Abstract
The utility of nitric oxide (NO)-releasing silica nanoparticles as a novel antibacterial is demonstrated against Pseudomonas aeruginosa. Nitric oxide-releasing nanoparticles were prepared via co-condensation of tetraalkoxysilane with aminoalkoxysilane modified with diazeniumdiolate NO donors, allowing for the storage of large NO payloads. Comparison of the bactericidal efficacy of the NO-releasing nanoparticles to 1-[2-(carboxylato)pyrrolidin-1-yl]diazen-1-ium-1,2-diolate (PROLI/NO), a small molecule NO donor, demonstrated enhanced bactericidal efficacy of nanoparticle-derived NO and reduced cytotoxicity to healthy cells (mammalian fibroblasts). Confocal microscopy revealed that fluorescently-labeled NO-releasing nanoparticles associated with the bacteria, providing rationale for the enhanced bactericidal efficacy of the nanoparticles. Intracellular NO concentrations were measurable when the NO was delivered from nanoparticles as opposed to PROLI/NO. Collectively, these results demonstrate the advantage of delivering NO via nanoparticles for antimicrobial applications.
Keywords: nitric oxide, silica nanoparticle, antibacterial, bactericidal, cytotoxicity, reactive nitrogen species, reactive oxygen species
Antibiotic resistance has resulted in bacterial infections becoming the most common cause of infectious disease-related death.1, 2 In the United States alone, nearly 2 million people per year acquire infections during a hospital stay, of which approximately 90,000 die.2 The primary culprits behind such deadly infections are antibiotic-resistant pathogens, which are responsible for approximately 70% of all lethal nosocomial infections. The growing danger of life-threatening infections and the rising economic burden of resistant bacteria have created a demand for new antibacterial therapeutics.
The use of nanoparticles as delivery vehicles for bactericidal agents represents a new paradigm in the design of antibacterial therapeutics. To date, most antibacterial nanoparticles have been engineered using traditional antibiotics that are either incorporated within the particle scaffold or attached to the exterior of the particle. In many cases, such particles have exhibited greater efficacy than their constituent antibiotics alone. For example, Gu et al. reported that vancomycin-capped gold nanoparticles exhibited a 64-fold improvement in efficacy over vancomycin alone.3 Similarly, silver nanoparticles have shown greater antibacterial activity than silver ion (Ag+) in solution due to the direct toxicity of the particles and tunable release of Ag+ based on nanocomposite size.4–6 Silica has also been used as an antibacterial. Lin and co-workers employed mesoporous silica nanoparticles to controllably release ionic liquids with proven bactericidal efficacy.7
While antibacterial nanoparticles have shown great promise, the use of conventional antibiotics (e.g., vancomycin) or classical antibacterial agents (e.g., Ag+) does not address bacterial resistance concerns.2, 8, 9 Nitric oxide (NO), a diatomic free radical that plays a key role in the natural immune system response to infection,10 may represent an alternative approach in the design of antibacterial nanoparticles. Macrophages and other inflammatory cells produce NO to battle foreign microbes.11 In fact, mice lacking the ability to endogenously produce NO are more susceptible to microbial infection.12 Nitric oxide has been shown to possess broad spectrum antibacterial activity, primarily due to its reactive byproducts such as peroxynitrite (ONOO−) and dinitrogen trioxide (N2O3).13 Both gram-positive and gram-negative bacteria have been found to be susceptible to gaseous NO, including methicillin-resistant Staphylococcus aureus (MRSA).14 The doses of NO required to kill bacteria proved non-toxic to human dermal fibroblasts. Using diazeniumdiolate small molecule NO donors, Raulli et al. reported the minimum inhibitory concentrations against several bacterial species.15 These initial studies illustrate NO’s tremendous potential as an antibacterial agent with broad spectrum activity.
Unfortunately, the utility of NO as an antibacterial agent is hindered by the lack of suitable vehicles for NO storage and delivery. Indeed, NO is an extremely reactive gas and difficult to administer as a therapeutic. To address delivery issues, Schoenfisch and coworkers have synthesized nanoparticle-based scaffolds capable of storing large payloads of NO.16–18 The nanoparticles spontaneously release tunable levels of NO under aqueous conditions at physiological temperature and pH, and thus represent attractive vehicles for delivering NO. Nanoparticle delivery of NO has two main advantages over previously-developed small molecule NO donor systems (e.g., diazeniumdiolates, nitrosothiols, and metal-NO complexes19, 20). First, the rate of NO release is easily modulated as a function of nanoparticle size, composition, and/or surface hydrophobicity, thereby allowing for control over the duration of NO release. Second, the versatility of the chemistry used to synthesize the nanoparticles allows for specific tailoring of particles with functional groups to minimize their toxicity and enable imaging and/or cell-specific targeting, while retaining the ability to deliver therapeutic levels of NO. Herein, we report the efficacy of NO-releasing silica nanoparticles against Pseudomonas aeruginosa, an opportunistic pathogen problematic in burn and chronic wound infections.21–23 Both the bactericidal efficacy and cytotoxicity of nanoparticle-derived NO is compared to NO release from a small molecule NO donor to illustrate the advantage of delivering NO from silica nanoparticles.
Results and Discussion
Characterization of NO-releasing silica nanoparticles and PROLI/NO
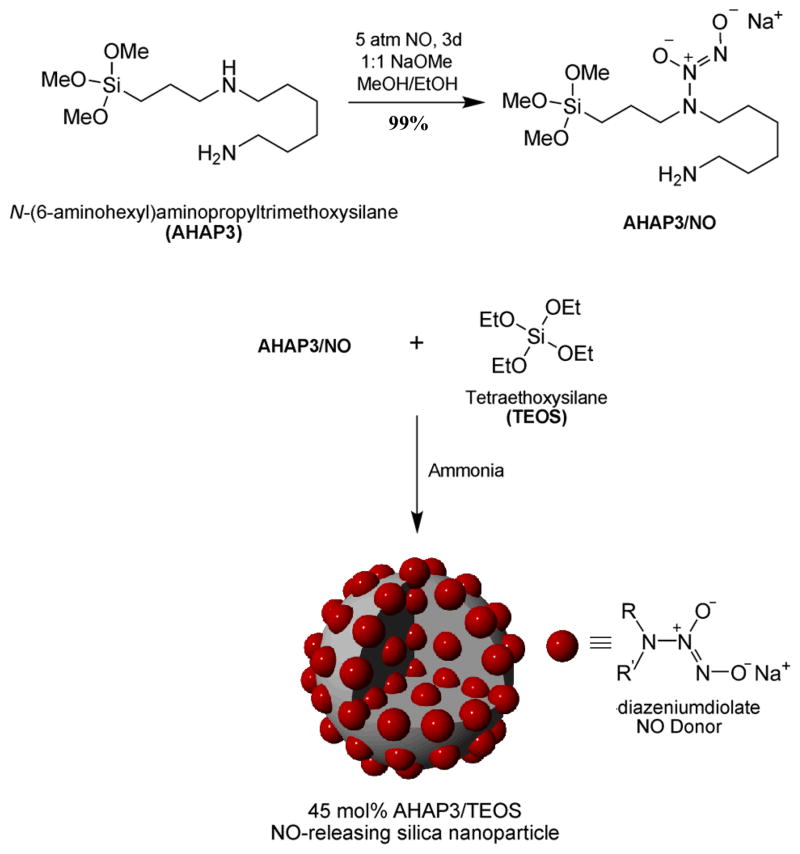
Shin et al. previously reported the synthesis and characterization of NO-releasing silica nanoparticles.24 Briefly, diazeniumdiolate NO donors were synthesized on aminoalkoxysilane precursors prior to nanoparticle construction (Scheme 1), enabling the formation of particles with superior NO release ability. By synthesizing diazeniumdiolate-modified aminoalkoxysilanes prior to nanoparticle synthesis, particle aggregation was reduced due to decreased hydrogen bonding interactions between amines. The synthesis resulted in ~99% amine-to-diazeniumdiolate conversion efficiency and greater yields of NO per mol of aminoalkoxysilane precursor compared to previous synthetic procedures used to generate NO-releasing silica nanoparticles.18 29Si NMR spectroscopy of the AHAP3/NO product was used to determine whether the presence of sodium methoxide would lead to self-condensation of the AHAP3/NO precursors during the diazeniumdiolate formation step (Step 1 of Scheme 1). Notably, no significant Tn peaks characteristic of organosilane polymerization were observed,18 indicating that the AHAP3/NO molecules did not pre-condense under such reaction conditions. 1H NMR of AHAP3/NO revealed that the protons adjacent to the secondary amine where diazeniumdiolate formation occurs became deshielded in the presence of the zwitterionic NO donor, shifting downfield from 2.45 ppm to 2.84 ppm. The presence of the diazeniumdiolate on AHAP3/NO was also confirmed via UV-Vis spectroscopy (λmax = 253 nm, characteristic of the diazeniumdiolate absorption maximum17, 25) and direct observation of the released NO via chemiluminescence.26
Scheme 1.
Synthesis of AHAP3 NO donor and co-condensation with TEOS to form NO-releasing silica nanoparticles. R=(CH2)3Si≡ and R′=H2N(CH2)6–.
