Abstract
The goal of this study was to measure spatially and temporally resolved effective diffusion coefficients (De) in biofilms respiring on electrodes. Two model electrochemically active biofilms, Geobacter sulfurreducens PCA and Shewanella oneidensis MR-1, were investigated. A novel nuclear magnetic resonance microimaging perfusion probe capable of simultaneous electrochemical and pulsed-field gradient nuclear magnetic resonance (PFG-NMR) techniques was used. PFG-NMR allowed noninvasive, nondestructive, high spatial resolution in situ De measurements in living biofilms respiring on electrodes. The electrodes were polarized so that they would act as the sole terminal electron acceptor for microbial metabolism. We present our results as both two-dimensional De heat maps and surface-averaged relative effective diffusion coefficient (Drs) depth profiles. We found that 1) Drs decreases with depth in G. sulfurreducens biofilms, following a sigmoid shape; 2) Drs at a given location decreases with G. sulfurreducens biofilm age; 3) average De and Drs profiles in G. sulfurreducens biofilms are lower than those in S. oneidensis biofilms—the G. sulfurreducens biofilms studied here were on average 10 times denser than the S. oneidensis biofilms; and 4) halting the respiration of a G. sulfurreducens biofilm decreases the De values. Density, reflected by De, plays a major role in the extracellular electron transfer strategies of electrochemically active biofilms.
Keywords: diffusion, diffusion coefficient, diffusivity, biofilm, electrochemically active, Geobacter, Shewanella, modeling, electron transfer, magnetic resonance
Introduction
The restricted diffusion of molecules through bacterial biofilms is a major physical aspect of what makes the biofilm mode of life distinct from the planktonic mode1. Constrained mass transport in biofilms, along with microbial metabolism, yields the microenvironments and physicochemical heterogeneities that ultimately lead to spatially divergent phenotypes and species diversification. Knowledge of diffusion coefficients in biofilms is critical for measuring and predicting the chemical fluxes in biofilms and ultimately for engineering systems to manipulate bacterial biofilms for our purposes. Examples in which diffusion coefficients play a practical role include 1) penetration of antibiotics into pathogenic biofilms2, 3, 2) heavy metal immobilization and reduction in environmental biofilms4, 5, and 3) penetration of organic molecules and oxygen into biofilm reactors used in wastewater treatment6-9.
Recently, mass transport limitations are being investigated for biofilms that respire on electrodes, known as electrochemically active biofilms (EABs)10-12. EABs are unique because of their ability to respire terminal electrons from metabolism onto solid electron-accepting electrodes10, 13. These biofilms are composed of microorganisms that are known as electricigens, exoelectrogenic bacteria, and anode-respiring, or anodophilic, species14, 15. EABs have been utilized in marine-based microbial fuel cells to power oceanographic sensors and monitoring devices16, 17; to extract energy and enhance chemical oxygen demand removal during wastewater treatment18, 19; as biosensors for nitrate/nitrite, glucose, and other chemicals20; to desalinate water while simultaneously generating electricity21-23; and also to produce hydrogen gas via microbially driven electrolysis24, 25. However, optimization of their electron transfer requires further fundamental knowledge of their internal chemical and electrical fluxes.
Effective diffusion coefficients (De) quantify the fluxes of molecules critical in several distinct processes required for EAB activity. The first is the transport of nutrients required for EAB growth and metabolism. EAB growth can be slowed when the rate at which the nutrients are consumed is higher than the rate at which the nutrients are replenished via diffusion, which may prevent regions of the biofilm from accessing nutrients. The second is the transport of electron equivalents generated during EAB growth, known as extracellular electron transfer (EET). EET is hypothesized to occur through conduction-based schemes, diffusion-based (mediated) schemes, or a mixture of the two26. G. sulfurreducens is thought to use conduction-based EET, while S. oneidensis MR-1 is thought to use a combination of the two schemes27-29. For EABs that depend on the diffusion of electron transfer-mediating molecules from the biofilm to the electrode surface, the generation of electron equivalents may exceed the flux of the mediators. Furthermore, diffusion of the electron mediators may control both the electrochemical potential loss through the formation of concentration gradients and the rate of electron mediator loss to the supernatant, ideas which have been explored by Torres et al. (2010)26. A third critical process dominated by diffusive transport is the movement of protons and pH-buffering compounds. The oxidation of substrates such as acetate or of electron mediators such as flavins is coupled to the production of protons, causing pH gradients to form as a consequence of biofilm metabolism30, 31. Proton buildup at the anode in a weakly buffered system, i.e. acidification, is a known problem in some microbial fuel cell designs32. Diffusion also controls the movement of charged electrolytes that act to maintain electroneutrality in the biofilm. While this list is not exhaustive, it is apparent that diffusive mass transport is influential in several processes that, under certain conditions, control the overall electron transfer rate of EABs respiring on electrodes. Therefore, it is critical to quantify De in EABs in order to develop our knowledge of internal chemical and electrical fluxes and how these fluxes relate to EET mechanisms.
To better understand EET mechanisms, mathematical models have been created to predict electron transfer rates from EABs to electrodes and to determine flux limitations in EABs. These models rely on De values to produce accurate representations and realistic predictions. Yet, there has been a trend in the literature to assume a constant De value within the biofilms, with no spatial or temporal variability. Van Wey et al. (2012)33 present several examples of biofilm models that assume a constant De throughout the biofilm34-39, and also examples of models that assume bulk liquid diffusion coefficients (Daq) instead of effective diffusion coefficients 34, 38-40. We found a similar trend in EAB models. Table 1 highlights the assumptions regarding diffusion coefficients in EAB models. Non-variable De assumptions are often selected to “reduce model complexity”41 or because some models yield insignificant changes in simulation outputs when more complex De parameterization is used42, 43.
Table 1. Diffusion coefficients in electrochemically active biofilm models.
Three diffusion coefficient assumptions are considered: 1) the diffusion coefficient is an effective value; 2) there is diffusion coefficient spatial variability; and 3) there is diffusion coefficient temporal variability. An effective diffusion coefficient (De) takes into account hindered diffusion caused by the porous nature of the biofilm matrix. De are smaller in magnitude than the free bulk liquid diffusion coefficient (Daq). Spatial variability takes into account biofilm structural heterogeneity and allows for the diffusion coefficients to change with location in the biofilm. Temporal variability takes into account the non-steady state nature of biofilms and allows the diffusion coefficients to change with time as the biofilm grows or changes morphology. This is a non-exhaustive list curated from the Thomson Reuters Web of KnowledgeSM database, ordered by publication date. Electrochemically active biofilm (EAB) models that do not rely on diffusive mass transport in the biofilm are not included (i.e., convection-only models).
| Biofilm Diffusion Coefficients | ||||||
|---|---|---|---|---|---|---|
| Citation | Article | Model Description | Dimensions | Effective |
Spatial
Variability |
Temporal
Variability |
| 57, 58 | Picioreanu et al., 2007 and 2008 | Biofilm-based microbial fuel cells | 1D,2D,3D | Yes | None | None |
| 59 | Marcus et al., 2007 | Conduction-based biofilm anode (Nernst-Monod) | 1D | Yes | None | None |
| 60 | Picioreanu et al., 2010 | pH and electrode geometry in microbial fuel cell | 2D | No | None | None |
| 61 | Marcus et al., 2010 | Electromigration impact on biofilm anode (PCBIOFILM) | 1D | Yes | Yes* | None |
| 62 | Marcus et al., 2011 | Biofilm-based microbial electrolysis cell (PCBIOFILM) | 1D | Yes | None | None |
| 63 | Strycharz et al., 2011 | Cyclic voltammetry of Geobacter biofilm | 1D | No† | None | None |
| 64 | Merkey and Chopp, 2012 | Anode geometry in microbial fuel cell | 2D | Yes | None | None |
This model provides qualitative analysis of EAB cyclic voltammograms and uses diffusion coefficient values several orders of magnitude larger than realistic values.
The consideration of migration causes an apparent change in the diffusion coefficient of charged solutes based on the electric field magnitude, which is a function of distance in this model.
