 1971, this war shows every sign of being as long-lasting as other wars waged against common nouns. As with the war on terror, it plays on a powerful emotion: fear. In both instances, some fear may be warranted; cancer is the second leading cause of death in the United States.
In the face of a daunting adversary, fear can be healthy, providing powerful motivation when action is needed. The war on cancer can point to some notable successes after 35 years — virtual cures for some forms of cancer and longer life expectancy for people with others. Still, 36 percent of U.S. cancer patients survive less than five years after a cancer diagnosis.
But fear can cloud rational decision making. For better or worse, it also has a way of opening wallets. And so, the latest barrage fired against cancer is a wad of cash — $100 million from the National Cancer Institute and the National Human Genome Research Institute for a three-year pilot to explore the feasibility of producing the Cancer Genome Atlas (previously known as the Human Cancer Genome Project). Tumor types to be studied remain to be determined. The project’s goal is pragmatic: to characterize specific tumors in such a way as to foster development of clinically meaningful applications for detecting, treating, and preventing cancer.
At the Univ. of Connecticut, molecular biologist and professor of medicine Marc F. Hansen, PhD, is cautiously optimistic about the Cancer Genome Atlas. “I’m worried it will turn out badly, but hoping it will turn out well.”
PHOTOGRAPH BY PETER HVIZDAK
In the context of annual direct and indirect costs associated with cancer — an estimated $69 billion and $120 billion, respectively, in 2004 — $100 million is a pittance. For that matter, so is the $1 billion or more the full-fledged atlas might cost. The fact that the endeavor is beginning as a couple of pilot projects reflects uncertainty in the scientific community about its merits and feasibility, along with worries that it will divert funds away from more deserving research projects (Elledge 2005, Miklos 2005).
The Cancer Genome Atlas seeks to build on the foundation established by the Human Genome Project, completed in 2003 at a cost of $2 billion (see “The Human Genome,” page 48), complementing programs such as the Cancer Genome Anatomy Project and the Cancer Biomedical Informatics Grid. The finished sequence generated by the Human Genome Project isn’t that of a single human but a composite of DNA sequences provided by several volunteers. It offers a starting point for studies comparing the human genome with those of other species, which can shed light into the evolution of various physiologic processes, with the expectation that improved therapies will follow. For the proposed atlas, the hope is that by comparing the healthy human genome with genomes from various kinds of tumors, new, precisely targeted molecular approaches can be devised to detect, treat, and prevent cancer.
The Human Genome.
The human genome refers to the entire sequence of DNA, which is carried in a set of 23 chromosomes. Except for gametes (sperm and ova), which are haploid and contain just one such set, every human cell is diploid, containing two complete sets of 23 chromosomes. Each set consists of 22 autosomes plus one sex chromosome.
Each chromosome contains one long double-stranded molecule of DNA comprising four different kinds of nucleotides, each built on a single- or double-ringed base (Table, page 50). The purine (double-ringed) bases in DNA are designated by the letters A (adenine) and G (guanine), the pyrimidine (single-ringed) bases by C (cytosine) and T (thymine). As first explained by Watson and Crick in their famous 1953 letter to Nature, the characteristics of the bases are such that a C in one strand of DNA always is matched by a G in the other, and an A in one strand always is matched by a T in the neighboring strand. These A–T and C–G pairs form the rungs in the DNA ladder, a double helix. Being complementary lets the strands be replicated with great fidelity so that the genetic code they contain is not corrupted when another molecule of DNA or RNA is produced.
Altogether, a single set of 23 human chromosomes contains about 3 billion pairs of nucleotide bases. Scattered among these are the coding sequences that constitute genes. Humans have about 22,000 genes, most of which encode one or more of the 100,000 proteins found in the human body. Some genes encode certain types of RNA or regulatory elements that turn genes on and off. Thanks to the regulatory elements, a single gene can produce several proteins.
The coding sequences (exons) in a gene are interrupted by non-coding sequences (introns). Before a protein can be expressed, the introns must be removed and the exons spliced together. The assembled exons serve as the template for synthesis of messenger RNA (mRNA), which travels from the cell nucleus to the protein-assembly apparatus in the cytoplasm, the ribosomes. In these organelles, short strands of transfer RNA (tRNA) fetch the amino acids that correspond to the various codons in the mRNA — triplets such as GGA (encoding glycine) or UUA (leucine) — which are strung together like a chain of pearls to form a protein.