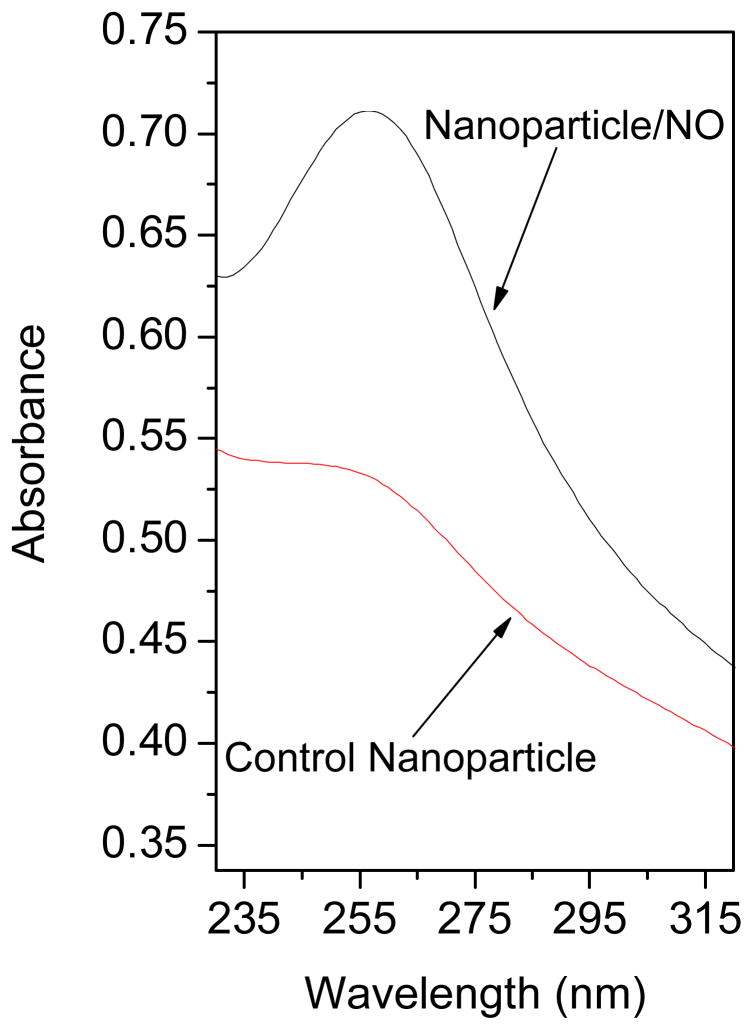
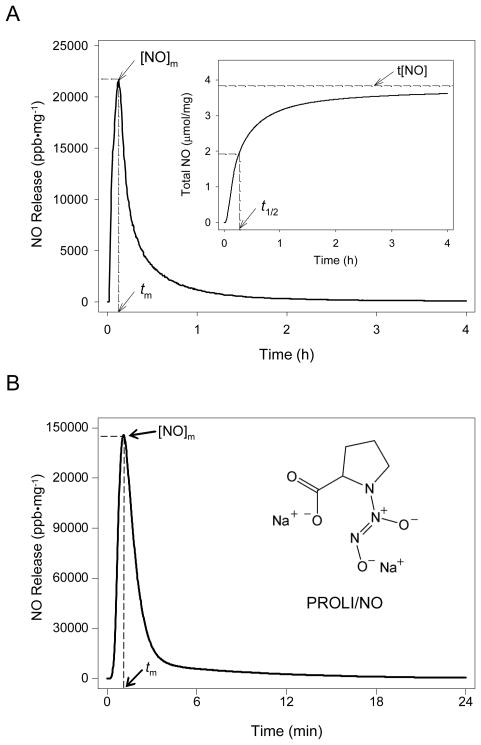
The presence of the diazeniumdiolate functional group in the nanoparticle scaffold was confirmed via UV absorbance spectroscopy and the direct measurement of NO release. As shown in Figure 1, diazeniumdiolate-modified 45 mol% AHAP3/TEOS silica nanoparticles (Nanoparticle/NO) exhibited a similar λmax of 257 nm. Nitric oxide release due to diazeniumdiolate decomposition27 was monitored in phosphate buffered saline (PBS; 10 mM, pH 7.4) at 37 °C using a chemiluminescence NO analyzer.26 As shown in Figure 2A, the total amount of NO released (t[NO]) and maximum NO flux ([NO]m) from the AHAP3 nanoparticle system were ~3.8 μmol·mg−1 and ~21700 ppb·mg−1, respectively. The NO release kinetics from the AHAP3 silica nanoparticles were relatively rapid compared to other NO-releasing silica nanoparticle systems,18 with a NO release half life (t1/2) of 18 min. As a result of the rapid NO release from the 45 mol% AHAP3 nanoparticles, the time required to reach the maximum NO flux (tm) of ~21700 ppb·mg−1 was only 8 min after immersion in buffer solution. The initial burst of NO allows for the relatively rapid delivery of micromolar quantities of NO that produce the reactive nitrogen and oxygen species that mediate NO’s bactericidal actions.28 As characterized by atomic force microscopy (AFM), the size and homogeneity of the 45 mol% AHAP3 nanoparticles were 136 ± 15 nm (Supporting Information). Both the size and NO-release properties of diazeniumdiolate-modified silica nanoparticles proved tunable based on the amount and identity of aminoalkoxysilane precursor employed in the synthesis (data not shown). A full systematic characterization of such properties as a function of NO donor is reported elsewhere.24
Figure 1.
UV absorbance of control and diazeniumdiolate-modified (Nanoparticle/NO) 45 mol% AHAP3/TEOS silica nanoparticles (concentration = 160 μg/mL in phosphate buffered saline).
Figure 2.
Nitric oxide release profiles of (A) 45 mol% AHAP3 silica nanoparticles and (B) PROLI/NO in PBS, pH 7.4, 37 °C. Inset of (A) represents total NO release. [NO]m = maximum NO flux; tm = time to reach maximum NO flux; t[NO] = total NO released; t1/2 = half life of NO release.
To facilitate comparison of the bactericidal efficacy of nanoparticle-derived NO with small molecule-derived NO, the amino acid proline was functionalized with diazeniumdiolate NO-donors as described by Saavedra et al.29 As shown in Fig. 2B, the release of NO from PROLI/NO was also extremely rapid, with a t1/2 of approximately 1.7 min. On a per mg basis, PROLI/NO released less than half as much total NO as the AHAP3 nanoparticles, with a t[NO] value of 1.8 μmol NO·mg−1. Due to its rapid NO release characteristics, however, the [NO]m for PROLI/NO (>145000 ppb·mg−1) was more than 6 times greater than the [NO]m generated by the nanoparticles per mg, with tm approximately 1 min after addition to buffer. Despite the large bolus of NO released by PROLI/NO, the extended duration of NO release from the AHAP3/TEOS nanoparticle system is more beneficial for antibacterial applications because its NO release capabilities are not immediately lost upon exposure to aqueous conditions. Indeed, effective NO-based antibacterial agents require NO release durations long enough to allow the NO donor vehicle to reach the intended site of action without becoming depleted of NO during transit, while still releasing bactericidal quantities of NO.
Bactericidal efficacy under static conditions
The bactericidal efficacy of the 45 mol% AHAP3 NO-releasing silica nanoparticles was evaluated against P. aeruginosa, an opportunistic gram-negative pathogen. Due to a multitude of virulence factors, P. aeruginosa is a common cause of burn wound infections leading to significant morbidity and mortality in burn wound victims.21,22 Additionally, P. aeruginosa plagues >30% of all leg and foot ulcers resulting in chronic wounds with impaired healing.23 Perhaps most alarming, however, is the emergence of multidrug-resistant P. aeruginosa that has been isolated from nosocomial burn wound patients.22, 30 P. aeruginosa clones resistant to both β-lactams and aminoglycosides, two classes of antibiotics that are commonly used to treat P. aeruginosa infections are documented.30 Thus, novel treatments for combating infections resulting from antibiotic-resistant pathogens are urgently needed. We hypothesized that the AHAP3/TEOS silica nanoparticle system, capable of storing multiple NO donors within each delivery vehicle, may represent an attractive new method for killing pathogenic bacteria due to its ability to release high localized doses of NO. To determine the influence of the nanodelivery vehicle on the antibacterial properties of NO, the bactericidal efficacy of 45 mol% AHAP3 nanoparticles was compared to that of PROLI/NO. Although a previous study evaluated the antibacterial properties of the small molecule NO-donor DETA/NO (diazeniumdiolate-modified diethylenetriamine),15 we observed that the diethylenetriamine backbone alone demonstrated considerable toxicity to P. aeruginosa (data not shown), which is consistent with the observations of others.31 Conversely, the backbone of PROLI/NO (the amino acid proline) exhibited no toxicity to P. aeruginosa up to 20 mg·mL−1, the highest concentrations tested.