However, De have been experimentally verified to be spatially and temporally variable and anisotropic1, 33, which is anticipated to be the case in most systems, owing to the heterogeneous nature of biofilms. Because of this understanding, and because diffusion resistance is a key defining feature of the biofilm mode of life, these complexities should increasingly become incorporated into mathematical models, as some researchers have already demonstrated44, 45. Furthermore, using experimental values of De measured in EABs respiring on electrodes in EAB models is important because biofilm structure and electron transfer capability in EABs have recently been shown to depend on whether the biofilm respired on a soluble electron acceptor or on an electrode acting as an electron acceptor46, 47. We have observed similar distinctions between biofilms respiring using these same two methods in our lab, and it is likely that these differences are associated with unique De profiles. Therefore, De measured for biofilms not respiring on electrodes cannot be used. However, to the best of our knowledge, there are no De data for EABs respiring on electrodes.
The lack of spatially and temporally resolved De data for EABs is the result of two historical factors. First, the current De measuring techniques were not designed to handle multi-dimensional and time-resolved in situ measurements in living biofilms. The preponderance of empirical De values that are available in the scientific literature are only 0- or 1-dimensional (that is, volume-averaged or surface-averaged). The primary techniques used to measure De are fluorescence recovery after photobleaching with scanning confocal laser microscopy48-50; porosity determination using microtomed slices51; injected dyes or tagged molecules52; and microelectrodes53. Regardless of the specific technique used, researchers generally measure De in distinct cell clusters (0-dimensional) or as depth profiles through the biofilm layers (1-dimensional), without monitoring how the values change with biofilm age. Note that microelectrodes have been used to measure multidimensional De, which are reported as 2D maps and 1D profiles54. However, this technique uses ferricyanide, which kills the biofilm and consequently does not allow for time series measurements. Second, the current techniques have not been tested for delivering De results for biofilms respiring on electrodes and may not be capable of doing so. New technology is required to handle the difficult task of measuring multidimensional De values over time in living biofilms that respire on electrodes.
In this study we used a novel nuclear magnetic resonance microimaging system capable of simultaneous electrochemical and pulsed-field gradient nuclear magnetic resonance (PFG-NMR) measurements of live biofilms. To make this possible we retrofitted our previously developed NMR biofilm reactor to act as an electrochemical cell while still allowing for NMR measurements55. This involved changing the radio-frequency coil geometry to avoid signal blocking by the conductive electrode and employing active polarization via a potentiostat. With the electrode polarized to a sufficiently positive potential (i.e., with the electrode potential fixed against a known reference), the EAB can actively respire terminal electrons onto the electrode during NMR measurements. Unlike traditional techniques for measuring De, PFG-NMR offers the ability to measure in situ De 1) in multiple spatial dimensions, 2) over extended time periods, 3) noninvasively, 4) nondestructively, 5) at high spatial resolution, and 6) in living biofilms. This allowed us to investigate the in situ De over time for two model EABs respiring on electrodes: Shewanella oneidensis MR-1 and Geobacter sulfurreducens PCA. To the best of our knowledge, this is the first time that De have been quantified for biofilms respiring on electrodes. We hypothesized that G. sulfurreducens would have relatively low De values to facilitate conductive electron transfer through a tight biofilm matrix, whereas S. oneidensis would have relatively high De values to ease the diffusion of electron transfer-mediating molecules. Furthermore, because a change in G. sulfurreducens biofilm morphology has been observed during changes to non-turnover conditions56, we also hypothesized that halting the respiration of the EABs by removing the polarization of the electrode would change the De values because of changes in the biofilm matrix in response to a changed metabolic state.
Materials and Methods
NMR microimaging biofilm reactor and bioelectrochemical cell
All NMR measurements were performed using a custom-made NMR biofilm reactor, shown in Figure 1 (also see Figure S1 in the Electronic Supplementary Information (ESI)). The reactor was machined from Torlon® polyamide-imide plastic, and the internal flow channel was 4 mm wide, 2 mm deep, and 40 mm long (320-μL volume). Polyetheretherketone (PEEK) plastic tubing (1/16” outer diameter, .030” inner diameter) was used for the inlet and effluent lines for the growth medium. Flow through the PEEK lines was set using a pulseless dual-syringe pump (Pharmacia P-500, Uppsala, Sweden) controlled remotely using a home-built, internet-accessible RS-232 pump controller based on a Lantronix XPort AR™ network processing module (Lantronix Inc, Irvine, CA, USA). Two additional PEEK tubing lines, connected adjacent to the inlet and effluent lines, were aseptically attached with 0.22-μm filters to a 4-channel IPC Ismatec® pump (IDEX Health & Science, Wertheim-Mondfeld, Germany). The pump was situated inside a plastic gas bag continuously purged with 100% N2 while the G. sulfurreducens experiments were run to ensure strict anaerobic conditions. This pump was activated to remove intractable bubbles from the reactor prior to the start of the experiment. After the experiment was initiated, these lines were sealed and remained unused during the remainder of the experiment. A 250-μm-diameter Ag/AgCl reference-electrode wire was placed inside the influent line using an epoxy-sealed T-junction allowing for it to protrude into the reactor. Similarly, a braided Pt counter electrode, made from 100-μm-diameter >99.99% pure Pt wire, was placed inside the effluent line. A 5-mm-diameter, 100-μm-thick 99.99% gold disc electrode (Gold foil SKU 265810, Sigma-Aldrich Corporation, St. Louis, MO, USA) was fixed with conductive silver epoxy (TIGA 901, Resin Technology Group LLC, South Easton, MA, USA) to the reactor door. A 250-μm-diameter 99.99% Cu wire (California Fine Wire Co., Grover Beach, CA, USA) was coiled in the epoxy between the gold disc electrode and the door; this provided a link for the gold disc electrode with the working electrode potentiostat connection. The potentiostat (Reference 600™, Gamry Instruments, Warminster, PA, USA) was located outside the NMR 5 -Gauss line.
Figure 1. Nuclear magnetic resonance microimaging system for studying EABs.
An illustration of the experimental arrangement for NMR used to study diffusion in EABs. The diagrams show (from left to right): the vertical bore superconducting magnet with perfusion lines leading to the bottom-loaded NMR probe (medium flowing against gravity); a cutaway view of the custom NMR probe, shown holding the NMR biofilm reactor; an external view of the NMR biofilm reactor; and a cutaway view of the NMR biofilm reactor containing a perfused EAB growing on a gold disc electrode.
Growth of G. sulfurreducens biofilm
The G. sulfurreducens PCA biofilms were grown inside the sealed NMR biofilm reactor. To sterilize the system, 30% H2O2 was pumped at 1 mL/hr for 24 hours, followed by a 12-hour rinse at 1 mL/hr of nanopure autoclaved water. For 24 hours prior to inoculation, the system was allowed to equilibrate at standard operating conditions to ensure that all oxygen was removed. This included 1) sparging the 2-L growth medium bottle with 20%/80% CO2/N2, 2) pumping growth medium (no fumarate) at 1 mL/hr into the biofilm reactor, 3) polarizing the electrode at +300 mVAg/AgCl, 4) passing 30 °C 100% N2 into the NMR bore and around the biofilm reactor, and 5) having 100% N2 flow into plastic gas bags encasing the syringe pump and flow breakers to avoid possible gas exchange with room air and temperature variation. Inoculum vials of G. sulfurreducens PCA (ATCC 51573) were prepared anaerobically using the Hungate technique65 in a serum vial without shaking, similar to the procedure described by Babauta et al. (2012)66. The growth medium in the vial consisted of potassium chloride, 0.38 g/L; ammonium chloride, 0.2 g/L; sodium phosphate monobasic, 0.069 g/L; calcium chloride, 0.04 g/L; magnesium sulfate heptahydrate, 0.2 g/L; sodium carbonate, 2 g/L; Wolfe’s vitamin solution, 10 mL/L; and modified Wolfe’s mineral solution, 10 mL/L. Acetate (20 mM) was provided as the electron donor, and 40 mM fumarate (inoculum only) was the electron acceptor. The headspace in the inoculum vial was 20%/80% CO2/N2. The medium used for growth in the biofilm reactor was identical, except that no fumarate or other soluble electron acceptor was provided. The gold disc electrode acted as the sole terminal electron acceptor for microbial metabolism, and is herein simply called the electrode.