The genotype is the complete set of genetic information possessed by an organism, through pairs of genes (one from each parent) found at specific points (genetic loci) along chromosomes. The phenotype is the set of observable traits of the organism that result from the interaction between the genotype and its environment. In a population, the same gene can take two or more different forms, known as alleles. If there are two alleles for a given gene (A, a), three different combinations are possible: AA, Aa, and aa. Depending on the property influenced by the gene in question, an individual with the AA genotype may have a phenotype that looks quite different from that produced by the aa genotype.
Marc F. Hansen, PhD, a molecular biologist and professor of medicine at the University of Connecticut Health Center, is cautiously optimistic about the pilot. “I’m worried it will turn out badly, but hoping that it will turn out well,” he says. Hansen views the atlas as an attempt to answer the age-old question, “Okay, now what?”
Because of the Human Genome Project, he notes, the United States has excessive sequencing capacity, acquired at a cost of about $200,000 per instrument (and 10–40 instruments per site). In that context, the atlas provides a way to keep expensive equipment put to what might turn out to be a good use. In the context of the War on Cancer, “The atlas is an attempt to finally make the Great Leap Forward,” Hansen says. “In the past, we’ve studied genes, one by one, but each represents only a small piece of the picture. Cancer is like a spider web — touch one small piece and everything changes. So, the idea here is to try to view everything at once, like a holograph.”
But Hansen is concerned that when an image comes into view, it won’t be even the tip of the iceberg, but a mere snowflake atop the iceberg’s tip.
“When we look at a DNA microarray from a tumor, we might see 1,000 genes that are different from those in normal tissue. But which are cause and which are effect?”
He thinks the project’s chances of succeeding depend on careful selection of tumor types most likely to identify critical molecular pathways. That would rule out the numerically big cancers (such as breast cancer), owing to the confounding effects of environmental and hormonal factors. Instead, he favors focusing on some of the more rare cancers, which tend to be more homogeneous and have yielded important discoveries in the past.
PROBLEM OF HETEROGENEITY
Creating a cancer atlas won’t be easy. Cancer is extremely heterogeneous at the macro and micro levels. Cancer isn’t a single disease. At the macro level, it’s a group of perhaps 200 different diseases, including about 50 major types that would be pursued in great detail if the pilot project deems it feasible. All are characterized by major malfunctioning of the genetic controls. Thanks to the characteristics of DNA, along with built-in damage-repair mechanisms, the human genome ordinarily is very stable. When that stability is lost, the manifestation can be cancer.
Only a small minority of cancers is inherited, and these tend to occur early in life. Yet the presence of oncogenic mutations does not indicate the inevitability of cancer. Neither can their absence be taken as assurance that cancer won’t develop. The vast majority of cancers occur in later life as a result of damage to somatic-cell DNA that accumulates as the person ages. Some inherited genetic mutations increase a person’s risk of developing a certain cancer (e.g., mutations of BRCA1 and BRCA2, which are associated with increased risk of breast and ovarian cancer). Epidemiological studies are required to tease out the circumstances under which a gene that carries an increased risk for cancer in fact results in cancer.
Prostate cancer illustrates the problems posed by heterogeneity among types of cancer, and is why the Cancer Genome Atlas might be beneficial, in theory. The prostate-specific antigen test is quite popular, notably among urologists (Barry 2006). Despite its widespread use to screen at-risk populations, the test has not yet translated into reductions in mortality (Concato 2006). Although it is good at detecting prostate cancer, especially at earlier stages, PSA testing can’t distinguish between the indolent tumor (which progresses so slowly the patient eventually dies of something else) and the virulent tumor (which poses an imminent threat). In addition, the PSA test lacks specificity, generating a substantial percentage of false-positive results that necessitate biopsies. A genetic profile — a personalized DNA biopsy — that can help distinguish a prostate tumor (or any tumor) that requires only “watchful waiting” from one that demands intensive therapy would be welcome.
HOW MANY CANCER TYPES?
One reason for the great heterogeneity among cancers is that, in addition to mutations in the genome, changes in the epigenome also come into play. The genome may be just the beginning of the story. It is becoming clear that the information contained in the genome, vast though it may be, is greatly extended by the epigenome, which controls the differential expression of genes in specific cells.
