Abstract
Colorectal cancers (CRCs) express the WNT effector protein β-catenin in a heterogeneous subcellular pattern rather than uniformly in the nucleus. In this study, we investigated this important aspect of molecular heterogeneity in CRCs by analyzing its basis and relationship with tumor initiating capability. CRC cells expressing the highest WNT expression showed only a marginal increase in tumor initiation capacity. Notably, high WNT activity correlated with a coincident activation of robust MAPK signaling, which when upregulated by KRAS expression or downregulated by EGFR inhibition elicited parallel effects on WNT activity. These findings suggested that on its own high WNT activity may not be a reliable signifier of tumor-initiating potential or stem-like potential. Further, they suggest that MAPK signaling is a critical modifier of intratumoral heterogeneity that contributes signiificantly to determining the impact of WNT activity on stemness phenotypes inCRC cells.
Keywords: Colon and colorectal cancer, beta-catenin, WNT signaling, KRAS and MAPK signaling, tumor-initiating cells
INTRODUCTION
Colorectal cancer (CRC) derives from normal colonic mucosa by stepwise accumulation of somatic genetic alterations, starting with inactivation of the tumor suppressor gene Adenomatous Polyposis Coli (APC) or activation of the oncogene β-catenin (CTNNB1) (1). Among additional mutations identified frequently in human CRC are those in the KRAS oncogene, which occur in over 40% of cases (2) and activate signaling through the Mitogen Activated Protein Kinase (MAPK) pathway. APC is a negative regulator of WNT signaling and its mutational inactivation leads to accumulation of β-catenin, which associates with transcription factors of the T-cell Factor (TCF) family to activate WNT target genes (3). Although this understanding implies that the WNT pathway is uniformly active in all tumor cells that carry mutant APC or CTNNB1, colon cancers show substantial heterogeneity in the accumulation of nuclear β-catenin, which is evident in about 60% of resected tumor specimens, often at the invasive front (4, 5). Such differential accumulation suggests that, in addition to APC and CTNNB1 mutations, other pathway alterations or stimulatory factors external to tumor cells influence β-catenin distribution and WNT pathway activity in CRC (6, 7). For example, CpG island hypermethylation (CIMP) is inversely associated with CTNNB1 activation (8) and APC mutation (9), and amplification of the CDK8 locus, which encodes a member of the mediator complex, contributes to CTNNB1-driven cell transformation (10).
Like other solid tumors, CRCs contain a subpopulation of cells that differ from the majority of tumor cells in displaying an enhanced potential to establish tumors in immune compromised mice; these are thought to represent the clonogenic tumor-initiating cells (11–13). Because some surface markers that have been used to enrich tumor-initiating cells, including CD133 and CD44, are proposed targets of WNT signaling (14, 15), one possibility is that cells showing nuclear β-catenin may harbor the highest tumorigenic potential. In support of this idea, a recent report showed that cell populations isolated from CRCs on the basis of high activity of a lentiviral WNT pathway reporter were more tumorigenic than cells with low or absent reporter activity (16). By contrast, we report here that our independent investigation of differential WNT activity in CRC cell lines and primary CRC xenografts revealed poor correlation between increased WNT activity and the potential to initiate tumors. In examining correlates of WNT signaling heterogeneity, we further found that nuclear β-catenin accumulated most in cells with active MAPK signaling and that nuclear β-catenin correlated with KRAS mutation in a large collection of surgical CRC cases. Moreover, gain- and loss-of-function studies revealed regulation of differential WNT activity by MAPK signaling. Thus, common mutations that activate MAPK signaling through the KRAS oncogene may be especially important in CRC in part by virtue of their effects on WNT pathway activity. One important feature of our study is the use of both cell lines and primary human CRCs, which may model tumor properties more accurately than do CRC cell lines alone.
MATERIALS AND METHODS
Cloning of lentiviral vectors
All template plasmids were obtained through Addgene (www.addgene.org). TOP-GFP was constructed by replacing the PGK promoter in the lentiviral vector pRRLSIN.cPPT.PGK-GFP. WPRE (Constructed in Didier Trono’s lab) with a 7xTCF/LEF optimal promoter cassette (7xTOP) from the M50 Super TOPFlash plasmid (17). To construct the double color vector TOP-GFP.mC, we inserted 4 additional TCF/LEF binding sites (GATCAAAGG) into a lentiviral TOP-dGFP reporter containing 3 such binding sites (18), yielding 7xTOP-dGFP. We then amplified this cassette using PCR and inserted it into the Hpa1 site of lentiviral PGK-H2BmCherry (19). To convert destabilized dGFP into enhanced eGFP, we used site directed mutagenesis (Stratagene) to insert a stop codon between GFP and the attached ornithine decarboxylase sequence, yielding lentiviral TOP-GFP.mC. To construct the control vectors FOP-GFP and FOP-GFP.mC, we replaced 7xTOP cassettes with synthetic 7xFOP cassettes (IDT-DNA), which carry mutated TCF/LEF binding sites (GgcCAAAGG). A mutant KRAS-expressing lentiviral vector UG2K was constructed by inserting KRASG12V from the pBabe K-Ras12V plasmid (20) into a pUltra lentiviral backbone (Constructed in Malcolm Moore’s lab) to yield lentiviral pUBC-GFP-P2A-KRASG12V (UG2K). Modified vector elements were verified by restriction analysis and sequencing.