During inoculation, the growth medium flow was stopped. Inoculum was taken anaerobically from the inoculum vials using a N2-filled gas sampling bag to provide a counter pressure as the inoculum was taken into a 20-mL syringe. The syringe was placed in the N2-purged gas bag encasing the syringe pump, and inoculum was pumped at 5 mL/hr into the reactor for 1 hr, and then at 0.7 mL/hr for another 10 hours. To improve cell attachment to the electrode, the inoculum pumping was halted and the reactor was kept horizontal (with the electrode normal aligned against gravity) with no flow for 24 hours. A NMR probe cover (constructed from 1.5” polyvinyl chloride pipe and cap and perfused with 30 °C 100% N2) was used to maintain anaerobicity and temperature while the sample was outside the magnet. After the 24 hours, the reactor was positioned vertically and the flow of growth medium (with no fumarate) was gradually increased up to 1 mL/hr, producing a laminar flow profile (Reynolds number of 0.1, dilution rate of 3.13 h−1). All heated N2 gas was temperature-controlled using a gas stream delivery unit (FTS Systems, Stone Ridge, NY). This kept the gas temperature at the NMR bore and clamp stand probe holder at 30 ± 0.6 °C. The sample was maintained under the NMR probe cover and on a clamp stand probe holder outside of and adjacent to the NMR magnet during growth; it was inserted into the NMR system for a few days at regular intervals for NMR measurements. The potentiostat was connected and active during the entire duration of the study, except for brief periods during which respiration onto the electrodes was halted to measure De under open circuit conditions.
Growth of S. oneidensis biofilm
S. oneidensis MR-1 biofilms were grown using a constant depth film fermentor (CDFF), after which the samples were transferred to the NMR biofilm reactor. Unlike the G. sulfurreducens biofilms, we had some difficulty growing S. oneidensis biofilms of an appropriate thickness for the NMR study. Because the spatial resolution for the depth profiles was 20 μm, our system required biofilms that were thicker than just a few tens of microns in order to produce relevant profiles. Therefore, we used a method that was identical to that described in Renslow et al. (2010), with the exception that electrodes were used in place of glass coverslips as the substrata. Although grown in a different way, these S. oneidensis biofilms allowed us to make interesting comparisons to G. sulfurreducens biofilms. Briefly, the electrodes were recessed to a depth of 400 μm using a custom gap-setting tool. S. oneidensis MR-1 inoculum and defined minimal growth medium with 25.4 mM lactate and 35 mM fumarate were introduced through ports located at the top of the CDFF. The medium was designed to be compatible with NMR measurements and was continuously purged with 100% N2 gas. Two Teflon® blades sheared away excess biomass to control the depth of the developing biofilm. The CDFF was operated in an incubator at 25 °C under anaerobic conditions. After 8-10 days, the biofilm samples were harvested aseptically through the sample port and transferred to the NMR. Prior to placement inside the NMR biofilm reactor, the reactor was cleaned and equilibrated similarly to the steps listed above for the G. sulfurreducens biofilm. The biofilms continued to grow in the NMR under continuous feed conditions at 0.5 mL/hr using the same defined medium as was used in the CDFF without the fumarate. The electrode upon which the S. oneidensis biofilms grew was polarized at +300 mVAg/AgCl, and was the sole terminal electron acceptor in the NMR biofilm reactor. The temperature was controlled by passing 25 °C 100% N2 into the NMR bore and around the biofilm reactor. The sample was maintained inside the NMR for the duration of the experiment.
Nuclear Magnetic Resonance Methods
The methods outlined here are similar to those discussed in our previous studies55, 67; however, parameter values and details have been changed to match the experimental conditions used in this study. Protons (1H) NMR experiments at 500 MHz were accomplished using a Bruker Avance III imaging spectrometer (Bruker Instruments, Billerica, MA, USA) with a 89-mm vertical bore, actively shielded superconducting magnet (11.7 T) employing a Bruker microimaging gradient insert (Micro2.5) with 150 gauss-per-centimeter triaxial imaging and diffusion gradients. The biofilm reactor was mounted on a custom perfusion probe with an Alderman-Grant-type radio frequency resonator with its linearly oscillating field direction aligned perpendicular to (and thus not shielded by) the electrode surface. To collect and process the data, ParaVision v5.1 imaging software (Bruker Biospin, Billerica, MA) was used. Measurements included: 1) 2D Fourier transform MRI; and 2) diffusion-mapping 2D Fourier transform MRI. Complete descriptions of the NMR methods are given in the Supplementary NMR Methods section in the ESI.
Computational Modeling
We developed a two-dimensional mathematical model to demonstrate how De assumptions can affect the predicted current output of an EAB. A complete description of the model is given in the Supplementary Computation Modeling section, and a graphical representation of the model geometry is shown in Figure S9 in the ESI.
Results and Discussion
We present the data here as both 2D De heat maps and Drs depth profiles. Daq are also provided to allow for direct applications in mathematical models using Fick’s law or related diffusion equations. De are measured inside biofilms and take into account the hindered molecular diffusion rate caused by biofilm structure and porosity. De are smaller in magnitude than Daq. Drs are a specific type of De generated by averaging De by depth inside the biofilm and normalizing the results against the Daq. Drs range from a value of 0 to 1, where 1 represents a Drs that is equal to the Daq. Figure S2 in the ESI gives a graphical representation of diffusion coefficients in biofilm systems, and Figure S3 shows example NMR measurement voxels, to scale, for both De and Drs. As discussed by Lewandowski and Beyenal (2007)68 and Van Wey et al. (2012)33, molecular transport parallel to the biofilm substratum is not as relevant as transport normal to the substratum, that is, through the depth of the biofilm. The major stratifications and microenvironments that appear in biofilm systems are caused by the concentration gradients that form along this axis68. The results obtained using PFG-NMR in our present study represent the De of water molecules normal to the electrode (and perpendicular to the flow direction). This is identical to our previous studies of De in biofilms grown on flat surfaces55, 68. The diffusion coefficients measured in our study are often called “self-diffusion” coefficients as a result of the diffusive transport of water molecules due to thermal energy without the presence of a concentration gradient required for detection by conventional Fickian methods. Effective Fickian diffusion coefficients of solutes at low concentrations, where the chemical activity of the solute is close to unity (i.e., as in an ideal solution), can be related to the self-diffusion coefficient of water69. This relationship is given by Equation 1:
| Equation 1 |
where Dei is the effective Fickian diffusion coefficient of solute i and Di is the bulk liquid Fickian diffusion coefficient of solute i. Note that the ratio De/Daq is referred to as the relative De and ranges from 0 to 1 within a biofilm; smaller values signify increased diffusion resistance. This equation provides a route for the direct use of results from this study in mathematical models, where the diffusion of each simulated solute can be governed by its own variable Dei.
G. sulfurreducens growth and De changes over time
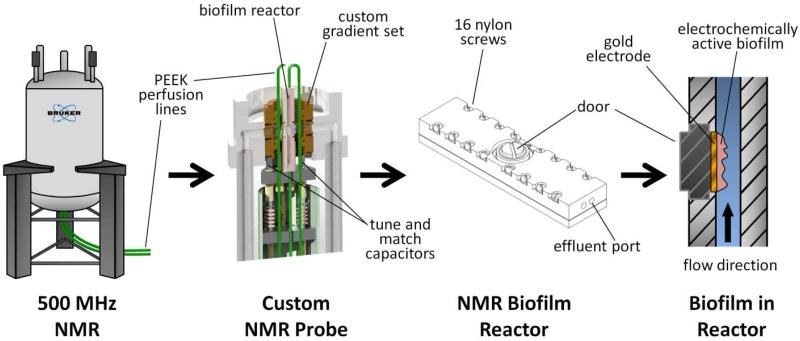
Figure 2 shows 2D MRI images of a G. sulfurreducens biofilm respiring on an electrode inside the NMR biofilm reactor, demonstrating its development over time. During the experiment growth medium was pumped against gravity, and the MRI images in this study are aligned identical to how the reactor was oriented during NMR measurements (the fluid flow was from the bottom of this page toward the top). The biofilm was maintained for nearly two months and grew to several hundred microns thick, similar to other G. sulfurreducens biofilms grown over long time periods in our lab31. As seen in Figure 2, the biofilm formed a continuous slab over the entire electrode. A “halo”-like artifact is visible above the biofilm in the three normal-plane 2D MRI images, forming alternating light and dark bands that culminate in a bright region directly over the top center of the biofilm. This artifact was only associated with the production of current, as it did not appear when no current passed; it was found to be a result of reduced spin-lattice (T1) relaxation rates. One large bubble is present in the upper-right (upstream) portion of the biofilm as shown in the face-plane view.