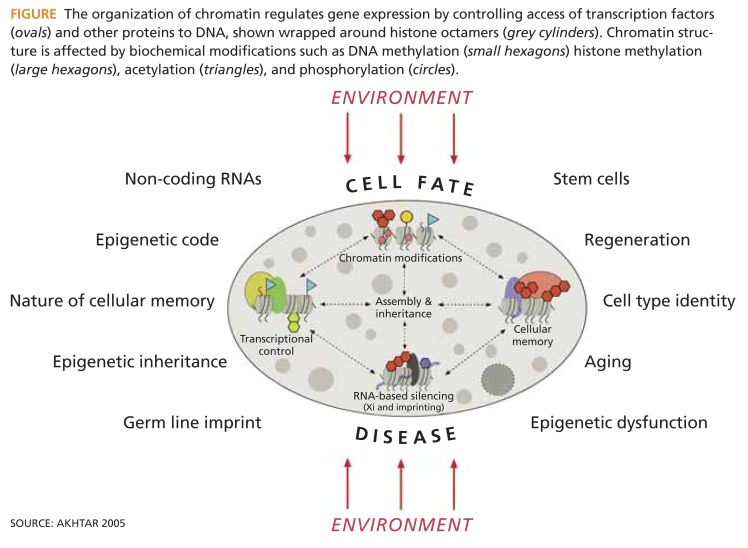
The concepts of genotype and phenotype can be applied to differentiated cells within an organism (Holliday 2005). Save for its gametes, every complex organism has the same double set of inherited genes in its cells once the zygote has been formed. And once cells begin to differentiate, cells with the same set of genes begin to express rather different proteins, reflecting the varying local environments within the cells. For a strand of DNA, the local environment is formed by the substances in which it is packed. Notable among these are small proteins known as histones. The DNA strand is wrapped twice around a core of histones, forming a tiny sphere known as a nucleosome, the basic unit of the chromatin polymer that cuts DNA down to a manageable size.
The tails of the histones can be altered through processes such as methylation, acetylation, and phosphorylation (Figure). These change the structure and function of the chromatin and alter access to the associated DNA sequence. It appears that changes to the chromatin structure play a major role in determining how DNA behaves in health and disease, and that epigenomic information is essential for understanding cancer. An important question to be answered through the cancer pilot is whether existing technologies for epigenomic analysis are adequate, and if not, whether the program should support their development.
FIGURE.
The organization of chromatin regulates gene expression by controlling access of transcription factors (ovals) and other proteins to DNA, shown wrapped around histone octamers (grey cylinders). Chromatin structure is affected by biochemical modifications such as DNA methylation (small hexagons) histone methylation (large hexagons), acetylation (triangles), and phosphorylation (circles).
SOURCE: AKHTAR 2005
The source of genetic instability that leads to cancer can range from the alteration or deletion of a single DNA nucleotide (a point mutation) to the deletion of a long sequence of nucleotides. Outright deletion of a gene is a certain method of “silencing” it — i.e., preventing it from expressing its product — but it is not the only way. Although gene silencing is being pursued as a means of shutting down genes involved in pathological processes (McCain 2004), in cancer the unwanted gene silencing that results from DNA hypermethylation contributes to pathological processes. Abnormal patterns of DNA hypermethylation are believed to disrupt the molecular pathways involved in DNA repair and maintenance of the normal cell cycle, leading to the transformation of a normal cell into a malignant one (Esteller 2001).
DNA methylation involves adding a methyl group to a cytosine base, producing methylcytosine, which acts like a fifth base (Herman 2003). Cytosine methylation is catalyzed by a family of three enzymes, the DNA methyltransferases (DNMTs), which provide an intriguing target for inhibition by small molecules like 5-aza-CdR and zebularine. Because cytosine methylation does not result in a permanent change to the nucleotide but instead needs to be sustained by assistance from DNMTs, it is thought that DNMT inhibition may allow demethylation to occur, restoring the gene’s normal function. On the other hand, there is concern that inhibition of DNA methylation might have the unwelcome consequence of inducing prometastatic genes through hypomethylation. The existence of multiple cancer methylomes (the set of DNA methylation modifications in a cell) alongside cancer genomes vastly complicates the task of those who would compile a Cancer Genome Atlas and employ it for practical purposes.