Tissue culture, lentivirus production, transduction and immunoblotting
Caco2 and HEK293T cells were purchased from the American Type Culture Collection. SW1222 cells were obtained from the Ludwig Institute for Cancer Research (New York, USA). Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS), 1% glutamine, and 1x Penicillin-Streptomycin. HEK293T cells, plated in 10-cm dishes, were co-transfected with 10 μg lentiviral vector, 10 μg pCMV-dR8.91, and 2 μg pMD2.G (21) in the presence of 60 μL LipoD293 (Signagen). Medium was replaced after 12 h with DMEM containing 30% FBS and 36 h later, conditioned medium was passed through 0.45 μm filters (Millipore), mixed 1:1 with DMEM, and used to infect cultured CRC cell lines or disaggregated primary colon cancer cells in the presence of 8 μg/mL polybrene (Sigma-Aldrich). Transduced tumor cells were washed in phosphate-buffered saline (PBS) and transplanted into NOD/SCID mice. For some experiments, single transduced GFP-positive tumor cells were sorted into 96-well plates and expanded in culture before xenotransplantation. Immunoblotting for UG2K vector tests were done using whole lysates of native or UG2K transduced Caco2 or SW1222 cells with rabbit phospho-p44/42 MAPK antibody (Ab), p44/42 MAPK Ab (both Cell Signaling; 1:1000) , and mouse β-Actin Ab (Santa Cruz; 1:200).
Primary colon cancers, tumor disaggregation, and cell sorting by flow cytometry
Primary tumor samples were obtained from Brigham and Women’s Hospital, Boston, following approval of an institutional review board and patients’ informed consent; to increase material, primary tumors were expanded subcutaneously in NOD/SCID mice (The Jackson Laboratory, Bar Harbor, ME). These tumor xenografts were minced with scalpels and single cell suspensions prepared by digestion with collagenase IV (Worthington Biomedical, Lakewood, NJ) and 0.01% DNase I (Sigma-Aldrich) at 37°C for 30–60 min. Red blood cells were lysed using ACK lysing buffer (Biowhittaker, Lonza, Walkersville, MD); debris were removed by Ficoll-Paque (GE Healthcare Bio-sciences, Uppsala, Sweden) gradient centrifugation. Cells were stained with APC-conjugated anti-mouse H-2Kd (clone SF1-1.1.1, eBioscience) antibody (Ab) and Hoechst 33258 (Sigma-Aldrich) to exclude mouse and dead cells, respectively, and with PerCP-eFluor 710-conjugated anti-human EpCAM (eBioscience) Ab to select human CRC cells. Samples were filtered through 40-μm strainers before analysis or sorting on a FACSAria II instrument (BD Biosciences). Population purities of >90% in each experiment were confirmed by flow cytometry immediately after sorting (Suppl. Fig. 4A).
Tumor induction and treatment in mice
Mice were housed in micro-isolator cages in a pathogen-free colony; housing and experiments followed protocols approved by an institutional Animal Care and Use Committee. Tumors were induced in 6-8 week old NOD/SCID (NOD.CB17-Prkdcscid, The Jackson Laboratory) mice. Transduced or control CRC cell lines, disaggregated primary tumor xenografts, or sorted tumor cell fractions were suspended in 100 μl of a 1:1 mixture of PBS and growth factor-depleted Matrigel (BD Bioscience) before subcutaneous injection. For comparative experiments, GFPhigh and GFPlow tumor cell subpopulations were injected into opposing flanks of the same mouse. Animals were monitored weekly and tumors were measured using calipers. Mice were sacrificed when tumors reached a size of 1.5 cm or in the event of earlier illness. In some experiments, 0.2 mg cetuximab (Bristol-Meyers Squibb) or PBS (control) was injected into the peritonea of tumor- bearing mice every other day, starting after subcutaneous tumors exceeded >0.5 cm; CTNNB1 staining was analyzed after 10 days of cetuximab treatment.
Gene expression analyses
RNA was isolated from flow-sorted GFPhigh and GFPlow tumor cells using TRIzol Reagent (Invitrogen), treated with DNase, and first strand cDNA was synthesized using a QuantiTect Reverse Transcription Kit (Qiagen). Quantitative real-time PCR applied 40 cycles of amplification at 95°C (15 sec) and 60°C (1 min) using Sybr green (Roche), a 7500 instrument (Applied Biosystems, Foster City, CA), and the primers listed in Suppl. Table 1. Real-time RT-PCR results were first normalized to Gapdh mRNA levels in the same sample and then to levels of the corresponding transcript in GFPlow tumor cells. For transcriptional profiling, total RNA was converted to cDNA, linearly amplified using the NuGEN Ovation V2 Amplification System (NuGEN, San Carlos, CA), and hybridized to Human Genome U133A 2.0 arrays (Affymetrix, Santa Clara, CA) according to the manufacturer’s protocols. Expression microarray data were analyzed using the dChip package (22), considering transcripts that achieved a minimum change of 1.2-fold between two groups as differentially expressed.