Figure 2. G. sulfurreducens biofilm growth.
Left) Time series of three normal-plane 2D MRI showing progression of the growth of a G. sulfurreducens biofilm. The white arrows indicate the top of the biofilm. The ages shown are 24, 35, and 52 days. In the initial image the biofilm is 170 μm thick, and in the final image the biofilm is 370 μm thick. Right) A face-plane 2D MRI of the biofilm. The white arrows indicate the edges of the biofilm on top of the electrode. A 1 mm scale bar is provided at the top of each MRI (the normal-plane and face-plane images have the identical scale).
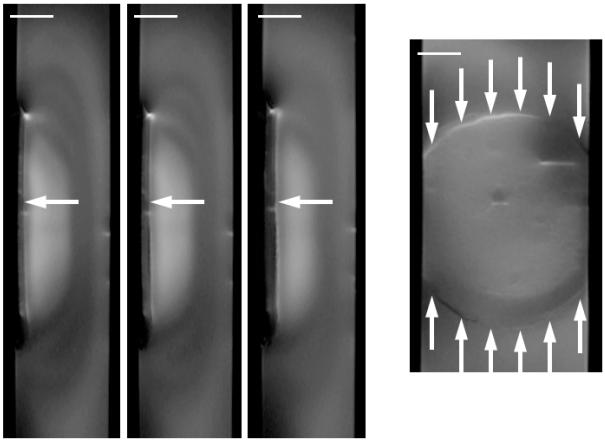
Both the 2D maps and 1D depth profiles of relative De of a G. sulfurreducens biofilm are shown in Figure 3. The Daq, used to generate the relative values, was 2.82 · 10−9 ms/s (σ: 0.10 · 10−9 ms/s) (for an extended discussion on the Daq and the effect of temperature, see the ESI). As in the MRI figures, the orientation of the 2D maps is identical to that of the experimental setup, with fluid flowing from the bottom of the page toward the top of the page. The 2D maps reveal that diffusion rates vary more with biofilm depth than they vary in the lateral directions. The top of the biofilm is nearly uniform along the length of the biofilm. See Figure S4 in the ESI for images demonstrating the development of De in finer time steps and also Figure S5, which shows the development of thickness over time. In the depth profiles, a sigmoid-shaped Drs profile is present regardless of biofilm age. However, the Drs decrease with biofilm age. The average Drs in the biofilm were 0.40, 0.33, and 0.34, and the minimum Drs in the biofilm were 0.21, 0.09, and 0.08 for 24, 35, and 52 days, respectively. Biofilm thicknesses and the tops of the biofilms as marked in the depth profiles were determined by image analysis of 2D MRI, identical to the method used in our previous studies55, 67. The top of the biofilm was defined as the tallest portion of the biofilm located in the 2 mm by 2 mm voxel used to generate the depth profiles. It is interesting to note that the Drs profiles begin to drop prior to the marked top of the biofilm. The 2D maps show dark features (lower diffusion regions) above the marked top, particularly as the biofilm becomes older. We believe this is a result of a low concentration of loose biomass, probably extracellular polymeric substances, protruding above the MRI visible top67.
Figure 3. Diffusion mapping of a G. sulfurreducens biofilm over time.
The top row shows two-dimensional De maps obtained using PFG-NMR, normalized against Daq, of the middle 2 mm of the biofilm. The dark regions represent low De. The bottom row shows Drs depth profiles derived by averaging the De of the middle 2 mm of the biofilm (shown in the maps). The top of the biofilm as determined using magnetic resonance imaging is indicated by the vertical lines. From left to right, the panels show the G. sulfurreducens biofilm at 24, 35, and 52 days old.
In this paper we also report biofilm dry weight densities (X, kg/m3), which can be approximated using the following equation, developed by Fan et al. (1990)70:
| Equation 2 |
Equation 2 gives us the ability to approximate the density of a biofilm when Drs is known. For the 24-day-old biofilm, the average density was calculated to be 41 kg/m3, corresponding to an average Drs of 0.40. The maximum density observed was 247 kg/m3, corresponding to an average Drs of 0.08. G. sulfurredens biofilms have been found to be denser near the electrode71 and are described in the literature as having a “top fluffy layer and dense inner core”56, which Equation 2 and Figure 3 quantitatively confirm for even the several-hundred-micron-thick biofilms reported here.
S. oneidensis growth and De changes over time
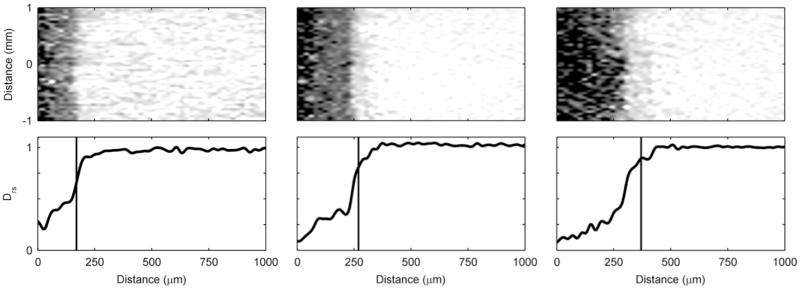
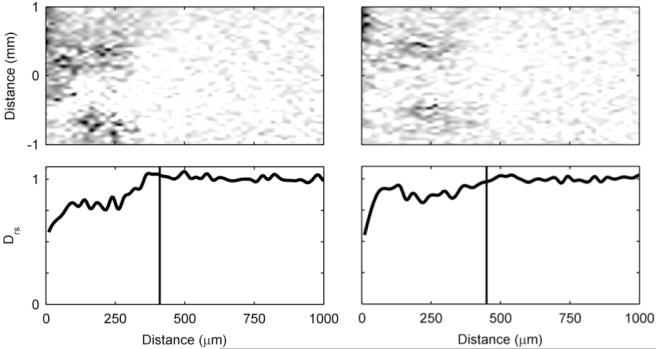
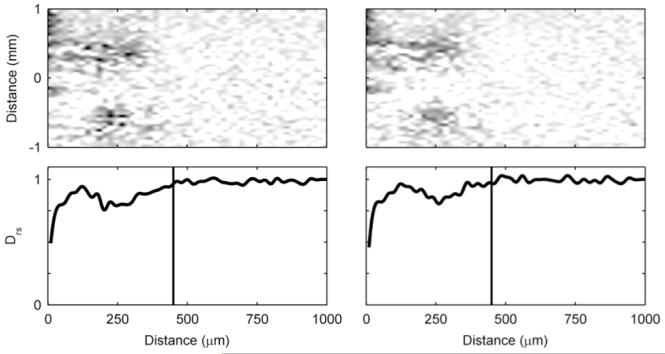
Figure 4 shows the progression over time of an S. oneidensis biofilm. Unlike the G. sulfurreducens biofilms, the S. oneidensis biofilms used in this study did not form a continuous slab that covered the entire electrode. Instead, towering clusters and heterogeneous colonies were observed. The 2D MRI shows that the biofilm only partially covered the electrode. In Figure 5, the corresponding De maps reveal further that the biofilm was comprised of individual clusters. The darker regions in the 2D map, which signify less diffusive regions, are sparse and disconnected. The maps reveal that the De in the S. oneidensis biofilms are much higher than those in the G. sulfurreducens biofilms. The Daq, used to generate the relative values, was 2.49 · 10−9 ms/s (σ: 0.06 · 10−9 ms/s). The average Drs in the S. oneidensis biofilm were 0.83 and 0.88 at 12 and 16 days, respectively, and the minimum Drs were 0.58 and 0.55 at 12 and 16 days, respectively. These values are nearly identical to those of S. oneidensis biofilms grown on glass coverslips in an earlier study by our group55. The profile shapes are also comparable. Growing the biofilms on electrodes did not appear to change the average or minimum De values. It is interesting to note, however, that in our previous study55 the De values steadily decreased and the biofilms grown on glass coverslips in a CDFF became thicker and denser over time. In this study, the S. oneidensis biofilms were very unstable on the electrode and readily sloughed off, especially as they aged. Even though the biofilm thickness increased over time, the biofilm surface coverage of the electrode decreased. This is shown in Figure S6 in the ESI. It was not possible to keep S. oneidensis biofilms intact for long periods of time, unlike the G. sulfurreducens biofilms. This may be caused by impeded growth of S. oneidensis biofilms on smooth gold surfaces in the NMR biofilm reactor, where the fluid flow is parallel to the electrode surface and is pumped against gravity. Anodic EAB morphology has been shown to be dependent on the electrode material72. Most likely a different geometry or graphite electrodes would work better, as demonstrated in the literature73; however, such a system would not be compatible with our NMR microimaging system. For the 16-day-old biofilm, the average density was calculated to be 4 kg/m3, corresponding to an average Drs of 0.88. This is nearly an order of magnitude lower than the average density of the G. sulfurreducens biofilms. The maximum density observed was 23 kg/m3, corresponding to an average Drs of 0.55. Again, this is nearly an order of magnitude lower than the maximum density observed in the G. sulfurreducens biofilms.