The emerging paradigm that combines genomic and epigenomic information may suggest the possibility of far more than 200 kinds of cancer — perhaps as many as there are people with the disease. In other words, by its nature, cancer is highly individualized. Mary’s ovarian cancer is different from Sue’s, and Sue’s differs from Sally’s. More than that, different tumors from the same patient can have different genetic profiles. And Mary’s cancer today may be different from her cancer last week, which may be different still from her cancer last month. When metastasis enters the picture, the genetic complexities within a single patient multiply.
On www.biotechnologyhealthcare.com.
A glossary of the terms used in this manuscript can be found by clicking on the link to this article in the April 2006 issue.
DAS KAISER WILHELM GESCHÜTZ?
In the waning days of a real war — the Great War — the Germans unveiled a new weapon, the Kaiser Wilhelm Geschütz (also known as the Paris Gun). It was intended to strike fear into the hearts of Parisians. From a distance of 70 miles, this long-range gun could lob shells 25 miles into the stratosphere. After a splendid journey of 170 seconds, the shells would drop on Paris, more or less. The Paris Gun indeed terrorized Parisians, but it did little else. It was wildly inaccurate — it couldn’t be aimed, just pointed in the right direction — and the payload it delivered was relatively puny. The Paris Gun was demanding and expensive to operate, needing considerable maintenance because of the massive powder charge employed.
The war ground to a halt, not because of the Kaiser Wilhelm Geschütz — let alone poison gas, machine guns, aeroplanes, or those newfangled tanks — but to a considerable extent because of prolonged and intensive application of an old-fashioned naval blockade. A blockade is neither dramatic nor rapid, but it can be effective, and the British employed it with devastating results. The blockade prevented raw materials, munitions, and food from reaching Germany. Ultimately, these preventive measures were a major factor in forcing Germany’s surrender.
In the War on Cancer, could the Cancer Genome Atlas be the equivalent of the Kaiser Wilhelm Geschütz? The atlas surely will provide extraordinary intellectual challenges and insights, but is it the most effective weapon? Would prolonged intensive application of preventive measures ultimately prove more effective than an all-out attack on multiple cancer genomes?
Instead of waging molecule-to-molecule combat, why not just make a concerted effort to end smoking? The American Cancer Society estimates that tobacco use accounted for more than 30 percent of the 570,000 U.S. cancer deaths in 2005. Substantial progress has been made on that front in males. Why not press ahead with smoking cessation efforts aimed at women?
The society also estimates that a third of U.S. cancer deaths last year were related to poor nutrition, physical inactivity, and excess weight. Considering the cancer deaths attributable to excessive alcohol consumption, or to excessive exposure to the sun and infectious diseases, it becomes clear that a substantial share of U.S. cancer deaths could be prevented by lifestyle modification.
To some critics, rather than offering the hope of improved methods for dealing with difficult problems, genomics itself constitutes a threat to our long-term well-being (Cooper 2003):
Molecular genetics, which has deeply influenced the philosophical framework of biology, often assumes that the primary threats to health are programmed in our DNA rather than our social environment, with disease being transmitted through abnormal physiology rather than food, air, [and] microorganisms.... Genomics may ... advance the claims of a science belief system over the pragmatic needs of the long-term movement toward prevention through creation of a healthier environment as the most effective means to control disease.
At the time the Germans put the Kaiser Wilhelm Geschütz into action, they had no hope of prevailing in the Great War. The United States, having entered the war by then, convoyed thousands of fresh troops across the Atlantic, providing welcome reinforcements for the exhausted British and French. Against this unrelenting onslaught, and in the context of the Allies’ suffocating naval blockade, the Kaiser Wilhelm Geschütz was nothing more than a grand gesture. But it surely must have been great fun to fire!
TABLE.