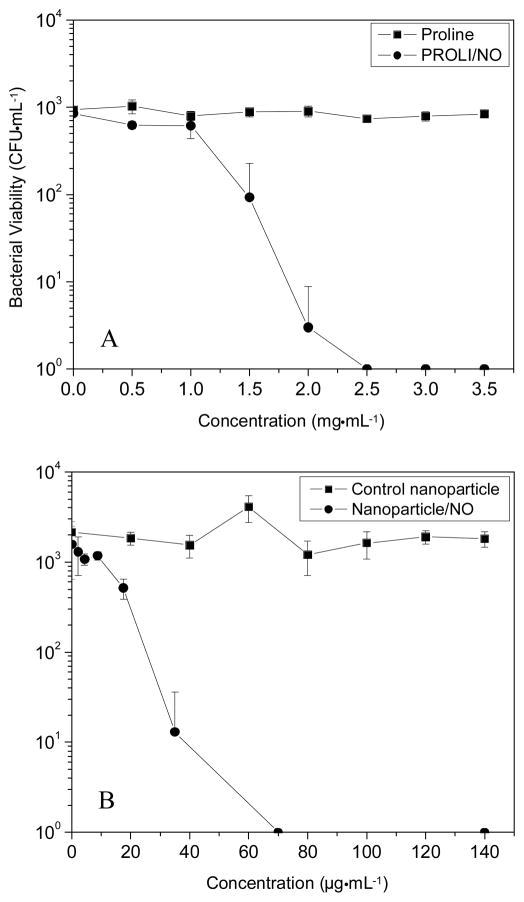
To facilitate direct comparison of the amount of NO necessary to kill P. aeruginosa, initial studies were conducted in PBS. The bacterial killing assays conducted in aqueous buffer demonstrate the bactericidal activity of NO under nutrient-free (“static”) conditions in which the bacteria were unable to replicate. In this manner, the data collected were not convoluted by the ability of the bacterial culture to proliferate in the medium during the experiment. Bacterial killing assays were performed instead of the more conventional minimum inhibitory concentration (MIC) assays in order to assess the extent to which NO actually kills P. aeruginosa as opposed to simply inhibiting its growth. An understanding of these parameters is important because it has been suggested that bactericidal agents are less likely to foster resistance among pathogens than those that are simply bacteriostatic.32 As shown in Figure 3, the concentrations of PROLI/NO and NO-releasing nanoparticles that proved completely bactericidal (3 logs of killing) to P. aeruginosa were 2.5 mg·mL−1 and 70 μg·mL−1, respectively. Thus, by mass, approximately 35 times more PROLI/NO was required than silica nanoparticles to completely kill all P. aeruginosa cells in the bacterial suspension. Both the proline and 45 mol% AHAP3 silica controls depleted of NO exhibited no killing of P. aeruginosa over the concentration ranges tested, indicating that the toxicity observed from the NO-releasing analogues was due entirely to NO. Real-time chemiluminescent detection of NO released from the two NO-donor systems (Fig. 2) allowed for a direct comparison of the amount of NO released into solution over the 1 h time course of the bactericidal assays. Of note, the amount of NO required per mL to elicit a 3 log reduction in bacterial viability was markedly less from the nanoparticle scaffold than from PROLI/NO (0.22 versus 4.5 μmol NO from nanoparticles and PROLI/NO, respectively). The amount of NO delivered is expressed as total μmol NO released instead of a concentration (e.g., mM) because the NO quickly reacts to form other reactive nitrogen and oxygen species. As such, the exact molar concentration of NO and the byproducts in solution are not known.
Figure 3.
Bactericidal efficacy of (A) proline and PROLI/NO and (B) control and NO-releasing 45 mol% AHAP3 silica nanoparticles (balance TEOS) against P. aeruginosa in phosphate buffered saline.
Time-based bactericidal assays under nutrient growth conditions
While the PBS-based bactericidal assays allow an uncomplicated comparison of the dose of NO from both systems required to kill P. aeruginosa, they do not demonstrate the temporal efficacy of each system, or accurately mimic a situation where the bacteria have the ability to replicate. To better understand such parameters, time-based killing assays were performed in tryptic soy broth (TSB) to test the bactericidal efficacy of NO-releasing silica nanoparticles in a culture medium where the bacteria had the capacity to proliferate and present a competition between the rate of bacterial cell killing and replication. Such time-kill studies offer valuable information regarding the temporal efficacy of antimicrobial agents.33 Conventional antibacterial susceptibility tests such as the MIC and minimum bactericidal concentration (MBC) assays do not allow for acute temporal studies. In the TSB nutrient medium, P. aeruginosa exposed to blank and control (proline and silica) solutions proliferated over the 2 h experiment (Figure 4). As expected, the concentration of both NO-releasing silica nanoparticles and PROLI/NO necessary to completely kill P. aeruginosa in TSB was greater than the dose necessary to achieve the same result in PBS. This increase is attributed to both the ability of P. aeruginosa to proliferate in TSB and the NO scavenging properties of the protein digest that comprises TSB. Indeed, chemiluminescent NO release measurements performed in TSB revealed that a significant amount of NO was scavenged by the TSB media itself (Supporting Information), effectively lowering the amount of NO able to act on the P. aeruginosa cells.
Figure 4.
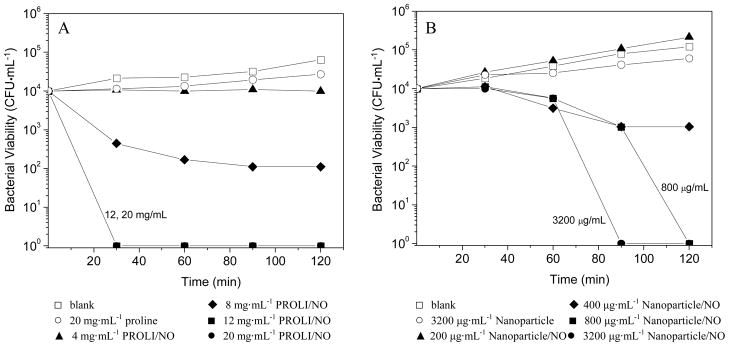
Time- and concentration-based bactericidal efficacy of (A) proline (control) and PROLI/NO and (B) control (Nanoparticle) and NO-releasing 45 mol% AHAP3 silica nanoparticles (Nanoparticle/NO).
Despite the scavenging of NO and bacterial proliferation in TSB, complete bacterial killing was still achieved, albeit at higher concentrations of both nanoparticles and PROLI/NO. Similar to the experiments performed in PBS, the amount (by mass) of PROLI/NO necessary to kill all P. aeruginosa was greater than that of NO-releasing nanoparticles. Figure 4 illustrates the dose- and time-dependent bactericidal activity of both PROLI/NO and NO-releasing silica nanoparticles. At a nanoparticle concentration of 400μg·mL−1, ~90% bactericidal efficacy was achieved after 2 h (one log reduction in viable P. aeruginosa; Fig. 4B). Doubling the particle concentration to 800 μg·mL−1 resulted in 100% bacterial killing over the same period (4 log reduction in viable P. aeruginosa). Complete bactericidal activity was achieved in a shorter period (90 min) using significantly greater concentrations of silica nanoparticles (3200 μg·mL−1). However, particle concentrations >3200 μg·mL−1 did not reduce the time necessary for 100% bacterial killing below 90 min (data not shown). In contrast, PROLI/NO achieved more rapid bacterial killing than the 45 mol% AHAP3 nanoparticles, but at significantly greater concentrations. For example, a concentration of 12 mg·mL−1 PROLI/NO resulted in complete killing after only 30 min. The difference in the rate of bacterial killing is attributed to the NO-release kinetics of each NO donor. The NO release from PROLI/NO is rapid with a half life (t1/2) of 1.7 min, resulting in rapid (≤30 min) bacterial killing at 12 and 20 mg·mL−1. In contrast, the NO release kinetics from 45 mol% AHAP3 silica nanoparticles, which were unchanged in the presence of TSB, are significantly longer (t1/2 = 18 min), thereby requiring longer incubation periods at 800 and 3200 μg·mL−1 to achieve complete bactericidal activity. Analogous to the results obtained in PBS, complete bacterial killing required a markedly greater amount of NO per mL from PROLI/NO (21.6 μmol) than from the NO-releasing silica nanoparticles (2.8 μmol).
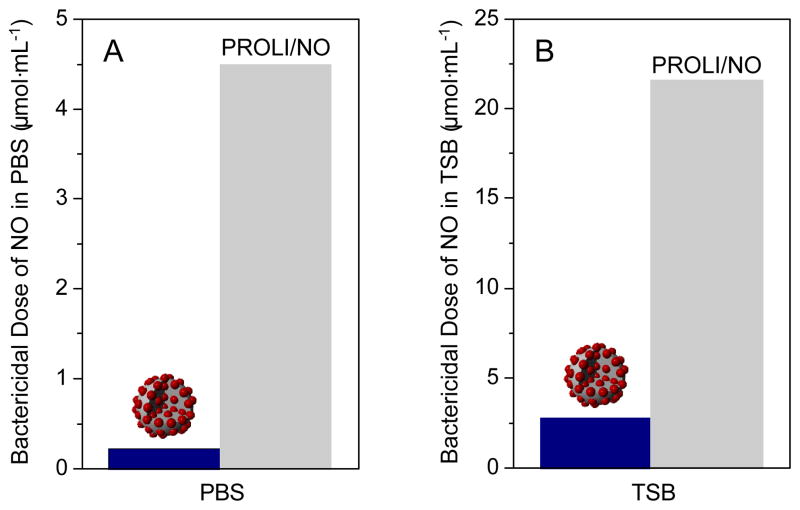
A direct comparison of the amount of NO required from each vehicle to achieve 100% bactericidal efficacy in both PBS and TSB is shown in Figure 5. Greater amounts of NO were necessary in TSB to achieve complete bactericidal activity than from the same vehicles in PBS due to both the ability of P. aeruginosa to proliferate in TSB and the NO-scavenging properties of TSB as noted above. Regardless of the media, NO delivered from the nanoparticles exhibited significantly greater bactericidal efficacy than NO delivered from the small molecule diazeniumdiolate (i.e., PROLI/NO). Indeed, the amount of NO required from PROLI/NO to completely kill P. aeruginosa was approximately one order of magnitude greater than that required from the 45 mol% AHAP3 nanoparticles. Since the reactivity of NO is largely dependent on its localized concentration and diffusion properties,34 NO derived from a small molecule dispersed throughout solution is expected to possess slower diffusion into bacterial cells and correspondingly lessened antibacterial activity compared to the high localized concentrations of NO delivered by silica nanoparticles.
Figure 5.
Comparison of the NO doses necessary from silica nanoparticles (dark blue) and PROLI/NO (grey) to achieve 100% bactericidal efficacy against P. aeruginosa in (A) PBS and (B) TSB.