Figure 4. S. oneidensis biofilm growth.
Left) Time series of two normal-plane 2D MRI showing the progression of the growth of an S. oneidensis biofilm. The white arrows indicate the top of the biofilm. The ages shown are 12 and 16 days. In the initial image the biofilm is 410 μm thick, and in the final image the biofilm is 450 μm thick. Right) A face-plane 2D MRI of the biofilm. The white arrows indicate the edges of the biofilm on top of the electrode. A 1 mm scale bar is provided at the top of each MRI (the normal-plane and face-plane images have the identical scale).
Figure 5. Diffusion mapping of S. oneidensis biofilms over time.
The top row shows two-dimensional De maps obtained using PFG-NMR, normalized against Daq, showing the middle 2 mm of the biofilm. Dark regions represent low De. The bottom row shows Drs profiles, derived by averaging the De of the middle 2 mm of the biofilm (shown in the maps). The top of the biofilm as determined by magnetic resonance imaging is indicated by the vertical lines. The panel on the lefts shows the S. oneidensis biofilm at 12 days, and the panel on the right shows it at 16 days.
Because of the cluster/colony nature of the S. oneidensis biofilms, we also wanted to see how De values changed over time in individual clusters in order to compare them better with the G. sulfurreducens biofilms, which had continuous cell coverage of the electrode. Table 2 provides the values for four distinct clusters (the locations of the clusters are shown in Figure S7 in the ESI). Even when comparing the average De of individual clusters, G. sulfurreducens has much lower De than S. oneidensis. For each cluster, the average De increased over time. The increase ranged from +9%, in cluster 3, up to +39%, in cluster 2. These results are different from all measurements performed on G. sulfurreducens biofilms, where the De decreased continuously over time. It is well known that cells in biofilms that are not under favorable growth conditions can undergo “seeding dispersal,” which decreases the biofilm density and increases De values74. We speculate that this type of event leads to increasing De values for both the individual clusters and the biofilm as a whole.
Table 2.
Average De of several example cell clusters in the S. oneidensis biofilms. The measurements were taken on clusters in the biofilm shown in Figure 4, and the specific clusters chosen are shown in Figure S7 in the ESI. De are given in 10−9 m2/s along with the standard deviation and the relative De in the adjacent column.
| Initial | End of experiment | |||
|---|---|---|---|---|
| De (sd) | Relative De | De (sd) | Relative De | |
| Cluster 1 | 2.02 (σ: 0.87) | 0.81 | 2.23 (σ: 0.53) | 0.90 |
| Cluster 2 | 1.53 (σ: 0.64) | 0.62 | 2.13 (σ: 0.66) | 0.86 |
| Cluster 3 | 2.05 (σ: 0.28) | 0.82 | 2.21 (σ: 0.29) | 0.89 |
| Cluster 4 | 1.54 (σ: 0.20) | 0.62 | 1.83 (σ: 0.23) | 0.74 |
Effect of halting respiration on De
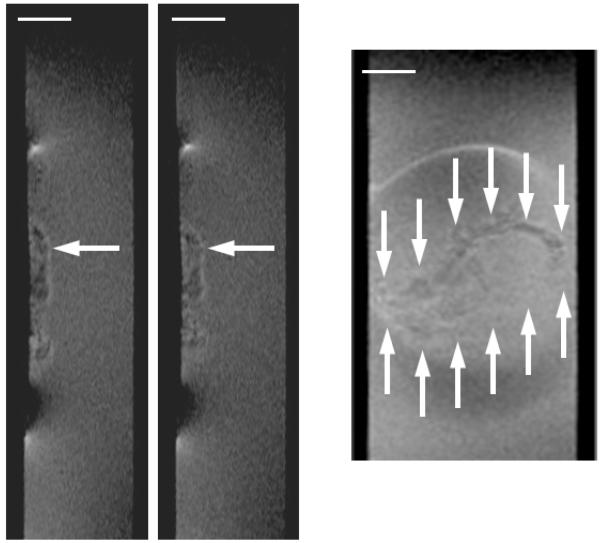
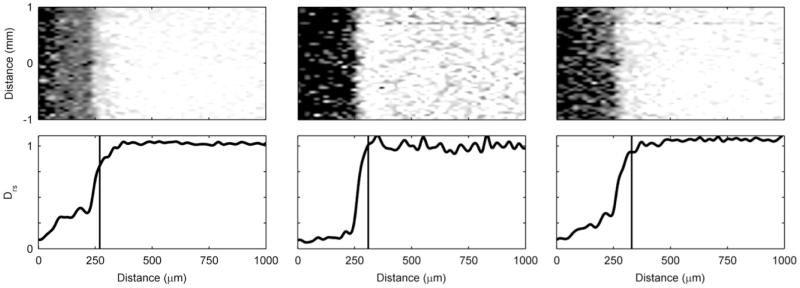
Figure 6 shows 2D De maps and Drs profiles of a G. sulfurreducens biofilm whose respiration was halted. We were interested in observing how halting respiration by removing access to the electrode as an electron acceptor would affect the De in the short term. With the polarization switched off, and the electrode at open circuit potential, would the De change in response to biofilm microstructure change? The Figure 6 image sequence reveals that the De are reduced when respiration is halted and recover once respiration is reinstated. The average Drs for the entire biofilm thickness went from 0.33 to 0.25 to 0.34, as the biofilm went from active respiration to halted respiration to active respiration. These differences are not statistically significant; however, if we consider only the bottom 200 microns of the biofilm, the differences are statistically significant (p<0.0001). Here the average Drs went from 0.26 to 0.09 to 0.18. Because the electrode is acting as the sole terminal electron acceptor for microbial metabolism, biofilm metabolic activity can be measured as electrical current passing through the electrode. During the three measurements shown in Figure 6, the current went from 3.3 A/m2 to 0.0 A/m2 to 2.3 A/m2 (see Figure S8 in the ESI to see the development of current over time). This allows us to directly verify that respiration was halted and then recovered. The change in De is not caused by electromigration, which alters the mobility of charged species, because the preponderance of the H1 interrogated are bound in uncharged water molecules (as the growth medium is near neutral pH, approximately 2 water molecules out of every 1 billion are split into hydronium and hydroxide). We found that preventing the electrode from acting as the electron acceptor by removing the polarization momentarily alters the morphology of the G. sulfurreducens biofilm and halts its metabolism. Previously Jain et al. (2011) showed that switching to non-turnover conditions (removal of the electron donor) caused a 35-μm-thick G. sulfurreducens biofilm to reduce to only 3.5 μm within 24 hours, purportedly because of biomass detachment. We did not observe such a change in thickness, although the change in De suggests that there were changes in the biofilm structure. This could be because in our case only the electron acceptor was removed, not the electron donor. Cao et al. (2012)67 also showed that microstructure changes in the biofilm can affect De. In that study, we demonstrated that exposure of an S. oneidensis biofilm to contaminants U(VI) and Cr(VI) reversibly altered both the metabolic rate and De, while not noticeably changing the biofilm macrostructure or thickness. The effects of altered metabolic states and electron donor/acceptor availability on biofilm microstructure are currently undetermined56. When we change the polarization state of the electrode for an S. oneidensis biofilm, this trend is not apparent. Figure 7 shows that the 2D De map and Drs profile do not change even after polarization is initiated. This may be because of the lower respiration rate of S. oneidensis compared to that of G. sulfurreducens. The current production during polarization only reached 18 mA/m2, which is at least two orders of magnitude less than the current produced by G. sulfurreducens biofilms. Thus, at least in the short term, the availability of the electron-accepting electrode does not affect S. oneidensis biofilm structure.