Bases, nucleosides, and nucleotides found in nucleic acids
| Purine bases (double ring) | Pyrimidine bases (single ring) | ||||
|---|---|---|---|---|---|
| Base | Adenine (A) | Guanine (G) | Cytosine (C) | Thymine (T) | Uracil (U) |
| Nucleoside | |||||
| Ribonucleoside* (base+ribose) | Adenosine | Guanosine | Cytidine | Uridine | |
| Deoxyribonucleoside† (base + deoxyribose) | Deoxyadenosine | Deoxyguanosine | Deoxycytidine | Deoxythymidine | |
| Nucleotide | |||||
| Ribonucleotide* (base+ribose+phosphoryl group) | Adenosine monophosphate (AMP) | Guanosine monophosphate (GMP) | Cytidine monophosphate (CMP) | Uridine monophosphate (UMP) | |
| Deoxyribonucleotide† (base+deoxyribose + phosphoryl group) | Deoxyadenosine monophosphate (dAMP) | Deoxyguanosine monophosphate (dGMP) | Deoxycytidine monophosphate (dCMP) | Deoxythymidine monophosphate (dTMP) | |
Blue shading=component of RNA.
Red shading=component of DNA.
Glossary
Words in italics appear in this glossary.
- Base.
A nitrogenous ring or double ring at the heart of a nucleoside or nucleotide. Single-ringed bases are the pyrimidines cytosine (C), thymine (T, found in DNA), and uracil (U, found in RNA). Double-ringed bases are the purines adenine (A) and guanine (G). Per Watson-Crick base pair rules, C binds only with G, and A binds only with U, in the case of RNA, or T, in the case of DNA.
- Base pair.
A complementary pair of nucleotide bases that connect either DNA or RNA into a double strand. The bases in DNA are abbreviated as A, C, G, and T. The bases in RNA are the same, except U replaces T. When double strands are formed, C binds only with G, and A binds only with U (or T).
- Chromatin.
The material found in chromosomes — a polymer consisting of doubled-stranded DNA, histones, nonhistone proteins, and some RNA. Basic unit of chromatin is the nucleosome.
- Codon.
A sequence of 3 nucleotides that dictates the synthesis of a specific amino acid. With 4 “letters” available, 64 codes are possible (4x4x4); 61 encode amino acids (most of the 20 amino acids are encoded by more than 1 codon), and 3 are stop codes (TAA, TAG, TGA).
- CpG island.
Clusters of CpG sites, most often associated with the gene promoter regions where transcription begins.
- CpG site.
A pair of nucleotides having its bases, cytosine (C) and guanine (G), connected by a phosphodiester bond (p). In mammals, nearly all DNA methylation occurs at CpG sites.
- DNA.
Deoxyribonucleic acid — repository of the genetic code. Found in the nucleus (and mitochondria).
- DNMTs.
DNA methyltransferases — family of three enzymes that catalyze methylation.
- Epigenetics.
In the context of cancer, heritable changes mediated by mechanisms other than alterations in the primary DNA sequence; methylation is epigenetic. Epigenetic events occur at the level of chromatin.
- Epigenome.
Mechanisms that control the differential expression of genes in specific cells, via methylation of cytosine in DNA and various modifications of histones.
- Expression.
Production of a product (protein or RNA) encoded by a gene.
- Gene expression.
Process by which a protein is produced from the information encoded in a gene. Within the nucleus, messenger RNA (mRNA) is transcribed from the DNA. The mRNA contains triplets of nucleotides (codons) that correspond to specific amino acids. After the mRNA is transported to ribosomes in the cytoplasm, short strands of transfer RNA (tRNA) translate the codons and gather the corresponding amino acids, which are assembled into proteins.
- Genetic locus.
The precise location of a gene along a chromosome. An individual has two genes at each genetic locus — one from each parent —the genotype.
- Genotype.
The combined genetic information present in the pair of genes found at a genetic locus. If both copies are identical, the genotype is a homozygote. If they are different, the genotype is a heterozygote. The physical manifestation of the genotype is the phenotype.
- Histones.
The proteins that package DNA within the chromosomes. Stretched out, the strand of DNA in a chromosome would be 1,000 times longer than the diameter of the cell’s nucleus; histones help condense the DNA into a compact form.
- Histone code.
Postulated set of modifications to histones, which control access to DNA.
- Metagene.
Pattern of multiple genes for stratifying patients.
- Methylation.
Adding a methyl group (–CH3) to a cytosine residue to convert it to 5-methyl-cytosine, which accounts for less than 1 percent of the bases in human DNA. DNA methylation occurs at CpG sites. Methylation of CpG islands is critical to gene activity and gene expression, with methylation being associated most often with the silencing of genes.