Confocal microscopy studies
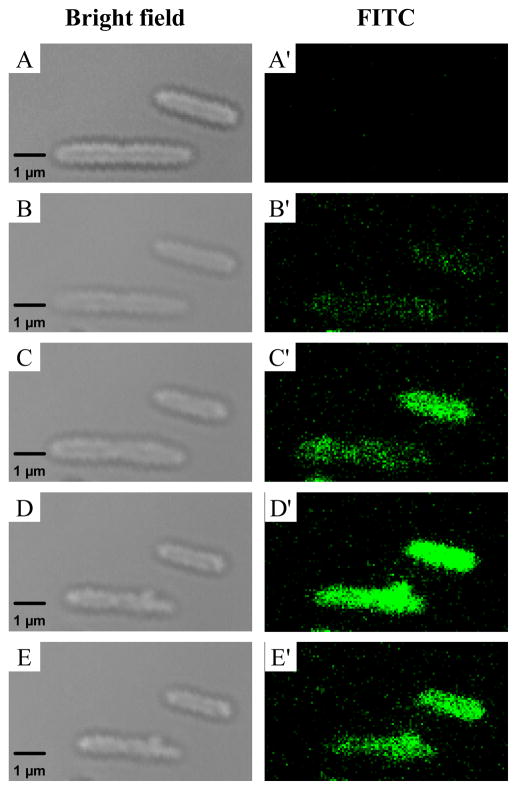
To understand the enhanced bactericidal efficacy of NO delivered from nanoparticles compared to PROLI/NO, fluorescein isothiocyanate (FITC)-modified silica nanoparticles were synthesized to visually determine if any nanoparticle interaction with P. aeruginosa cells existed. After synthesis of the nanoparticles, characteristic FITC fluorescence was observed at 500 – 530 nm when the particles were excited at 488 nm. Incorporation of FITC into the silica nanoparticle scaffold did not significantly alter the NO-release properties of the nanoparticles (data not shown) or the particle diameter (124 ± 13 nm vs. 136 ± 15 nm with and without FITC, respectively). Using the FITC-modified silica nanoparticles, confocal fluorescence microscopy studies were conducted to determine if the enhanced bactericidal efficacy of the nanoparticles was due to nanoparticle interaction with P. aeruginosa cells. As shown in Figure 6, nanoparticles began to associate with the P. aeruginosa cells as early as 10 min post-injection. The possible mechanism by which this association occurs is not entirely understood, but most likely is attributed to electrostatic35 and/or hydrophobic36 interactions between the particles and bacterial membrane.
Figure 6.
Scanning confocal microscopy images of FITC-modified NO-releasing silica nanoparticle association with P. aeruginosa cells. Images were acquired in bright-field mode (A–E) and on the FITC fluorescence channel (A′–E′) before injection (A, A′) of 100 μg·mL−1 NO-releasing FITC-modified AHAP3 silica nanoparticles, and 10 (B, B′), 20 (C, C′), 30 (D, D′), and 60 (E, E′) minutes post-injection.
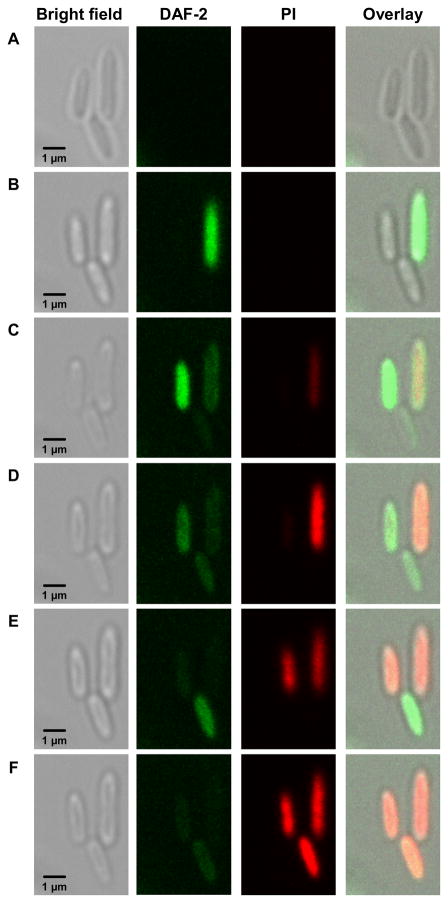
A NO-sensitive fluorescence probe, 4,5-diaminofluorescein diacetate (DAF-2 DA),37 was employed to determine if the association between NO-releasing silica nanoparticles and P. aeruginosa cells resulted in high local concentrations of NO and more efficient delivery of NO to the bacterial cells. Once DAF-2 DA permeates the bacterial cell membrane, intracellular esterases hydrolyze the acetate groups to generate the membrane-impermeable DAF-2.37 In the presence of NO, DAF-2 is nitrosated by reactive nitrogen species (e.g., N2O3) and exhibits bright green intracellular fluorescence.37 Cells loaded with DAF-2 were imaged in the presence of propidium iodide (PI), a nucleic acid viability dye that only enters cells with compromised membranes. Inside the cell, PI exhibits strong red fluorescence upon interaction with nucleic acid material. Red fluorescence due to PI entering cells with disrupted plasma membranes thus indicates cell death.38 Prior to introduction of NO-releasing silica nanoparticles, no autofluorescence was observed from either DAF-2 DA or DAF-2. However, P. aeruginosa cells loaded with DAF-2 exposed to 100 μg·mL−1 NO-releasing nanoparticles exhibited strong DAF-2 fluorescence (Fig. 7B–E), indicative of a high localized concentration of NO in close proximity to the bacterial cells. As more NO was released from the nanoparticles, the DAF-2 green fluorescence in each cell increased progressively, indicating that the NO level inside each cell was increasing. After reaching a peak intracellular intensity of DAF-2 fluorescence, PI then rapidly entered the bacterial cells due to membrane disruption and cell death. The increase in PI fluorescence coincided with a decrease in DAF-2 fluorescence (Fig. 7C–F), suggesting that the DAF-2 fluorophore leaked from the cytosol through the damaged cell membrane that allowed PI to enter the cells.
Figure 7.
Intracellular DAF-2 (green) and propidium iodide (red) fluorescence from P. aeruginosa bacterial cells incubated with 100 μg·mL−1 45 mol% AHAP3/TEOS NO-releasing silica nanoparticles. DAF-2 fluorescence indicates the presence of NO and reactive nitrogen species, while propidium iodide fluorescence indicates membrane destruction and cell death. Images were acquired (A) 30 min, (B) 83 min, (C) 113 min, (D) 124 min, (E) 132 min, and (F) 140 min after nanoparticle addition.
In contrast to the strong intracellular green fluorescence observed from DAF-2 in the presence of 100 μg·mL−1 NO-releasing silica nanoparticles, DAF-2 fluorescence was not observed when an equal amount of NO was delivered with PROLI/NO (data not shown). As indicated by the absence of any PI fluorescence from the bacterial cells over the same period, P. aeruginosa cell death was not observed with this dose of NO from PROLI/NO, thus reaffirming that doses of NO delivered from nanoparticle delivery vehicles were more efficient at killing P. aeruginosa cells compared to similar doses from small molecule NO donors. When the amount of PROLI/NO was increased to bactericidal levels (5 mg·mL−1), rapid cell death was observed as evidenced by bright red intracellular PI fluorescence in the confocal microscopy images (Supporting Information). However, DAF-2 fluorescence was still not observed prior to cell death (in contrast to the nanoparticles), indicating that the NO concentration surrounding the cells was not high enough to induce intracellular DAF-2 fluorescence. These data reveal that the delivery of NO is significantly more efficient from silica nanoparticles. As such, lower doses of NO delivered from silica nanoparticles effectively kill the bacteria.
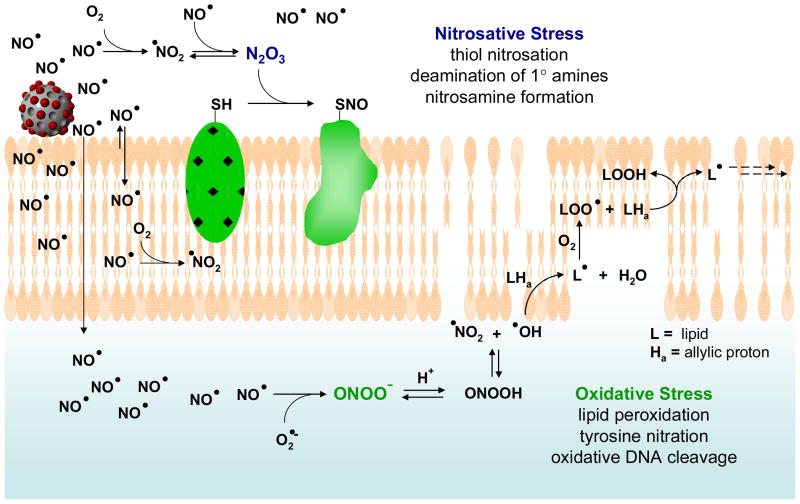
As shown in Figure 8, the ability of the nanoparticles to deliver appreciable NO payloads in close proximity to the bacterial cells allows the NO to more efficiently target cellular components (e.g., cell membrane, DNA, proteins, etc.) critical to cell function, circumventing the need for NO to diffuse across large distances in solution to reach the cell. As a lipophilic molecule, NO is capable of rapidly crossing cell membranes.13 The release of high levels of NO at or near the cell membrane would be expected to lead to high intracellular concentrations of NO. The antibacterial properties of NO and its reactive byproducts have been thoroughly reviewed,13, 28, 39–41 and are typically ascribed to either nitrosative or oxidative stress. In addition to causing DNA deamination, nitrosative species such as N2O3 may nitrosate thiols (S-nitrosation) on proteins and initiate disulfide bridging with other thiols on the protein,42 thereby directly altering protein function.13 Due to their lipophilic nature, NO and O2 tend to concentrate in cell membranes, accelerating NO’s oxidation to N2O3 and creating greater nitrosative stress within and near the bacterial membrane.41 Nitrosation of both cell surface proteins and intracellular proteins (including enzymes) has been shown to cause bacterial cell death.13 Oxidative stress is driven primarily by peroxynitrite (ONOO−), which forms via the reaction of NO with superoxide endogenously derived from the bacterial cellular respiration process.13, 43–46 Thus, oxidative damage is expected to occur predominantly inside the cell, since superoxide does not readily cross cell membranes.47 A significant antibacterial process related to oxidative stress is peroxynitrite-dependent lipid peroxidation, which stems from OH and NO2 radicals derived from peroxynitrous acid (HOONO; Fig. 8).48 The production of NO2 radical from NO and O2 is also accelerated in membranes, leading to even greater NO-mediated oxidative stress within bacterial cell membranes.41 Membrane destruction via lipid peroxidation has been proposed as one of the major mechanisms of NO-mediated bactericidal activity. As a peroxynitrite-dependent process requiring superoxide, NO released in close proximity to a bacterial cell would be expected to exert greater bactericidal effects than NO released diffusely throughout solution by generating a larger intracellular NO concentration.13 Indeed, we observed direct evidence of membrane destruction during the confocal microscopy experiments by the rapid appearance of intracellular PI fluorescence (Fig. 7) in cells treated with NO-releasing silica nanoparticles. By virtue of the extended NO-release half-life of the silica nanoparticles relative to PROLI/NO, a significant portion of the NO is retained until after particle association with the P. aeruginosa cells. Such high localized NO release in close proximity to the bacterial cells may then facilitate delivery of greater concentrations of NO and other reactive species to the cell membrane and into the cell itself, leading to enhanced bactericidal efficacy of NO delivered from nanoparticles.