Histology and immunohistochemistry
Tumor tissues were fixed in 4% paraformaldehyde, dehydrated in ethanol and xylenes, and embedded in paraffin. Sections were stained with hematoxylin and eosin (H&E). For immunohistochemistry, antigens were retrieved in 10 mM Na citrate, pH 6, for 10 min in a pressure cooker (Biocare Medical, Concord, CA). Slides were blocked with 0.5% hydrogen peroxide in methanol for 20 min, followed by 5% FBS in PBS for 30 min. Slides were incubated first with mouse β-catenin (BD Transduction Laboratories; 1:250), mouse active-β-Catenin (Millipore; 1:100), rabbit GFP (Cell Signaling; 1:250), or rabbit phospho-p44/42 MAPK Ab (Cell Signaling; 1:250) Ab overnight at 4°C, washed with PBS, and then with biotinylated goat anti-mouse or anti-rabbit Ab (Vector Laboratories, Burlingame, CA; 1:300) for 1 h at room temperature. Staining was detected using the Vectastain Elite ABC kit (Vector) and diaminobenzidine as the substrate. For single staining, slides were counterstained with hematoxylin. For double staining, dephospho-β-Catenin stained slides were incubated with rabbit GFP Ab (Cell Signaling; 1:250) for 1 h and developed with a MACH 3 Rabbit AP-Polymer kit (Biocare medical) and Permanent Red (Dako). For p-ERK/β-catenin and GFP/β-catenin double staining, slides were incubated first with rabbit GFP or phospho-p44/42 MAPK Ab (Cell Signaling; 1:250) for 1 h at room temperature, washed in PBS, and then with goat anti-rabbit Alexa Fluor 488- or Alexa Fluor 546-conjugated secondary Ab (Invitrogen; 1:500). Fluorescence images were taken on a Nikon TE300 microscope or a Nikon inverted Ti microscope with a Yokogawa spinning disk confocal system. Slides were then washed in PBS and stained withβ-catenin Abas described above.
CRC specimencollections and mutational analysis
A total of 301 paraffin-embedded CRC tissues were analyzed. KRAS mutations were determined on DNA extracted from the xenograft tumors and archived tissues by targeted pyrosequencing for codons 12 and 13 (23). Xenograft tumors were additionally tested for mutations in NRAS codons 12, 13 and 61, and BRAF codon 600 (24). Frequency data were analyzed using the Chi-squared-test. Statistical procedures applied SPSS version 19.0 (SPSS Inc.). Human subjects committees at the respective hospitals approved the study.
RESULTS
Isolation of CRC cell populations with differing levels of WNT pathwayactivity
Caco2 and SW1222 human CRC cells carry classical APC mutations (25) and form well-differentiated glandular tumors in NOD/SCID mice, with heterogeneous intratumoral expression and distribution of β-catenin (Suppl. Fig. 1A–B), similar to observations in primary CRCs (5). To mark tumor cells showing high intrinsic WNT activity, we transduced these cell lines with a lentiviral vector carrying TOP-GFP, a WNT-pathway reporter in which multimerized TCF/LEF promoter elements drive expression of green fluorescent protein (GFP, Fig. 1A). To test reporter specificity, we used FOP-GFP, a control vector with mutant TCF-binding sites (Suppl. Figure 2A). Cells were expanded clonally to exclude the non-transduced population and then xenografted into NOD/SCID mice (Fig. 1B). These clones showed high GFP in all cultured cells (Suppl. Fig 2B), whereas their xenografts showed differential GFP expression (Figure 1C, Suppl. Figure 2C), pointing to an environmental influence on WNT heterogeneity. In tumors expanded from Caco2TOP-GFP cells, the GFP signal co-localized with nuclear staining for β-catenin (Fig. 1C) and in SW1222TOP-GFP tumors, GFP and nuclear β-catenin largely overlapped (Suppl. Figure 2C). Moreover, GFP expression marked tumor cells with active, dephospho-β-Catenin (Suppl. Figure 2D).
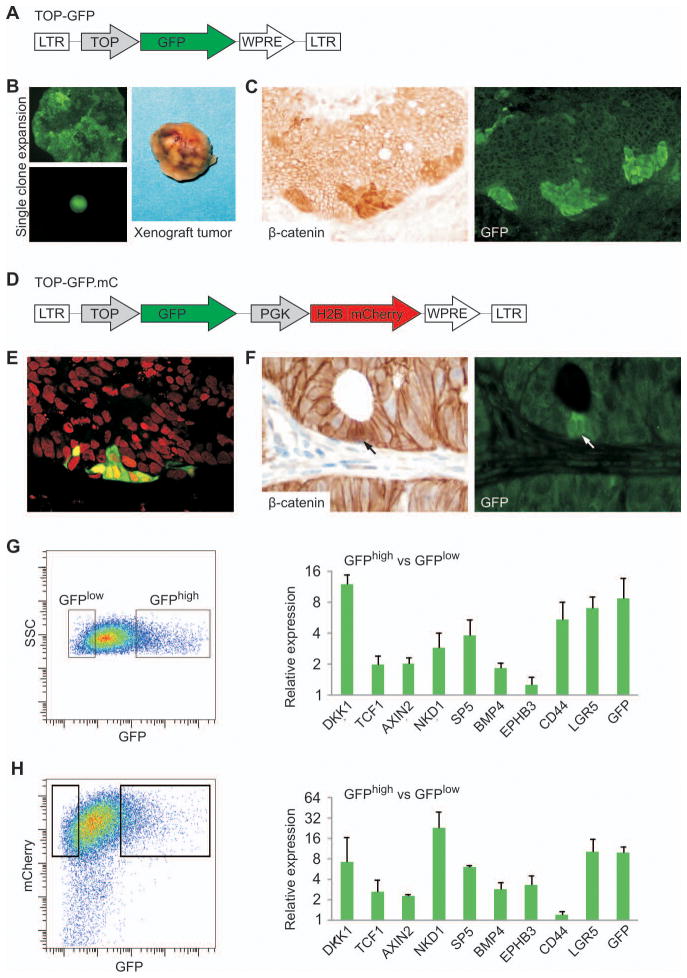
Figure 1. Use of a fluorescent reporter to mark the subpopulation of cells with highest WNT activity in CRC cell line xenografts and primary tumors.