Figure 6. Diffusion mapping of a G. sulfurreducens biofilm with and without a polarized electrode.
De for a ~41-day-old G. sulfurreducens biofilm. Left) Electrode polarized to +300 mVAg/AgCl. Middle) Polarization switched off: open circuit potential. Right) Polarized again to +300 mVAg/AgCl.
Figure 7. Diffusion mapping of an S. oneidensis biofilm with and without a polarized electrode.
Diffusion mapping of an S. oneidensis biofilm with and without a polarized electrode. Left) Polarization off: open circuit potential. Right) Polarized to +300 mVAg/AgCl.
Does De reflect the type of electron transfer strategy used by a biofilm?
So far, we have looked at two distinct species of EABs. The De of G. sulfurreducens and S. oneidensis biofilms are strikingly different, suggesting that De could reflect the ability of different species to generate high current on electrodes. While the De reported in our study are only a measurement of the self-diffusion of water within EABs, they are likely indicative of and influential on each species’ unique EET strategy.
For biofilms that rely solely upon conduction-based EET, such as G. sulfurreducens75, diffusion is only required for the transport of nutrients, metabolic products, electrolytes to maintain electroneutrality, and buffering components. EET in these systems is dependent upon a coherent conductive pathway that allows electrons to reach the electrode. A tightly bound biofilm matrix, measured as a low De, could reduce electron transfer resistance by providing increased extracellular polymeric substances to carry current. This idea is supported by evidence from G. sulfurreducens variant KN400, which has a higher protein and pili concentration and a higher cell density compared to G. sulfurreducens DL163. Despite the thinner biofilms, KN400 produces a higher current density. Currently the precise form of conduction involved in EET is yet to be unequivocally proven76-78. Regardless of form, a high-density biofilm will lead to reduced electrical resistance, either by shrinking the spacing and increasing the density of electron superexchange mediators or by increasing the density of components that exhibit metal-like conduction.
We hypothesize that spatial differences in density may also be indicative of conduction-based EET. The current passing through the bottom layers of the biofilm is cumulative from the electrons being passed from the top layers. Therefore, it is logical that we see a much higher biofilm density at the bottom of the G. sulfurreducens biofilm, as others have also reported56, 71. Future experiments will determine whether high biofilm conductivity and a high enough De to allow for electron donor penetration are mutually exclusive. It is possible that if De is too low near the bottom of the biofilm and metabolic activity at the top of the biofilm is sufficiently fast, nutrient delivery could be prevented to regions of the biofilm. However, the biofilm may continue to grow and respire through the dense regions of the biofilm that are metabolically dormant yet still conducting. That is, if the biofilm matrix has a high enough conductivity, then the biofilm thickness may not be limited to only a few tens of microns, because nutrients will only need to diffuse into the top, metabolically active, portion of the biofilm.
A commonly described potential bottleneck for EAB respiration is proton buildup at the anode, especially for G. sulfurreducens biofilms, because of their higher metabolic rates and tighter matrices. This acidification is linked to De because the pH in the biofilm is a function of proton diffusion, diffusion of buffering species, and a large set of reaction rates and equilibriums62. Some studies have suggested that engineering cells to form more porous biofilms with higher De may help alleviate the metabolic inhibition that happens as the pH drops to near 6 or lower79. Based on our hypothesis that a higher biofilm density will increase conduction-based EET, we believe this approach may not be the most efficient route to increase current production. Furthermore, while some studies have shown pH to be a limiting factor because of low diffusion80, a recent study by our group demonstrated for a 200-μm-thick G. sulfurreducens biofilm that pH (as low as 6.3) was not limiting current production31. Thus pH limitations are not an issue for all biofilms, even if they are very thick, and the diffusion of protons may not always be a bottleneck, even during high current production. Future experiments and mathematical models will need to determine the balance between the diffusion of metabolic products and the density that decreases electrical resistance.
For biofilms that rely upon diffusion-based EET, the role of De on overall electron transfer ability is more complicated. Evidence suggests that S. oneidensis uses this type of EET mechanism28, 81. Note, however, that there is also evidence that S. oneidensis uses both conduction- and diffusion-based electron transfer27, 82, in particular the presence of electrically conductive nanowires. Several papers have discussed the energetics related to the use of mediating molecules to gain access to insoluble electron acceptors81, 83, 84. While mediated electron transfer offers an evolutionary advantage by allowing access to normally inaccessible electron acceptor sources, there is a metabolic cost associated with producing endogenous extracellular mediators. De controls not only the rate of transport of reduced mediators from the cell to the electrode, a potential metabolic bottleneck, but also the rate at which these molecules are transported out of the biofilm and lost to the environment. In practical terms, De controls the average number of reduction-oxidation cycles per mediator, which is critical for determining the metabolic energy gained by investing in mediator production84. If the mediators do not undergo enough recycles, they are no longer mutually beneficial for the individual cell and community. Thus there is a dichotomy: a low De limits the rate of transport of mediators to the electrode, but a low De also slows the loss of mediators and keep mediators localized in the biofilm community for successive redox cycling. Perhaps for G. sulfurreducens biofilms, using conduction-based EET is a more metabolically efficient strategy than diffusion-based EET because their density, which we found to be an average of 10 times higher than that of S. oneidensis biofilms, may be too high to allow for metabolically relevant mediator recycling rates. Conversely, S. oneidensis has a lower respiration rate and forms less dense biofilms, making it appear ideally suited to exploit diffusion-based EET. It is hypothesized that the biofilm mode of life, which lowers the local De per cell, is necessary for diffusion-based EET in order to prevent convection, which would rapidly deplete any endogenous mediators in typical environments81.
A low diffusion rate in a dense EAB would offer further benefits if electrons can undergo chained electron transport via mediators, analogous to the proton in the Grotthuss mechanism85, 86. Protons are known to have an unexpectedly high diffusion coefficient due to the Grotthuss mechanism (i.e., hopping of protons between water molecules, prototropic mobility), thus providing a high net flux of protons compared to other molecules. Likewise, mediators such as flavins may also allow electron hopping between themselves because of their rapid equilibrium of the flavoquinone and flavohydroquinone forms and flavosemiquinone and also their propensity to form temporary dimer and higher polymer complexes87-89. De in biofilms with this capability would have a different role because a mediating molecule would only need to diffuse far enough to interact with another flavin, as opposed to diffusing the entire distance between the cell and the insoluble electron acceptor. In our opinion, flavins and similar molecules may be better described as mediators, as opposed to the common misnomer of “shuttles,” because the transport is neither active nor merely between clearly defined locations (a cell and the electron acceptor). Ultimately a charge-transport diffusion coefficient for electrons which accounts for the net flux of electrons caused by both the self-diffusion of mediators and hopping will need to be determined for EAB systems that rely on diffusion-based EET.