Figure 8.
Proposed mechanisms by which NO acts as an antibacterial agent (adapted from references 13, 28, 39–41; not to scale). NO’s antibacterial properties are attributed to both nitrosative and oxidative stress exerted by reactive byproducts such as N2O3 and ONOO− (peroxynitrite). Nitrosative stress leads in part to nitrosation of thiols on proteins as well as DNA deamination, while oxidative stress is responsible for membrane destruction via lipid peroxidation. Notably, increased NO and O2 concentrations in lipid membranes leads to enhanced production of both nitrosative and oxidative species such as N2O3 and NO2 in the membrane.
Cytotoxicity of AHAP3 nanoparticles and PROLI/NO against L929 mouse fibroblasts
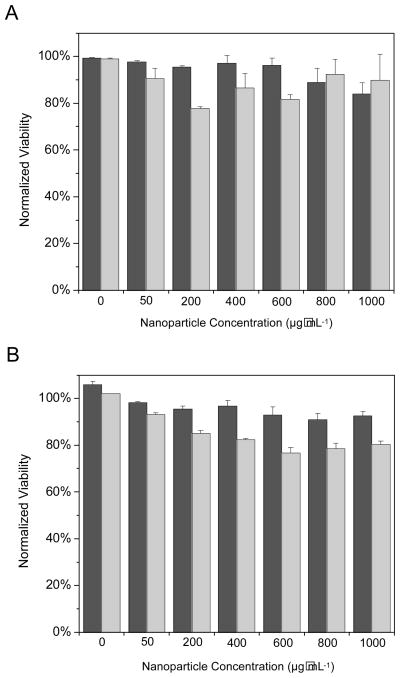
The significant toxicity that NO-releasing 45 mol% AHAP3 silica nanoparticles exhibited against P. aeruginosa cells demands study of their effect on healthy mammalian cells as well. Although Ghaffari et al. have demonstrated that NO gas (200 ppm for 4 h) was not toxic to human dermal fibroblasts,14 similar studies were conducted to determine the combined effects of NO and the silica nanoparticle scaffold on L929 mouse fibroblast cells. Such cells represent the standard for cytotoxicity testing of novel therapeutic agents.49, 50 Survival of the L929 cells in the presence of control and NO-releasing silica nanoparticles was monitored via both propidium iodide (PI) and lactate dehydrogenase (LDH) viability assays over 2 h to mimic the time-based bactericidal assays described above. As discussed above, healthy cells with uncompromised membranes exclude PI in the buffer solution, while disrupted plasma membranes allow PI to diffuse into the cell and emit characteristic fluorescence after complexation with intracellular nucleic acids.38 Positive detection of LDH in the culture medium also indicates compromised cellular membranes that allow larger proteins to leak out of the cell, further indicating membrane disruption and cell death. Both assays thus monitor membrane permeability to assess cell viability, a suitable method to assay for the destructive properties of reactive NO byproducts that are known to form in greater quantities at lipid membranes.41 A range of nanoparticle concentrations was tested to encompass the bactericidal concentrations of 45 mol% AHAP3 silica in the PBS and TSB assays (70 μg·mL−1 and 800 μg·mL−1, respectively). As shown in Figure 9, both control and NO-releasing 45 mol% AHAP3 silica nanoparticles were found to present minimal toxicity to the L929 fibroblasts. Remarkably, when exposed to the same concentration of NO-releasing silica nanoparticles required to induce 4 logs of bacterial killing (800 μg·mL−1; Fig. 4B), L929 cells maintained 92% viability as measured by the PI assay. Thus, P. aeruginosa appears to be extremely susceptible to NO-releasing silica nanoparticles, while such delivery vehicles pose minimal threat to healthy mammalian fibroblasts.
Figure 9.
Toxicity of 45 mol% AHAP3 control (dark gray) and NO-releasing (light gray) silica nanoparticles to L929 mouse fibroblasts as measured by (A) membrane permeability to propidium iodide and (B) lactate dehydrogenase leaching.
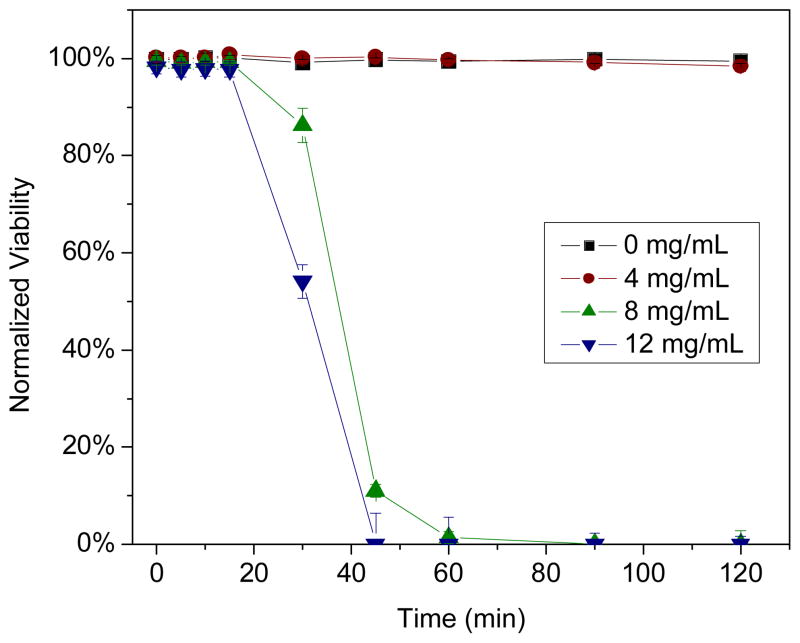
In contrast, PROLI/NO proved toxic to L929 fibroblasts when administered at concentrations required to kill P. aeruginosa. When L929 cells were exposed to the dose of PROLI/NO required to induce 4 logs of bacterial killing in TSB within 2 h (12 mg·mL−1; Fig. 4A), 100% cell death (i.e., 0% viability) was observed within 45 min (Figure 10). While proline, (the precursor of PROLI/NO) exhibited no toxicity to L929 fibroblasts at concentrations up to 16 mg·mL−1 (data not shown), 100% fibroblast cell death was observed within 90 min at a PROLI/NO concentration of 8 mg·mL−1. Upon reducing the PROLI/NO concentration to 4 mg·mL−1, 100% fibroblast viability was maintained through 2 h, but at the expense of bactericidal efficacy (Fig. 4A). These results reinforce the advantages of delivering NO via nanoparticle scaffolds, as both the amount of NO necessary to kill bacteria and toxicity to healthy mammalian cells are reduced.
Figure 10.
Cytotoxicity of PROLI/NO against L929 fibroblast cells as measured by propidium iodide viability assay. The backbone of PROLI/NO (the amino acid proline) demonstrated no toxicity to L929 cells at concentrations up to 16 mg/mL.
Conclusions
Nitric oxide delivered from silica nanoparticles was shown to be significantly more effective at killing pathogenic P. aeruginosa than NO derived from the small molecule NO donor PROLI/NO. Indeed, significantly less NO was required from the nanoparticles to kill P. aeruginosa than from PROLI/NO, even though the initial NO release from PROLI/NO was 6-fold greater than from 45 mol% AHAP3 silica nanoparticles. In vitro cytotoxicity experiments conducted with L929 mouse fibroblasts confirmed that NO-releasing silica nanoparticles are largely non-toxic to mammalian fibroblast cells at concentrations capable of killing P. aeruginosa, while PROLI/NO presents significant toxicity to such cells when administered at bactericidal concentrations. Such results demonstrate the promise that NO holds as a new strategy for battling bacterial infection. Confirmation of particle association with P. aeruginosa cells and the measurement of intracellular NO levels helped elucidate the differential toxicity observed between macromolecular and small molecule NO donors. The versatility in the synthesis of NO-releasing silica scaffolds allows for both tuning of size and exterior functionality that may further enhance their use as antibacterial agents. Future studies are aimed at identifying the intracellular location of the reactive radical species formed upon NO release and establishing a mechanistic understanding of the interactions between NO-releasing nanoparticles and bacterial cell membranes. As well, experiments are underway to evaluate the antibacterial properties of NO-releasing silica nanoparticles against other species of pathogenic bacteria including gram-positive strains and those that exhibit resistance to conventional antibiotics.