(A) Lentiviral TOP-GFP vector used to transduce CRC cell lines. LTR, long terminal repeat; TOP, 7xTCF/LEF-responsive promoter element; WPRE, woodchuck hepatitis post-transcriptional regulatory element. (B) We expanded single fluorescent GFP+ clones in culture and injected them into NOD/SCID mice to form tumor xenografts. (C) Section of a Caco2TOP-GFP xenograft, stained for β-catenin (left) and GFP (right), demonstrating co-localization of the two signals. (D) Lentiviral double-color construct TOP-GFP.mC used to transduce primary colon cancers, containing the additional nuclear red fluorescence signal histone H2B-coupled mCherry, driven by the PGK promoter. (E) Confocal image of a TOP-GFP.mC-transduced primary colon cancer xenograft showing ubiquitous red nuclear fluorescence, with additional green fluorescence in a subset of tumor cells. (F) Section of a TOP-GFP.mC transduced primary colon cancer xenograft, stained with β-catenin Ab (left) and GFP (right); arrows indicate co-localization of the two markers. (G–H) Flow cytometric sorting of disaggregated tumor cell suspensions from Caco2TOP-GFP (G) and TOP-GFP.mC primary colon cancer xenografts (H) based on differential GFP fluorescence (left scatter plots) and qRT-PCR expression analysis of a panel of WNT-target genes (right graphs) in the sorted cell populations.
To mark tumor cells with differential WNT activity in primary CRCs, we first propagated fresh tumors as xenografts in mice, noting that they preserved their original morphology and displayed differential expression of nuclear β-catenin (Suppl. Fig. 1B–C). Because primary tumors have a heterogeneous composition, we preferred to avoid in vitro expansion of single transduced cells, which might select atypical clones. Instead we constructed a two-color reporter, TOP-GFP.mC, which allows constitutive nuclear mCherry expression in infected cells and regulated GFP expression in response to WNT pathway activity (Fig. 1D, Suppl. Fig. 2E). Fluorescence microscopy revealed nuclear mCherry expression in transduced primary CRC cells and a subset of these cells expressed GFP (Fig. 1E). Thus, after excluding non-transduced, mCherry-negative tumor cells, GFP again marked rare tumor cells that showed nuclear β-catenin (Fig. 1F).
We isolated GFPhigh and GFPlow cell fractions by flow cytometric sorting of disaggregated xenografts from TOP-GFP transduced cell lines and TOP-GFP.mC transduced primary CRCs. The GFPhigh population showed significantly higher mRNA levels of known WNT target genes, including the intestinal stem cell marker LGR5 (Fig. 1G–H, Suppl. Figure 3A). Gene expression profiles of GFPhigh and GFPlow cells from Caco2TOP-GFP revealed differential expression of 692 transcripts. Of these, 11 are annotated components of the WNT pathway and 9 of the 11 transcripts showed higher expression in the GFPhigh cell fraction (Suppl. Fig. 3B). Thus, TOP-GFP based constructs reliably marked a WNT-active subpopulation of CRC cells with nuclear β-catenin and allowed their isolation from bulk xenografts. However, the lesser enrichment for nuclear β-catenin and lower differential expression of WNT target genes among GFPhigh cells from SW1222TOP-GFP suggest that CRCs vary in the degree of heterogeneity in WNT activity.
Tumor cell fractions with different levels of WNT activity are equally tumorigenic
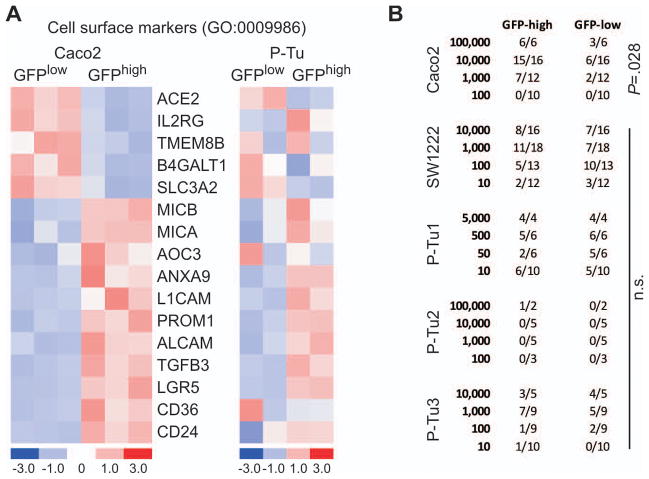
To characterize tumor cells with high and low WNT activity, we examined differential expression of all cell surface marker genes, as defined by Gene Ontology criteria, in GFPhigh and GFPlow cells isolated from Caco2TOP-GFP xenografts. Among the 16 differentially expressed surface markers were several that are reported or suspected to mark tumorigenic CRC cells, including CD133 (PROM1), CD166 (ALCAM), and LGR5; expression of these transcripts was consistently higher in the GFPhigh than in the GFPlow cell fraction (Fig. 2A). mRNA expression profiles of TOP-GFP.mC-transduced GFPhigh and GFPlow primary tumor cells also showed increased levels of these candidate stem and progenitor cell markers (Fig. 2A). These results suggested the possibility that CRC cells with the highest WNT activity might harbor increased tumorigenic potential, as others report (16). To test this hypothesis, we injected limiting dilutions of GFPhigh and GFPlow colon cancer cells into both flanks of NOD/SCID mice and followed the appearance of tumors. Indeed, GFPhigh cells from Caco2TOP-GFP xenografts were more tumorigenic than the GFPlow cell fraction and the tumors grew faster (Fig. 2B, Suppl. Fig. 4B). Despite extensive testing on many animals, however, we did not observe such differences for cells isolated from SW1222TOP-GFP tumors or from 3 primary CRCs transduced with TOP-GFP.mC (Fig. 2B); one primary tumor manifested low tumorigenic potential in mice irrespective of cellular GFP expression levels. These data show that CRC cells with high WNT activity and high expression of candidate cancer stem cell markers do not necessarily have an exclusive or enhanced capacity to spawn new tumors.