A final consideration relating De to diffusion-based EET is the possible hindering effect of high biomass densities on cellular surface exposure. Bouhenni et al. (2010) demonstrated that the exposure of outer membrane c-type cytochromes MtrC and OmcA is crucial for EET in S. oneidensis90. They found that it was not the mediator concentration that was limiting, but rather the surface exposure of the cytochrome proteins. It is currently unclear whether an increased biofilm density would sterically restrict access to these outer membrane proteins, which have been experimentally confirmed to interact with flavin molecules91, 92. The evidence for outer membrane cytochromes playing a significant role in diffusion-based EET keeps mounting91, 93. Therefore, a low-density biofilm (and, by extension, a high De) may provide more access to the exterior surfaces of cells that use diffusion-based EET. It may be sufficient for biofilms merely to prevent convection to preserve higher concentrations of mediators while maintaining a high surface area exposure to allow mediator-cytochrome interactions.
The high density of EABs, reflected in De, plays a major role in the EET strategies used by biofilms. Measuring the De values in EABs is important because of the complex connection of each EET mechanism to diffusive processes and biofilm density. As discussed here, each EET scheme offers an ability to optimize for electron transfer with association to De. This will be especially true for biofilms that use a combination of EET mechanisms, such as S. oneidensis and mixed-species biofilms. De will be a critical parameter for fully understanding both chemical and electrical fluxes in EAB systems, and determining the exact role of diffusion in these biofilms will provide a crucial piece of the EET big picture.
Effects of different Drs assumptions on predicted current output
We generated a 2D mathematical model simulating the 52-day-old G. sulfurreducens biofilm (shown in the rightmost panel of Figure 3). We assumed conduction-based extracellular electron transfer dictated by the Nernst-Monod equation59. Using this model we calculated the pseudo-steady state current output of the G. sulfurreducens biofilm. Three cases of the model were tested: a constant Drs in the biofilm, a linearly decreasing Drs, and a Drs profile derived from the empirical data taken from the measurements of the 52-day-old G. sulfurreducens biofilm. Figure S10 shows the three Drs profiles. See the complete details of the model in the Supplementary Computational Modeling section in the ESI. By calculating the theoretical current for the G. sulfurreducens biofilm, it is possible to see that the Drs plays an important role in determining biofilm metabolic activity. For example, the current produced using a linearly decreasing profile was 18% higher than that produced assuming a constant Drs (2.65 A/m2, compared to 3.12 A/m2). Using the empirical profile, a current of 2.88 A/m2 was calculated (8% higher than a constant Drs). Using Equation 2, we also compared current production when the biofilm density was related to the Drs profile. In these cases, linearly decreasing and empirical Drs profiles resulted in a current decrease of 4-5% instead of the increase seen with a non-variable biofilm density. Acetate concentration profiles were also affected by the treatment of Drs in the biofilm. This can be seen in Figure S11 in the ESI. Bulk measurements such as electrical current production do not capture how the Drs assumptions affect the biofilm microenvironment.
Table 3 provides a summary of the effect of Drs assumptions on pseudo-steady state current, acetate flux at the top of the biofilm, and acetate concentration at the bottom of the biofilm. In more complex simulations, the changes seen in the biofilm microenvironment could lead to model output differences, as local concentrations can impact biofilm growth, utilization rates, gene expression, etc. Furthermore, the effect of the Drs assumption will also be seen in an electron-mediated biofilm such as S. oneidensis, where the current produced is dependent upon not only the electron donor transport, but also the electron transfer-mediating molecules. Our model confirmed that the use of different Drs profiles can lead to different predicted electrical current production from an EAB, even when only the electron donor is considered, which is just one of the processes controlled by diffusion in EABs.
Table 3.
Effect of Drs assumptions on simulated current output and substrate utilization. For each Drs assumption case, total current, acetate flux, and acetate concentration are given. Total current (A/m2) is based on conducted electron transfer from the entire biofilm. Acetate flux (mmol/cm2 day) is a measure of the average flux at the top of the biofilm in the 2 mm × 2 mm surface area NMR measurement voxel. Acetate concentration is given for the acetate available at the bottom of the biofilm in the same NMR measurement voxel. These three measurements were chosen to exemplify the effects of different Drs assumptions.
| De Assumption | Total Current (A/m2) |
Top of Biofilm Acetate Flux (mmol/cm2 day) |
Bottom of Biofilm Acetate Concentration (mM) |
|---|---|---|---|
| Non-variable density | |||
| Constant Drs | 2.65 | 0.28 | 1.55 |
| Linearly Decreasing Drs | 3.12 | 0.30 | 2.33 |
| Empirical Profile Drs | 2.88 | 0.29 | 1.18 |
| With variable density | |||
| Constant Drs | 2.65 | 0.28 | 1.55 |
| Linearly Decreasing Drs | 2.54 | 0.24 | 0.05 |
| Empirical Profile Drs | 2.53 | 0.25 | 0.21 |
Conclusions
In this study we quantified 2D effective diffusion coefficient maps and surface-averaged relative effective diffusion coefficient profiles in electrochemically active biofilms respiring on electrodes. We found that:
Drs decreases with depth in G. sulfurreducens biofilms, following a sigmoid shape
Drs at a given location decreases with G. sulfurreducens biofilm age
Average De and Drs profiles in G. sulfurreducens biofilms are lower than those in S. oneidensis biofilms—the G. sulfurreducens biofilms studied here were on average 10 times denser than the S. oneidensis biofilms
Halting the respiration of a G. sulfurreducens biofilm decreases the De values.
Supplementary Material
Broader Context.
Biofilms that can use solid external electron acceptors for respiration are now known to be ubiquitous in nature. In the past two decades, a significant amount of research has been performed to understand how extracellular electron transfer occurs and how this phenomenon can be applied for beneficial purposes. Despite their unique form of respiration, it has been common for researchers to assume that electrochemically active biofilms have similar properties to typical biofilms, which rely on soluble electron acceptors. However, as demonstrated in this study, electrochemically active biofilms have distinct properties that are indicative of the biofilms’ respiration strategies. Elucidating these distinctions will allow for the engineering of tailored systems and techniques, enhancing our ability to exploit the energy generated by extracellular electron transfer.
Acknowledgements
This research was supported by the U.S. Office of Naval Research (ONR), grant #N00014-09-1-0090. All NMR experiments were performed at EMSL, a national scientific user facility sponsored by the Department of Energy’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory. We thank Hardeep Mehta (EMSL Instrument Development Lab) for developing a compatible resonator and Kenneth Swanson and James Follansbee (EMSL Instrument Development Lab) for developing the networked pump controller. Ryan Renslow and Jerome Babauta acknowledge NIH Training Grant (T32-GM008336).