Methods
Materials
Tetraethoxysilane (TEOS) and sodium methoxide (NaOCH3) were purchased from Fluka (Buchs, Switzerland). N-(6-Aminohexyl)aminopropyltrimethoxysilane (AHAP3) and 3-aminopropyltrimethoxysilane (APTMS) were purchased from Gelest (Tullytown, PA). Methanol (MeOH), ethanol (EtOH), and ammonia solution (NH4OH, 30 wt% in water) were purchased from Fisher Scientific (Fair Lawn, NJ). Tryptic soy broth (TSB, soybean-casein digest) was purchased from Becton, Dickinson and Company (Sparks, MD). Nitric oxide (NO, 99.5%) was obtained from Linde (Raleigh, NC), and argon (Ar) and nitrogen (N2) gases were purchased from National Welders (Raleigh, NC). P. aeruginosa (ATCC #19143) and L929 mouse fibroblast cells were purchased from American Type Culture Collection (Manassas, VA). 4,5-Diaminofluorescein diacetate (DAF-2 DA) was purchased from Calbiochem (San Diego, CA). Fluorescein isothiocyanate (FITC), proline and reagents for the propidium iodide and lactate dehydrogenase cytotoxicity assays were purchased from Sigma (St. Louis, MO). Other solvents and chemicals were analytical-reagent grade and used as received. A Millipore Milli-Q UV Gradient A-10 System (Bedford, MA) was used to purify distilled water to a final resistivity of 18.2 MΩ·cm and a total organic content of ≤6 ppb.
Synthesis of NO-releasing silica nanoparticles
The synthesis and characterization of NO-releasing silica nanoparticles has been described previously.18 Briefly, an aminoalkoxysilane solution was prepared by dissolving AHAP3 (2.3 mmol) in 20 mL of EtOH and 4 mL of MeOH in the presence of NaOCH3 (2.3 mmol). The solution was then placed into 10 mL vials equipped with stir bars. The vials were placed in a Parr bottle, connected to an in-house NO reactor, and flushed with Ar six times to remove O2 in the solution. The reaction bottle was pressurized to 5 atm NO for 3 d with continuous stirring of the silane solution. Prior to removing the diazeniumdiolate-modified AHAP3 silane sample (AHAP3/NO), unreacted NO was purged from the chamber with Ar. Silane solutions were prepared by mixing TEOS (2.8 mmol) and AHAP3/NO (2.3 mmol; corresponding to 45 mol%, balance TEOS) in the EtOH/MeOH solution for 2 min (Scheme 1). The silane solution was then added into 22 mL of EtOH and 6 mL ammonia catalyst (30 wt% in water), and mixed vigorously for 30 min at 4 °C. The precipitated nanoparticles were collected by centrifugation (5000 rpm, 5 min), washed with EtOH several times, dried under ambient conditions for 1 h, and stored in a sealed container at −20 °C until used. Diazeniumdiolate incorporation into the nanoparticle scaffold was confirmed by UV absorbance spectroscopy. The UV absorbance spectra of nanoparticles (both NO-releasing and controls depleted of diazeniumdiolates) suspended in phosphate buffered saline (PBS) at 160 μg·mL−1 were recorded on a Perkin-Elmer Lambda 40 UV/Vis Spectrometer.
Characterization of diazeniumdiolate-modified silane (AHAP3/NO)
1H NMR (CD3OD, δ): 0.61 (br, SiCH2), 1.32 (qt, NRCH2CH2CH2CH2CH2CH2NH2), 1.43 (m, SiCH2CH2CH2, NRCH2CH2), 1.56 (m, CH2CH2NH2), 2.56 (m, CH2CH2NH2), 2.84 (br,CH2NRCH2), 4.88 (s, Si(OCH3)3) where R = NONO−Na+. UV-Vis (EtOH): λmax = 253 nm. 29Si nuclear magnetic resonance (NMR) spectroscopy was also employed to characterize AHAP3/NO. Cross-polarization/magic angle spinning (CP/MAS) 29Si NMR spectra of the AHAP3/NO were obtained at 20 °C on a Bruker 360 MHz DMX spectrometer (Billerica, MA) equipped with wide-bore magnets (triple-axis pulsed field gradient double-resonance probes). The alcoholic solution of diazeniumdiolate-modified silane (i.e., AHAP3/NO) was loaded into 4 mm rotors (double-resonance frequency of 71.548 MHz) and spun at a speed of 8.0 kHz. The chemical shifts were determined in ppm relative to a TMS external standard.
Synthesis of fluorescently-labeled NO-releasing silica nanoparticles
The synthesis of fluorescently-labeled NO-releasing silica nanoparticles was adapted from a previously reported literature procedure.51 Briefly, FITC (10 μmol) was reacted with neat APTMS (200 μmol) overnight in the dark to yield the FITC-modified silane. 1H NMR (CD3OD, δ): 0.32 (br, SiCH2), 1.52 (qt, SiCH2CH2CH2NH-FITC), 2.73 (t, SiCH2CH2CH2NH-FITC), 3.69 (m, SiCH2CH2CH2NH-FITC), 4.81 (s, Si(OCH3)3), 6.46 (m, aromatic), 7.01 (m, aromatic) 7.11 (d, aromatic) 7.45 (d, aromatic) 7.57 (d, aromatic). UV-Vis (EtOH): λmax = 493 nm.
Next, 100 μL of the FITC-modified silane solution was co-condensed with AHAP3/NO (2.3 mmol) and TEOS (2.8 mmol) in the EtOH/ammonia solution as described above to yield FITC-labeled NO-releasing silica nanoparticles (Supporting Information). Incorporation of FITC was confirmed by exciting the particles at 488 nm and observing the characteristic fluorescence due to FITC at 500 – 530 nm.
Size characterization of silica nanoparticles
The size of control, NO-releasing, and FITC-modified silica nanoparticles was characterized via atomic force microscopy (AFM). Prior to analysis, the particles were suspended in toluene, deposited on a freshly cleaved mica surface (SPI; West Chester, PA), and dried under ambient conditions for 3 h. Contact mode AFM images were obtained in air using a Molecular Force Probe 3D AFM (Asylum Research; Santa Barbara, CA) controlled with MFP-3D software running under Igor Pro (Wavemetrics; Lake Oswego, OR). Triangular silicon nitride cantilevers with a nominal spring constant of 0.12 N·m−1 and resonance frequency of 20 kHz (Veeco; Santa Barbara, CA) were used to acquire height/topography images at a scan rate of 1.0 Hz.
Synthesis of 1-[2-(carboxylato)pyrrolidin-1-yl]diazen-1-ium-1,2-diolate (PROLI/NO) (adapted from a previously reported procedure29)
Proline (300 mg, 2.6 mmol) was dissolved in a 50:50 mixture of methanol:ether and treated with 281 mg (5.2 mmol) of NaOCH3. The basic solution was placed in a glass hydrogenation bomb and stirred. The bomb was copiously flushed with Ar to remove atmospheric O2, followed by introduction of NO gas at 5 atm. After 3 d, the glass vial was removed from the vessel after thorough flushing with Ar. The solution was treated with cold ether to precipitate the product (PROLI/NO). The NO donor precipitate was then filtered, and dried under vacuum at −70 °C (dry ice/acetone bath) to yield 299 mg PROLI/NO.
Nitric oxide release measurements
Nitric oxide release from both the diazeniumdiolate-modified silica nanoparticles and PROLI/NO was measured in deoxygenated phosphate-buffered saline (PBS, 0.01 M; 37 °C) at pH 7.4 using a Sievers NOA 280i chemiluminescence NO analyzer (Boulder, CO). Nitric oxide released from the donors was transported to the analyzer by a stream of N2 (70 mL·min−1) passed through the reaction cell. The instrument was calibrated with air passed through a NO zero filter (0 ppm NO) and a 24.1 ppm NO standard gas (balance N2).
Bactericidal assays under static conditions
To test the bactericidal properties of PROLI/NO and NO-releasing 45 mol% AHAP3/TEOS silica nanoparticles under non-growth (“static”) conditions, P. aeruginosa was cultured to a concentration of 108 colony-forming-units (CFUs) per mL in tryptic soy broth (TSB), resuspended in sterile PBS, and adjusted to a concentration of 103 CFU·mL−1. Silica nanoparticles (NO-releasing and control), PROLI/NO, and proline were added to separate aliquots of the bacterial suspension over a concentration range optimized for each system. After 1 h incubation at 37 °C with gentle agitation, 100-μL aliquots from each suspension were plated on tryptic soy agar. After overnight incubation at 37 °C, the colonies on each plate were counted, allowing for calculation of the number of viable P. aeruginosa cells in each vial at the time of plating.
Time-based bactericidal assays under nutrient growth conditions
To test the temporal efficacy of the NO-releasing silica nanoparticles, time-based antibacterial assays were conducted in TSB nutrient media. P. aeruginosa was cultured in TSB to a concentration of 108 CFU·mL−1 and diluted to 104 CFU·mL−1 in additional TSB. Silica nanoparticles (control and NO-releasing), PROLI/NO, and proline were added to separate aliquots of the 104 bacterial suspension over concentration ranges optimized for each system. Every 30 min for 2 h, 100-μL aliquots of each suspension were removed, diluted 10-fold in PBS, and plated on tryptic soy agar. Bacterial viability was determined as described above after incubating the plates overnight at 37 °C.