Figure 2. Lack of tumor-initiating capacity of WNT/GFPhigh tumor cell subpopulations despite enriched expression of suggested cancer stem cell markers.
(A) Heat maps depicting differential expression of cell surface markers in GFPhigh and GFPlow cells isolated from Caco2TOP-GFP (left) and from TOP-GFP.mC-transduced primary tumor (P-Tu, right) xenografts. Columns represent samples, and rows represent genes. Expression is represented in a pseudocolor scale (−3 to +3), with red denoting high and blue denoting low relative expression. (B) Frequencies of tumor formation upon subcutaneous injection of 10 to 100,000 flow-sorted GFPhigh and GFPlow tumor cells into NOD/SCID mice. Caco2TOP-GFP, SW1222TOP-GFP, and 3 independent TOP-GFP.mC transduced primary colon cancer xenografts (P-Tu1-3) were tested, with at least 3 different tumor xenografts from each source. The fractions indicate numbers of tumors formed (numerator) per experimental replicate (denominator). The p-value refers to a significant difference in tumor frequency between GFPhigh and GFPlow cells (Chi-square test). n.s. = non-significant.
High WNT activity is associated with MAPK pathway activation in colon cancer cells
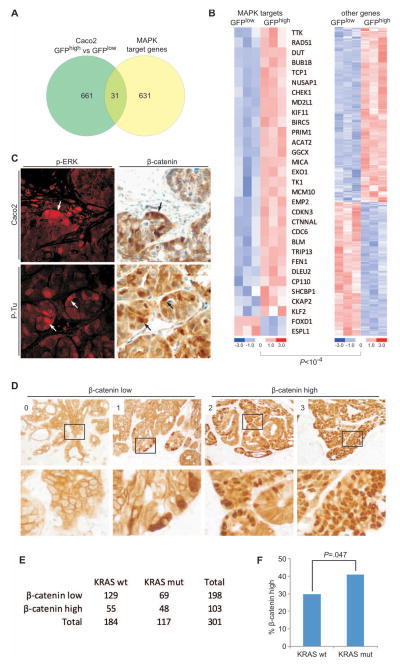
Recent data from zebrafish embryos suggest a role for Mitogen Activated Protein Kinase (MAPK) signaling in nuclear accumulation of β-catenin in the presence of mutant APC (26). To investigate if MAPK activity has a role in differential WNT activity in CRC cell subpopulations, we first compared the profile of transcripts that are altered in GFPhigh or GFPlow tumor cells to a canonical MAPK target gene signature (27). Of the 31 differentially expressed genes common to the two sets (Fig. 3A), 29 were expressed at higher levels in WNT-active, GFPhigh cells compared to GFPlow cells (p < 10−14, Fig. 3B); only 2 genes were expressed at lower levels, indicating activation of MAPK targets in the GFPhigh cell fraction. We then tested our xenograft sources for mutations most likely to affect MAPK signaling, in KRAS, BRAF and NRAS. Both cell lines were wild type for all 3 genes but primary Tu1 carried the V600E mutation in BRAF and primary Tu3 had a G12A mutation in KRAS. Surprisingly, although MAPK activation is considered a global feature of CRCs carrying such mutations, the activity marker phospho-ERK appeared only in a fraction of the cells in every tumor source (Fig. 3C and data not shown). Importantly, immunostaining of Caco2 and primary colon cancer xenografts revealed co-localization of p-ERK and nuclear β-catenin (Fig. 3C). Thus, MAPK signaling is heterogeneous in CRC and increased in the tumor cell fraction with high WNT activity and nuclear accumulation of β-catenin.
Figure 3. Independent lines of evidence correlating MAPK activation with nuclear β-catenin expression.
(A) Venn diagram showing the overlap of differentially expressed genes in GFPhigh and GFPlow cells of Caco2TOP-GFP tumors and a set of recently reported MAPK target genes (27). (B) Heat maps depicting the relative levels of differentially expressed MAPK target genes (left) and non-MAPK targets (right) in GFPhigh and GFPlow cells isolated from Caco2TOP-GFP tumors. The p-value denotes significantly enriched overexpression of MAPK targets in GFPhigh cells (Chi-square test). (C) Double immune staining for phospho-ERK (pERK, red) and β-catenin (brown), demonstrating heterogeneous expression and co-localization (arrows) of the two proteins in Caco2 (upper panel) and primary colon cancer (P-Tu, lower panel) xenografts. (D) Assessment of nuclear β-catenin staining in a collection of 301 primary colon cancers. Tumors were assigned scores from 0 (no nuclear β-catenin) to 3 (most tumor cells with strong nuclear β-catenin) and accordinglycategorized as β-cateninlow (score 0–1) and β-cateninhigh (score 2–3). Images in the lower panels show higher magnifications of the areas boxed in the upper panels. (E–F) Cross tabulation of nuclear β-catenin expression and KRAS mutational status (mut, activating mutation; wt, wild type) revealed significantly (Chi-square test) increased frequencies of β-cateninhigh among cases with activating KRAS mutations.