- Mitochondria.
Organelles containing respiratory enzymes, used to generate ATP from food molecules. Mitochondria have their own DNA and are believed to have originated as free-living bacteria that, perhaps 3.5 billion years ago, entered into a symbiotic relationship with another prokaryote, thus forming a eukaryote.
- mRNA.
Messenger RNA — a long, single-stranded molecule of RNA that is transcribed from a gene on DNA. mRNA is able to move from the nucleus through the cytoplasm to carry the gene’s message to a ribosome, where the message is translated into a protein with assistance from tRNA.
- Nucleoside.
Base + sugar. In RNA, the sugar is ribose; in DNA, deoxyribose. RNA nucleosides: adenosine, guanosine, cytidine, uridine. DNA nucleosides: deoxyadenosine, deoxyguanosine, deoxycytidine, deoxythymidine.
- Nucleosome.
The basic unit of the chromatin polymer.
- Nucleotide.
The building blocks of RNA and DNA, composed of a base + sugar + phosphate. In the DNA double helix, the bases form the cross-supports, while the sugars and phosphates provide lengthwise structure. RNA nucleotides: adenosine monophosphate (AMP), guanosine monophosphate (GMP), cytidine monophosphate (CMP), uridine monophosphate (UMP). DNA nucleotides: deoxyadenosine monophosphate (dAMP), deoxyguanosine monophosphate (dGMP), deoxycytidine monophosphate (dCMP), deoxythymidine monophosphate (dTMP or TMP).
- Oncogene.
Genes that, on activation, promote unchecked cell growth and multiplication.
- Phenotype.
An organism’s observable traits, produced through the interaction of its genotype with the environment.
- Ribosome.
The intracellular machinery where proteins are assembled, using the codes carried in mRNA.
- RNA.
Ribonucleic acid. Once thought to serve primarily as a messenger for DNA, carrying DNA’s genetic code in the process of gene expression, now known to play other roles, including enzymatic and regulatory functions.
- Sequence.
A continuous series of nucleotides.
- Somatic cell.
A cell in the body of an organism, in contrast with a gamete (sperm or ova). A somatic cell genetic mutation is one that was not inherited and cannot be passed along to offspring (unlike genetic mutations in gametes).
- Transcription.
Formation of a strand of RNA from a DNA template, at the start of the process of translating the coding of a gene into a protein.
- Translation.
Expression of protein from mRNA.
- tRNA.
Transfer RNA — small units of RNA that recognize codons in mRNA.
- Tumor suppressor gene.
Recessive gene that suppresses tumor formation, such as p53. Proper function of the gene can be blocked by mutation or methylation of its promoter.
REFERENCES
- Akhtar A, Cavalli G. The epigenome network of excellence. PLoS Biol. 2005;3:e177. doi: 10.1371/journal.pbio.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry MJ. The PSA conundrum. Arch Intern Med. 2006;166:7–8. doi: 10.1001/archinte.166.1.7. [DOI] [PubMed] [Google Scholar]
- Concato J, Wells CK, Horwitz RI, et al. The effectiveness of screening for prostate cancer. A nested case-control study. Arch Intern Med. 2006;166:38–43. doi: 10.1001/archinte.166.1.38. [DOI] [PubMed] [Google Scholar]
- Cooper RS, Psaty BM. Genomics and medicine: distraction, incremental progress, or the dawn of a new age? Ann Intern Med. 2003;138:576–580. doi: 10.7326/0003-4819-138-7-200304010-00014. [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Hannon GJ. An open letter to cancer researchers. Science. 2005;310:439–440. doi: 10.1126/science.310.5747.439b. [DOI] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Holliday R. DNA methylation and epigenotypes. Biochemistry. 2005;70:612–617. doi: 10.1007/s10541-005-0144-x. [DOI] [PubMed] [Google Scholar]
- McCain J. Renewing the assault on mRNA. Biotechnol Healthcare. 2004;1(1):44–53. [PMC free article] [PubMed] [Google Scholar]
- Miklos GL. The Human Cancer Genome Project — one more misstep in the war on cancer. Nat Biotechnol. 2005;23:535–537. doi: 10.1038/nbt0505-535. [DOI] [PubMed] [Google Scholar]