Interaction between nanoparticles and bacterial cells
P. aeruginosa was cultured in TSB to 108 CFU·mL−1, pelleted by centrifugation, resuspended in PBS, and adjusted to a concentration of 106 CFU·mL−1 in PBS. The bacterial suspension was seeded onto a glass microscope slide where the bacteria were allowed to adhere to the slide for 30 min. The microscope slide was placed on the stage of a Zeiss LSM 510 confocal fluorescence microscope (Chester, VA) and bright-field and fluorescence images of the bacteria were acquired with a 63x N.A. 1.4 planapochromat oil immersion lens. Next, an aliquot of FITC-modified NO-releasing silica nanoparticles (final concentration = 100 μg·mL−1) was added and bright-field and fluorescence images of the same field were captured after 0, 10, 20, 30, and 60 min. The FITC fluorophores were excited with the 488 nm line of an Ar laser and the fluorescence was collected using a BP 500–530 nm bandpass filter.
Confocal fluorescence microscopy for detection of intracellular NO and cell killing
Confocal microscopy experiments were conducted to simultaneously monitor intracellular concentrations of NO within P. aeruginosa cells and the kinetics of cell killing using 4,5-diaminofluorescein diacetate (DAF-2 DA, a NO-sensitive fluorescence probe), and propidium iodide (PI, a fluorescence viability dye that enters only cells with compromised membranes and emits bright red fluorescence after binding to DNA38). An aliquot of P. aeruginosa, cultured as described above, was resuspended at 106 CFU·mL−1 in PBS supplemented with DAF-2 DA (10 μM) and incubated at 37 °C for 30 min. 500 μL of the cells loaded with DAF-2 were transferred onto a glass microscope slide affixed in a circular microscope cell to allow capture of initial images. 500 μL of PBS containing 10 μM DAF-2 DA, 60 μM PI and either NO-releasing silica nanoparticles (200 μg·mL−1) or PROLI/NO (388 μg·mL−1) was then introduced. Images were immediately collected with a 63x objective every minute for 2.5 h. The fluorescent reaction product of NO and DAF-2 (excitation λmax = 495 nm, emission λmax = 515 nm)37 was excited with the 488 line of an argon laser. Fluorescence was collected using a BP 500–530 nm bandpass filter. PI fluorescence was excited with the 543 nm line of a HeNe laser and collected with a long-pass (LP 560) filter. All confocal microscopy experiments were performed at 25 °C. At 25 °C, the half life of NO release from 45 mol% AHAP3 silica nanoparticles increased to >1 h. Thus, the initial level of NO release was reduced, extending the duration of NO release.
Propidium iodide cytotoxicity assay
L929 mouse fibroblasts were plated on 24-well tissue culture treated dishes (BD Bioscience #353047) at a density of 3.0 × 105 cells·mL−1 (150 × 103 cells per well) and incubated overnight at 37 °C in 5% CO2/95% air. For the PI assay, the incubation buffer (Minimum Essential Medium) was aspirated from each of the wells and replaced with 500 μL of Krebs-Ringer-HEPES (KRH) buffer containing 115 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM KH2PO4, 1.2 mM MgSO4, 25 mM HEPES, pH 7.4, supplemented with 30 μM PI.52 Control or NO-releasing silica nanoparticles were added to the wells at 0, 50, 200, 400, 600, 800, or 1000 μg·mL−1, or PROLI/NO was added at 0, 4, 8, or 12 mg·mL−1. The fluorescence resulting from PI complexation with intracellular nucleic acid material38 in cells with compromised membranes was acquired for a total of 120 min. Upon completion of these measurements, the cells were incubated with digitonin (40 μM) for 20 min to completely permeabilize the plasma membranes and achieve a maximum PI fluorescence. Cell viability is presented as the increase in PI fluorescence from each well expressed as the percentage of maximal fluorescence obtained from cells treated with digitonin (100% cell death).
Lactate dehydrogenase cytotoxicity assay
The lactate dehydrogenase (LDH) cytotoxicity assay was performed concomitantly with the same cells used for the PI assay described above. Every 15 min, 20-μL aliquots of KRH buffer were removed from the plate used for the PI assay and stored at −20 °C in black 96-well plates (Greiner; Monroe, NC) for subsequent LDH analysis. The 96-well plates containing aliquots of incubation buffer were warmed to 37 °C. Lactate dehydrogenase activity was measured from the rate of NADH production after adding 180 μL of KRH buffer containing 0.22 mM NAD+, 11.1 mM sodium lactate and 11.1 mM hydrazine, pH 8.0 into each well.53 The NADH fluorescence was monitored with a FluoStar Galaxy plate reader using 340 nm excitation and 460 nm emission filters. The LDH activity is expressed as the change in relative fluorescence per min per well. The data are normalized to maximal LDH activity in each well obtained from samples treated with 40 μM digitonin for 20 min.
Statistics
For the bactericidal assays conducted in PBS, n = 3 and data are expressed as mean values ± standard deviation. Data from both the PI and LDH cytotoxicity assays are presented as mean values ± standard error of the mean.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH EB000708). Graduate research fellowships from Pfizer (EMH, NAS) are gratefully acknowledged. CBJ and DAW acknowledge undergraduate research grants from the E. C. Markham Research Fund and the Frances C. and William P. Smallwood Foundation, respectively.
Footnotes
Supporting Information Available: Synthesis of FITC-modified silica nanoparticles, AFM analysis of nanoparticle dimensions, scavenging of NO by TSB, and confocal fluorescence microscopy images of PROLI/NO-treated P. aeruginosa cells. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Robson MC. Wound Infection: A Failure of Wound Healing Caused by an Imbalance of Bacteria. Surg Clin N Am. 1997;77:637–650. doi: 10.1016/s0039-6109(05)70572-7. [DOI] [PubMed] [Google Scholar]
- 2.Bad Bugs. No Drugs: As Antibiotic Discovery Stagnates, a Public Health Crisis Brews. Infectious Diseases Society of America. 2004:1–35. [Google Scholar]
- 3.Gu H, Ho PL, Tong E, Wang L, Xu B. Presenting Vancomycin on Nanoparticles to Enhance Antimicrobial Activities. Nano Lett. 2003;3:1261–1263. [Google Scholar]
- 4.Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Ramirez JT, Yacaman MJ. The Bactericidal Effect of Silver Nanoparticles. Nanotechnology. 2005;16:2346–2353. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- 5.Sambhy V, MacBride MM, Peterson BR, Sen A. Silver Bromide Nanoparticle/Polymer Composites: Dual Action Tunable Antimicrobial Materials. J Am Chem Soc. 2006;128:9798–9808. doi: 10.1021/ja061442z. [DOI] [PubMed] [Google Scholar]
- 6.Panacek A, Kvitek L, Prucek R, Kolar M, Vecerova R, Pizurova N, Sharma VK, Nevecna T, Zboril R. Silver Colloid Nanoparticles: Synthesis, Characterization, and their Antibacterial Activity. J Phys Chem B. 2006;110:16248–16253. doi: 10.1021/jp063826h. [DOI] [PubMed] [Google Scholar]
- 7.Trewyn BG, Whitman CM, Lin VSY. Morphological Control of Room-Temperature Ionic Liquids Templated Mesoporous Silica Nanoparticles for Controlled Release of Antibacterial Agents. Nano Lett. 2004;4:2139–2143. [Google Scholar]
- 8.Silver S. Bacterial Silver Resistance: Molecular Biology and Uses and Misuses of Silver Compounds. FEMS Microbiol Rev. 2003;27:341–353. doi: 10.1016/S0168-6445(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 9.Li XZ, Nikaido H, Williams KE. Silver-Resistant Mutants of Escherichia coli Display Active Efflux of Ag+ and Are Deficient in Porins. J Bacteriol. 1997;179:6127–6132. doi: 10.1128/jb.179.19.6127-6132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marletta MA, Tayeh MA, Hevel JM. Unraveling the Biological Significance of Nitric Oxide. BioFactors. 1990;2:219–225. [PubMed] [Google Scholar]
- 11.MacMicking J, Xie Q, Nathan C. Nitric Oxide and Macrophage Function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 12.MacMicking J, Nathan C, Hom G, Chartrain N, Fletcher DS, Trumbauer M, Stevens K, Xie Q, Sokol K, Hutchinson N, Chen H, Mudgett JS. Altered Response to Bacterial Infection and Endotoxic Shock in Mice Lacking Inducible Nitric Oxide Synthase. Cell. 1995;81:641–650. doi: 10.1016/0092-8674(95)90085-3. [DOI] [PubMed] [Google Scholar]
- 13.Fang FC. Mechanisms of Nitric Oxide-Related Antimicrobial Activity. J Clin Invest. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghaffari A, Miller CC, McMullin B, Ghahary A. Potential Application of Gaseous Nitric Oxide as a Topical Antimicrobial Agent. Nitric Oxide. 2006;14:21–29. doi: 10.1016/j.niox.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Raulli R, McElhaney-Feser G, Hrabie JA, Cihlar RL. Antimicrobial Properties of Nitric Oxide Using Diazeniumdiolates as the Nitric Oxide Donor. Rec Res Devel Microbiology. 2002;6:177–183. [Google Scholar]