To assess this association in clinical samples, we examined MAPK signaling and nuclear β-catenin accumulation in 301 primary CRCs with known KRAS mutation status. Similar to the xenografts, phospho-ERK immunostaining in 118 of these CRC specimens confirmed nonuniform MAPK signaling, irrespective of the underlying KRAS gene status (Suppl. Figure 5A). We then scored nuclear β-catenin in tissue sections on a scale from 0 (no nuclear β-catenin) to 3 (most tumor cells showing strong nuclear β-catenin), hence classifying cases as β-cateninlow (scale 0–1) or β-cateninhigh (scale 2–3, Fig. 3D), and observed that KRAS mutant tumors are significantly enriched for high nuclear β-catenin (Fig. 3E–F). Taken together, these data support the idea that active KRAS might contribute to nuclear accumulation of β-catenin in CRC, where both MAPK signaling and WNT activity show intra-tumor heterogeneity. These findings suggested roles for KRAS and MAPK signaling in differential WNT activity.
Functional relationship of MAPK and WNT pathway activities
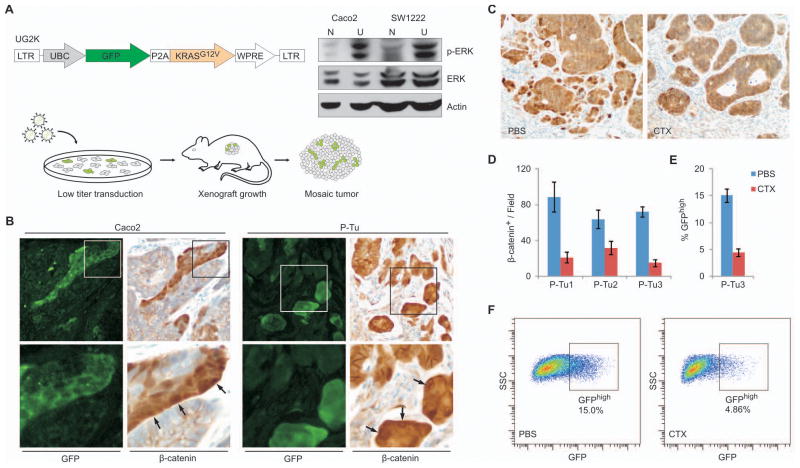
To test if MAPK pathway activation influences WNT pathway activity, we constructed a lentiviral vector to express constitutively active KRAS and, as a marker of cell infection, GFP (Fig. 4A). Increased phospho-ERK levels in transduced cells verified that introduction of this vector activates MAPK signaling, as expected (Fig. 4A). We infected CRC cell lines and primary tumors with low-titer lentivirus and grafted the cells in NOD/SCID mice to obtain tumors containing a mix of infected and uninfected tumor cells (Fig. 4A). Immunostaining of tumor sections revealed increased β-catenin levels and its nuclear localization in transduced, GFP+ tumor cells (Fig. 4B, Suppl. Fig. 5B). The levels of WNT target transcripts were also increased selectively in the transduced cell fractions (Suppl. Fig. 5C). Thus, forced activation of MAPK signaling through high levels of constitutively active KRAS leads to nuclear accumulation of β-catenin and increased WNT activity in CRC.
Figure 4. KRAS activation increases, and interference with EGFR signaling decreases the WNT-active cell fraction in Caco2 and primary tumor xenografts.
(A) Vector schema: Lentiviral vector UG2K for Ubiquitin-C promoter (UBC) driven overexpression of mutant KRASG12V together with GFP, separated by a P2A cleavage peptide. Right panel: Immunoblot showing increased phospho-ERK (p-ERK) and unchanged total-ERK (ERK) expression in UG2K-transduced (U), serum starved cells compared to native (N) Caco2 and SW1222 cells. Actin served as a loading control. Bottom: Experimental scheme to generate colon cancer xenografts with mosaic UG2K transduction. Colon cancer cell lines and primary tumor xenografts were exposed for short periods to low titers of UG2K lentivirus and injected subcutaneously into NOD/SCID mice for tumor formation. (B) Sections of mosaic UG2K-transduced tumor xenografts double stained for GFP and β-catenin, showing increased nuclear β-catenin in GFP-expressing tumor cells (arrows). Images in the lower panels show higher magnifications of areas boxed in the upper panels. (C–D) Representative β-catenin immune staining of a primary colon cancer xenograft (C) and quantitation of the β-catenin positive cell fraction in 3 primary colon cancer xenografts (D) treated in vivo with cetuximab (CTX) or PBS. (E–F) Reduced GFPhigh tumor cell fraction in TOP-GFP.mC transduced primary colon cancer xenografts following cetuximab (CTX) treatment, as measured by flow cytometry of disaggregated tumor cells (F). Error bars indicate the standard error of the mean from 3 experiments.
To test if inhibition of MAPK signaling opposes β-catenin accumulation, we treated cultured CRC cells and mice bearing primary CRC xenografts with the epidermal growth factor receptor (EGFR) antibody cetuximab, which inhibits cellular MAPK activity (28). Treated tumors showed significantly fewer cells with nuclear β-catenin (Fig. 4C–D). Likewise, treatment of cultured Caco2TOP-GFP cells reduced expression of GFP and WNT target genes (Suppl. Fig. 5D). Moreover, primary tumor xenografts carrying the WNT reporter TOP-GFP.mC showed reduced GFP fluorescence and depletion of the GFPhigh cell fraction (Fig. 4E–F). Thus, EGFR antibody-mediated blockade of MAPK signaling reduces WNT pathway activity in CRC cells. Surprisingly, these effects were evident despite the presence of a KRAS mutation in one primary tumor.
DISCUSSION
APC or CTNNB1 mutations in nearly all CRCs are presumed to activate WNT signaling in all clonally derived tumor cells (29, 30). Heterogeneity in nuclear β-catenin levels in most CRCs and differential expression of WNT target genes therefore present a paradox and suggest that additional factors contribute to WNT pathway activity (6, 7). To isolate viable tumor cells showing differential WNT activity, we transduced xenografts of primary tumors and CRC cell lines with fluorescent reporters of canonical WNT activity (18). Although cultured CRC cells expressed GFP uniformly, all xenografts displayed highly heterogeneous GFP expression, similar to a recent study (16). Thus, the local environment or the 3-dimensional structure in xenografts seem necessary to reveal this latent heterogeneity. In tumor xenografts, nuclear β-catenin co-localized with high GFP expression as well as with enriched expression of WNT pathway components and WNT target genes in GFPhigh tumor cells. These data confirmed differential WNT pathway activity within CRCs and provided a reliable means to isolate cells with different levels of WNT activity. However, even though differentially expressed genes included the candidate human CRC stem-cell markers CD133, CD44, and CD166 (11–13), and the intestinal stem cell marker LGR5 (31), WNT/GFPhigh cells from only 1 of the 5 tested sources showed a modest increase in tumor-initiating capacity. These data from a small number of primary CRC specimens indicate that high WNT activity does not signify particular tumorigenic potential in CRC and is not a reliable marker for tumor initiation in xenografts. Accordingly, they question the generality of a recent argument that high WNT activity is a distinguishing feature of CRC stem cells (16). The contrast between our findings and those of Vermeulen (16) mirror the conflicting results from different laboratories’ use of CD133 as a surface marker of tumor-initiating CRC cells (32). We suggest that the differing outcomes of such studies reflect highly variable genetic or phenotypic properties of tumors. Differences in experimental design, such as whether primary tumor cells were first expanded in spheroid cultures (16) or cultivated directly in xenografts, may also contribute to the discrepancy. Of course, it is still unclear how accurately tumor xenografts model tumor-initiating cell subpopulations (33, 34).
Despite the lack of an obvious association with tumor initiation, heterogeneous WNT activity correlates with disease progression in CRC (5, 6, 26), adding to the importance of identifying the causes. Brabletz et al and Vermeulen et al have separately proposed that tumor cell stimulation by the microenvironment may modulate β-catenin accumulation and WNT pathway activity (6, 16), and studies in zebrafish embryos and a single CRC cell line recently implicated tumor cell MAPK signaling in the underlying mechanism (26). Reinforcing this idea, we observed co-localization of nuclear β-catenin and the MAPK pathway indicator phospho-ERK in cell line and primary tumor xenografts, and expression of MAPK target genes is enriched in highly WNT-active CRC cells. We also detected a statistical association between nuclear β-catenin and the presence of activating mutations in the MAPK pathway gene KRAS. Our data are consistent with the observation that intestinal tumors in KrasG12V; Apc+/1638N mice have more cells with nuclear β-catenin than do their Apc+1638N littermates (35). Notably both, KRAS mutant and wild-type tumors showed heterogeneous MAPK activity, implying that signaling remains responsive to exogenous factors even when cells carry activating mutations. Although this observation begs an explanation for variable MAPK pathway activity among cells in the same tumor, our data begin to explain why MAPK and WNT pathway activities are high in the same cells.
KRAS mutations that activate MAPK signaling are especially common in selected cancers, including about 40% of CRCs (2), where they occur early in the disease course, soon after APC or CTNNB1 mutations (36). KRAS mutations in the intestinal epithelium are oncogenic only when accompanied by deregulation of the WNT pathway (37). The full significance of this association and of the sequence of mutations is unclear and our data shed useful light. Forced expression of mutant KRAS in CRCs enhanced nuclear β-catenin accumulation and increased the levels of WNT target genes, whereas blocking the tyrosine kinase receptor EGFR, an activator of MAPK signaling (38), had the converse effects. Thus, MAPK signaling does not merely coincide with nuclear β-catenin but also regulates it. In line with these observations, insertional mutagenesis in ApcMin mice recently uncovered 4 new candidate positive modifiers of canonical Wnt signaling (39) and one of these, EGF receptor kinase substrate 8, transduces EGF and RAS signals. Thus, modulation of canonical Wnt signaling through MAPK is a factor in colorectal tumorigenesis and our study highlights this effect in tumor cell subpopulations. Phelps et al recently proposed that KRAS and RAF1 but not MEK1 induce nuclear accumulation in APC deficient cells (26). Further study is necessary to elucidate the detailed basis for this pathway cross-talk.
The fact that EGF, and perhaps additional signals from the tumor microenvironment, contribute to heterogeneous WNT activity in CRC also suggests new considerations for treatment of this disease. There are intensive ongoing efforts to design drugs that suppress WNT activity in CRC; these drugs may synergize with those that target MAPK signaling, such as cetuximab. Conversely, investigators recently described the use of transcriptionally active β-catenin to induce apoptosis in APC-deficient tumors (40). If growth factor signaling enhances WNT activity, then simultaneous targeting of growth factor pathways may down-regulate the intended target and hence diminish the effects of WNT-directed therapy. Our observation that cetuximab reduced intra-tumoral heterogeneity of nuclear β-catenin also raises the possibility that the degree of heterogeneity in WNT pathway activity may help predict the therapeutic response to agents that target MAPK signaling. Thus, a fuller understanding of the interactions between WNT and MAPK signaling will better inform the design and interpretation of therapeutic trials.