- 16.Rothrock AR, Donkers RL, Schoenfisch MH. Synthesis of Nitric Oxide-Releasing Gold Nanoparticles. J Am Chem Soc. 2005;127:9362–9363. doi: 10.1021/ja052027u. [DOI] [PubMed] [Google Scholar]
- 17.Stasko NA, Schoenfisch MH. Dendrimers as a Scaffold for Nitric Oxide Release. J Am Chem Soc. 2006;128:8265–8271. doi: 10.1021/ja060875z. [DOI] [PubMed] [Google Scholar]
- 18.Shin JH, Metzger SK, Schoenfisch MH. Synthesis of Nitric Oxide-Releasing Silica Nanoparticles. J Am Chem Soc. 2007;129:4612–4619. doi: 10.1021/ja0674338. [DOI] [PubMed] [Google Scholar]
- 19.Hrabie JA, Keefer LK. Chemistry of the Nitric Oxide-Releaseing Diazeniumdiolate (“Nitrosohydroxylamine”) Functional Group and its Oxygen-Substituted Derivatives. Chem Rev. 2002;102:1135–1154. doi: 10.1021/cr000028t. [DOI] [PubMed] [Google Scholar]
- 20.Wang PG, Xian M, Tang X, Wu X, Wen Z, Cai T, Janczuk AJ. Nitric Oxide Donors: Chemical Activities and Biological Applications. Chem Rev. 2002;102:1091–1134. doi: 10.1021/cr000040l. [DOI] [PubMed] [Google Scholar]
- 21.Pruitt BA, McManus AT, Kim SH, Goodwin CW. Burn Wound Infections: Current Status. World J Surg. 1998;22:135–145. doi: 10.1007/s002689900361. [DOI] [PubMed] [Google Scholar]
- 22.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa Infection: Lessons from a Versatile Opportunist. Microbes and Infection. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 23.Howell-Jones RS, Wilson MJ, Hill KE, Howard AJ, Price PE, Thomas DW. A Review of the Microbiology, Antibiotic Usage and Resistance in Chronic Skin Wounds. J Antimicrob Chemother. 2005;55:143–149. doi: 10.1093/jac/dkh513. [DOI] [PubMed] [Google Scholar]
- 24.Shin JH, Schoenfisch MH. Inorganic/Organic Hybrid Silica Nanoparticles as Nitric Oxide Delivery Scaffolds. Chem Mater. :CM702526Q. doi: 10.1021/cm702526q. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hrabie JA, Klose JR, Wink DA, Keefer LK. New Nitric Oxide-Releasing Zwitterions Derived from Polyamines. J Org Chem. 1993;58:1472–1476. [Google Scholar]
- 26.Beckman JS, Conger KA. Direct Measurement of Dilute Nitric Oxide in Solution with an Ozone Chemiluminescent Detector. Methods Enzymol. 1995;7:35–39. [Google Scholar]
- 27.Davies KM, Wink DA, Saavedra JE, Keefer LK. Chemistry of the Diazeniumdiolates. 2. Kinetics and Mechanism of Dissociation to Nitric Oxide in Aqueous Solution. J Am Chem Soc. 2001;123:5473–5481. doi: 10.1021/ja002899q. [DOI] [PubMed] [Google Scholar]
- 28.Wink DA, Mitchell JB. Chemical Biology of Nitric Oxide: Insights into Regulatory, Cytotoxic, and Cytoprotective Mechanisms of Nitric Oxide. Free Radical Biol Med. 1998;25:434–456. doi: 10.1016/s0891-5849(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 29.Saavedra JE, Southan GJ, Davies KM, Lundell A, Markou C, Hanson SR, Adrie C, Hurford WE, Zapol WM, Keefer LK. Localizing Antithrombotic and Vasodilatory Activity with a Novel, Ultrafast Nitric Oxide Donor. J Med Chem. 1996;39:4361–4365. doi: 10.1021/jm960616s. [DOI] [PubMed] [Google Scholar]
- 30.Hsueh PR, Teng LJ, Yang PC, Chen YC, Ho SW, Luh KT. Persistence of a Multidrug-Resistant Pseudomonas aeruginosa Clone in an Intensive Care Burn Unit. J Clin Microbiol. 1998;36:1347–1351. doi: 10.1128/jcm.36.5.1347-1351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dukelow AM, Weicker S, Karachi TA, Razavi HM, McCormack DG, Joseph MG, Mehta S. Effects of Nebulized Diethylenetetraamine-NONOate in a Mouse Model of Acute Pseudomonas aeruginosa Pneumonia. Chest. 2002;122:2127–2136. doi: 10.1378/chest.122.6.2127. [DOI] [PubMed] [Google Scholar]
- 32.Stratton CW. Dead Bugs Don’t Mutate: Susceptibility Issues in the Emergence of Bacterial Resistance. Emerg Infect Dis. 2003;9:10–16. doi: 10.3201/eid0901.020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li RC, Zhu M, Schentag JJ. Achieving an Optimal Outcome in the Treatment of Infections: The Role of Clinical Pharmacokinetics and Pharmacodynamics of Antimicrobials. Clinical Pharmacokinet. 1999;37:1–16. doi: 10.2165/00003088-199937010-00001. [DOI] [PubMed] [Google Scholar]
- 34.Lancaster JR. A Tutorial on the Diffusibility and Reactivity of Free Nitric Oxide. Nitric Oxide. 1997;1:18–30. doi: 10.1006/niox.1996.0112. [DOI] [PubMed] [Google Scholar]
- 35.Shai Y. From Innate Immunity to de-Novo Designed Antimicrobial Peptides. Curr Pharmaceut Design. 2002;8:715–725. doi: 10.2174/1381612023395367. [DOI] [PubMed] [Google Scholar]
- 36.Poelstra KA, van der Mei HC, Gottenbos B, Grainger DW, van Horn JR, Busscher HJ. Pooled Human Immunoglobulins Reduce Adhesion of Pseudomonas aeruginosa in a Parallel Plate Flow Chamber. J Biomed Mater Res. 2000;51:224–232. doi: 10.1002/(sici)1097-4636(200008)51:2<224::aid-jbm11>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 37.Kojima H, Nakatsubo N, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T. Detection and Imaging of Nitric Oxide with Novel Fluorescent Indicators: Diaminofluoresceins. Anal Chem. 1998;70:2446–2453. doi: 10.1021/ac9801723. [DOI] [PubMed] [Google Scholar]
- 38.Tas J, Westerneng G. Fundamental Aspects of the Interaction of Propidium Diiodide with Nucleic Acids Studied in a Model System of Polyacrylamide Films. J Histochem Cytochem. 1981;29:929–936. doi: 10.1177/29.8.6168679. [DOI] [PubMed] [Google Scholar]
- 39.Stamler JS, Lamas S, Fang FC. Nitrosylation: The Prototypic Redox-Based Signaling Mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 40.Fang FC. Antimicrobial Reactive Oxygen and Nitrogen Species: Concepts and Controversies. Nat Rev Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 41.Moller MN, Li Q, Lancaster JR, Denicola A. Acceleration of Nitric Oxide Autoxidation and Nitrosation by Membranes. IUBMB Life. 2007;59:243–248. doi: 10.1080/15216540701311147. [DOI] [PubMed] [Google Scholar]
- 42.Stamler JS. S-Nitrosothiols and the Bioregulatory Actions of Nitrogen Oxides through Reactions with Thiol Groups. Curr Top Microbiol. 1995;196:19–36. doi: 10.1007/978-3-642-79130-7_4. [DOI] [PubMed] [Google Scholar]
- 43.Polack B, Dacheux D, Delic-Attree I, Toussaint B, Vignais PM. Role of Manganese Superoxide Dismutase in a Mucoid Isolate of Pseudomonas aeruginosa: Adaptation to Oxidative Stress. Infect Immun. 1996;64:2216–2219. doi: 10.1128/iai.64.6.2216-2219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hassett DJ, Cohen MS. Bacterial Adaptation to Oxidative Stress: Implications for Pathogenesis and Interaction with Phagocytic Cells. FASEB J. 1989;3:2574–2582. doi: 10.1096/fasebj.3.14.2556311. [DOI] [PubMed] [Google Scholar]
- 45.Fridovich I. The Biology of Oxygen Radicals. Science. 1978;201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 46.Hassett DJ, Schweizer HP, Ohman DE. Pseudomonas aeruginosa sodA and sodB Mutants Defective in Manganese- and Iron-Cofactored Superoxide Dismutase Activity Demonstrate the Importance of the Iron-Cofactored Form in Aerobic Metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nunoshiba T, deRojas-Walker T, Wishnok JS, Tannenbaum SR, Demple B. Activation by Nitric Oxide of an Oxidative-Stress Response that Defends Escherichia coli Against Activated Macrophages. Proc Natl Acad Sci US A. 1993;90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukuto JM, Cho JY, Switzer CH. The Chemical Properties of Nitric Oxide and Related Nitrogen Oxides. In: Ignarro LJ, editor. Nitric Oxide: Biology and Pathology. Academic Press; San Diego: 2000. [Google Scholar]
- 49.Turner TD, Spyratou O, Schmidt RJ. Biocompatibility of Wound Management Products: Standardization of and Determination of Cell Growth Rate in L929 Fibroblast Cultures. J Pharm Pharmacol. 1989;41:775–780. doi: 10.1111/j.2042-7158.1989.tb06364.x. [DOI] [PubMed] [Google Scholar]
- 50.Nablo BJ, Schoenfisch MH. In Vitro Cytotoxicity of Nitric Oxide-Releasing Sol-Gel Derived Materials. Biomaterials. 2005;26:4405–4415. doi: 10.1016/j.biomaterials.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 51.Lin YS, Tsai CP, Huang HY, Kuo CT, Hung Y, Huang DM, Chen YC, Mou CY. Well-Ordered Mesoporous Silica Nanoparticles as Cell Markers. Chem Mater. 2005;17:4570–4573. [Google Scholar]
- 52.Bielinska AU, Chen C, Johnson J, Baker JR. DNA Complexing with Polyamidoamine Dendrimers: Implications for Transfection. Bioconj Chem. 1999;10:843–850. doi: 10.1021/bc990036k. [DOI] [PubMed] [Google Scholar]
- 53.Puranam KL, Boustany R-M. Assessment of Cell Viability and Histochemical Methods in Apoptosis. In: Hannun YA, Boustany R-M, editors. Apoptosis in Neurobiology. CRC Press LLC; Boca Raton, FL: 1998. pp. 129–152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.