Supplementary Material
Acknowledgments
We thank Dr. Michael Zinner and members of the Departments of Surgery and Pathology at Brigham & Women’s Hospital for generously providing freshly resected CRC specimens and the Ludwig Institute for Cancer Research for the gift of SW1222 cells. Supported by a grant from the Harvard Stem Cell Institute (R.A.S.), a fellowship from the Deutsche Forschungsgemeinschaft (D.H.), and grants R01 CA151993 (S.O.) and P50CA127003 from the National Institutes of Health.
Financial support
Supported by a grant from the Harvard Stem Cell Institute (R.A.S.), a fellowship from the Deutsche Forschungsgemeinschaft (D.H.), and grants R01 CA151993 (S.O.) and P50CA127003 from the National Institutes of Health.
Footnotes
Disclosures: No conflicts of interest.
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer. 2011;50:307–12. doi: 10.1002/gcc.20854. [DOI] [PubMed] [Google Scholar]
- 3.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Brabletz T, Jung A, Hermann K, Gunther K, Hohenberger W, Kirchner T. Nuclear overexpression of the oncoprotein beta-catenin in colorectal cancer is localized predominantly at the invasion front. Pathol Res Pract. 1998;194:701–4. doi: 10.1016/s0344-0338(98)80129-5. [DOI] [PubMed] [Google Scholar]
- 5.Horst D, Reu S, Kriegl L, Engel J, Kirchner T, Jung A. The intratumoral distribution of nuclear beta-catenin is a prognostic marker in colon cancer. Cancer. 2009;115:2063–70. doi: 10.1002/cncr.24254. [DOI] [PubMed] [Google Scholar]
- 6.Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz-Schughart LA, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A. 2001;98:10356–61. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19:150–8. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Dehari R, et al. Correlation of beta-catenin localization with cyclooxygenase-2 expression and CpG island methylator phenotype (CIMP) in colorectal cancer. Neoplasia. 2007;9:569–77. doi: 10.1593/neo.07334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samowitz WS, Slattery ML, Sweeney C, Herrick J, Wolff RK, Albertsen H. APC mutations and other genetic and epigenetic changes in colon cancer. Mol Cancer Res. 2007;5:165–70. doi: 10.1158/1541-7786.MCR-06-0398. [DOI] [PubMed] [Google Scholar]
- 10.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–51. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 13.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 14.Katoh Y, Katoh M. Comparative genomics on PROM1 gene encoding stem cell marker CD133. Int J Mol Med. 2007;19:967–70. [PubMed] [Google Scholar]
- 15.Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R, et al. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol. 1999;154:515–23. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermeulen L, De Sousa EMF, van der Heijden M, Cameron K, de Jong JH, Borovski T, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–76. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 17.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13:680–5. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 18.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–14. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 19.Kita-Matsuo H, Barcova M, Prigozhina N, Salomonis N, Wei K, Jacot JG, et al. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS One. 2009;4:e5046. doi: 10.1371/journal.pone.0005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khosravi-Far R, White MA, Westwick JK, Solski PA, Chrzanowska-Wodnicka M, Van Aelst L, et al. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol Cell Biol. 1996;16:3923–33. doi: 10.1128/mcb.16.7.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naldini L, Blomer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A. 1996;93:11382–8. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–21. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irahara N, Baba Y, Nosho K, Shima K, Yan L, Dias-Santagata D, et al. NRAS mutations are rare in colorectal cancer. Diagn Mol Pathol. 2010;19:157–63. doi: 10.1097/PDM.0b013e3181c93fd1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ilyas M, Tomlinson IP, Rowan A, Pignatelli M, Bodmer WF. Beta-catenin mutations in cell lines established from human colorectal cancers. Proc Natl Acad Sci U S A. 1997;94:10330–4. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phelps RA, Chidester S, Dehghanizadeh S, Phelps J, Sandoval IT, Rai K, et al. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137:623–34. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurchott K, Kuban RJ, Krech T, Bluthgen N, Stein U, Walther W, et al. Identification of Y-box binding protein 1 as a core regulator of MEK/ERK pathway-dependent gene signatures in colorectal cancer cells. PLoS Genet. 2010;6:e1001231. doi: 10.1371/journal.pgen.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang S, Armstrong EA, Benavente S, Chinnaiyan P, Harari PM. Dual-agent molecular targeting of the epidermal growth factor receptor (EGFR): combining anti-EGFR antibody with tyrosine kinase inhibitor. Cancer Res. 2004;64:5355–62. doi: 10.1158/0008-5472.CAN-04-0562. [DOI] [PubMed] [Google Scholar]
- 29.Fearon ER, Hamilton SR, Vogelstein B. Clonal analysis of human colorectal tumors. Science. 1987;238:193–7. doi: 10.1126/science.2889267. [DOI] [PubMed] [Google Scholar]
- 30.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 31.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 32.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–20. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–9. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 34.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–9. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 35.Janssen KP, Alberici P, Fsihi H, Gaspar C, Breukel C, Franken P, et al. APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology. 2006;131:1096–109. doi: 10.1053/j.gastro.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 36.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 37.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–8. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim Biophys Acta. 2007;1773:1161–76. doi: 10.1016/j.bbamcr.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.March HN, Rust AG, Wright NA, Ten Hoeve J, de Ridder J, Eldridge M, et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nature genetics. 2011;43:1202–9. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Ren X, Alt E, Bai X, Huang S, Xu Z, et al. Chemoprevention of colorectal cancer by targeting APC-deficient cells for apoptosis. Nature. 2010;464:1058–61. doi: 10.1038/nature08871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.