Nomenclature
- Daq
bulk liquid diffusion coefficients
- De
effective diffusion coefficient
- Dei
effective Fickian diffusion coefficient of solute i
- Di
bulk liquid Fickian diffusion coefficient of solute i
- Drs
surface-averaged relative effective diffusion coefficient
Works Cited
- 1.Stewart PS. Journal of Bacteriology. 2003;185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pibalpakdee P, Wongratanacheewin S, Taweechaisupapong S, Niumsup PR. International Journal of Antimicrobial Agents. 2012;39:356–359. doi: 10.1016/j.ijantimicag.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 3.Stewart PS. Antimicrobial Agents and Chemotherapy. 1996;40:2517–2522. doi: 10.1128/aac.40.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phoenix VR, Holmes WM, Ramanan B. Mineralogical Magazine. 2008;72:483–486. [Google Scholar]
- 5.Teitzel GM, Parsek MR. Applied and Environmental Microbiology. 2003;69:2313–2320. doi: 10.1128/AEM.69.4.2313-2320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabije AH, Agapay RC, Tampus MV. Asia-Pacific Journal of Chemical Engineering. 2009;4:735–743. [Google Scholar]
- 7.Lin Y-H, Hsien T-Y. Water Science and Technology. 2009;59:1703–1711. doi: 10.2166/wst.2009.203. [DOI] [PubMed] [Google Scholar]
- 8.Wichern M, Lindenblatt C, Lubken M, Horn H. Water Research. 2008;42:3899–3909. doi: 10.1016/j.watres.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 9.Zhou XH, Liu J, Song HM, Qiu YQ, Shi HC. Environmental Engineering Science. 2012;29:466–471. [Google Scholar]
- 10.Borole AP, Reguera G, Ringeisen B, Wang ZW, Feng YJ, Kim BH. Energy & Environmental Science. 2011;4:4813–4834. [Google Scholar]
- 11.Lovley DR. Nature Reviews Microbiology. 2006;4:497–508. doi: 10.1038/nrmicro1442. [DOI] [PubMed] [Google Scholar]
- 12.Lovley DR. Current Opinion in Biotechnology. 2008;19:564–571. doi: 10.1016/j.copbio.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Logan BE. Nature Reviews Microbiology. 2009;7:375–381. doi: 10.1038/nrmicro2113. [DOI] [PubMed] [Google Scholar]
- 14.Babauta J, Renslow R, Lewandowski Z, Beyenal H. Biofouling. 2012 doi: 10.1080/08927014.2012.710324. In Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsili E, Sun J, Bond DR. Electroanalysis. 2010;22:865–874. [Google Scholar]
- 16.Donovan C, Dewan A, Heo D, Beyenal H. Environmental Science & Technology. 2008;42:8591–8596. doi: 10.1021/es801763g. [DOI] [PubMed] [Google Scholar]
- 17.Zhang F, Tian L, He Z. Journal of Power Sources. 2011;196:9568–9573. [Google Scholar]
- 18.Du Z, Li H, Gu T. Biotechnology Advances. 2007;25:464–482. doi: 10.1016/j.biotechadv.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Ramnarayanan R, Logan BE. Environmental Science & Technology. 2004;38:2281–2285. doi: 10.1021/es034923g. [DOI] [PubMed] [Google Scholar]
- 20.Erable B, Duteanu NM, Ghangrekar MM, Dumas C, Scott K. 736 Biofouling. 2010;26:57–71. doi: 10.1080/08927010903161281. [DOI] [PubMed] [Google Scholar]
- 21.Forrestal C, Xu P, Ren ZY. Energy & Environmental Science. 2012;5:7161–7167. [Google Scholar]
- 22.Kim Y, Logan BE. Environmental Science & Technology. 2011;45:5840–5845. doi: 10.1021/es200584q. [DOI] [PubMed] [Google Scholar]
- 23.Qu YP, Feng YJ, Wang X, Liu J, Lv JW, He WH, Logan BE. Bioresource Technology. 2012;106:89–94. doi: 10.1016/j.biortech.2011.11.045. [DOI] [PubMed] [Google Scholar]
- 24.Geelhoed JS, Stams AJM. Environmental Science & Technology. 2011;45:815–820. doi: 10.1021/es102842p. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Grot S, Logan BE. Environmental Science & Technology. 2005;39:4317–4320. doi: 10.1021/es050244p. [DOI] [PubMed] [Google Scholar]
- 26.Torres CI, Marcus AK, Lee H-S, Parameswaran P, Krajmalnik-Brown R, Rittmann BE. Fems Microbiology Reviews. 2010;34:3–17. doi: 10.1111/j.1574-6976.2009.00191.x. [DOI] [PubMed] [Google Scholar]
- 27.Gorby YA, Yanina S, McLean JS, Rosso KM, Moyles D, Dohnalkova A, Beveridge TJ, Chang IS, Kim BH, Kim KS, Culley DE, Reed SB, Romine MF, Saffarini DA, Hill EA, Shi L, Elias DA, Kennedy DW, Pinchuk G, Watanabe K, Ishii S. i., Logan B, Nealson KH, Fredrickson JK. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11358–11363. doi: 10.1073/pnas.0604517103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3968–3973. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Canstein H, Ogawa J, Shimizu S, Lloyd JR. Applied and Environmental Microbiology. 2008;74:615–623. doi: 10.1128/AEM.01387-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babauta JT, Nguyen HD, Beyenal H. Environmental Science & Technology. 2011;45:6654–6660. doi: 10.1021/es200865u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babauta JT, Nguyen HD, Harrington TD, Renslow R, Beyenal H. Biotechnol. Bioeng. 2012 doi: 10.1002/bit.24538. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sleutels T, Hamelers HVM, Buisman CJN. Environmental Science & Technology. 2010;44:8259–8263. doi: 10.1021/es101858f. [DOI] [PubMed] [Google Scholar]
- 33.Van Wey AS, Cookson AL, Soboleva TK, Roy NC, McNabb WC, Bridier A, Briandet R, Shorten PR. Biotechnol. Bioeng. 2012;109:1280–1292. doi: 10.1002/bit.24390. [DOI] [PubMed] [Google Scholar]
- 34.Alpkvist E, Klapper I. Bulletin of Mathematical Biology. 2007;69:765–789. doi: 10.1007/s11538-006-9168-7. [DOI] [PubMed] [Google Scholar]
- 35.Chang I, Gilbert ES, Eliashberg N, Keasling JD. Microbiology-Sgm. 2003;149:2859–2871. doi: 10.1099/mic.0.26211-0. [DOI] [PubMed] [Google Scholar]
- 36.Hermanowicz SW. Mathematical Biosciences. 2001;169:1–14. doi: 10.1016/s0025-5564(00)00049-3. [DOI] [PubMed] [Google Scholar]
- 37.Rahman NK, Abu Bakar MZ, Uzir MH, Kamaruddin AH. Mathematical Biosciences. 2009;218:130–137. doi: 10.1016/j.mbs.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Rajagopalan S, Rockstraw DA, MunsonMcGee SH. Bioresource Technology. 1997;61:175–183. [Google Scholar]
- 39.Xavier JB, Picioreanu C, van Loosdrecht MCM. Water Science and Technology. 2004;49:177–185. [PubMed] [Google Scholar]
- 40.Kreft JU, Picioreanu C, Wimpenny JWT, van Loosdrecht MCM. Microbiology-Sgm. 2001;147:2897–2912. doi: 10.1099/00221287-147-11-2897. [DOI] [PubMed] [Google Scholar]
- 41.Boltz JP, Morgenroth E, Brockmann D, Bott C, Gellner WJ, Vanrolleghem PA. Water Science and Technology. 2011;64:930–944. doi: 10.2166/wst.2011.709. [DOI] [PubMed] [Google Scholar]
- 42.Radu AI, Vrouwenvelder JS, van Loosdrecht MCM, Picioreanu C. Chemical Engineering Journal. 2012;188:30–39. [Google Scholar]
- 43.Taherzadeh D, Picioreanu C, Horn H. Biophysical Journal. 2012;102:1483–1492. doi: 10.1016/j.bpj.2012.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindley B, Wang Q, Zhang TY. Physical Review E. 2012:85. doi: 10.1103/PhysRevE.85.031908. [DOI] [PubMed] [Google Scholar]
- 45.Zhang TY. Bulletin of Mathematical Biology. 2012;74:1427–1447. doi: 10.1007/s11538-012-9719-z. [DOI] [PubMed] [Google Scholar]
- 46.Inoue K, Leang C, Franks AE, Woodard TL, Nevin KP, Lovley DR. Env Microbiol Rep. 2011;3:211–217. doi: 10.1111/j.1758-2229.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 47.Nevin KP, Kim BC, Glaven RH, Johnson JP, Woodard TL, Methe BA, DiDonato RJ, Covalla SF, Franks AE, Liu A, Lovley DR. Plos One. 2009:4. doi: 10.1371/journal.pone.0005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bryers JD, Drummond F. Biotechnol. Bioeng. 1998;60:462–473. [PubMed] [Google Scholar]
- 49.deBeer D, Stoodley P, Lewandowski Z. Biotechnol. Bioeng. 1997;53:151–158. doi: 10.1002/(SICI)1097-0290(19970120)53:2<151::AID-BIT4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 50.Lawrence JR, Wolfaardt GM, Korber DR. Applied and Environmental Microbiology. 1994;60:1166–1173. doi: 10.1128/aem.60.4.1166-1173.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bishop PL, Zhang TC, Fu YC. Water Science and Technology. 1995;31:143–152. [Google Scholar]
- 52.Stewart PS, Davison WM, Steenbergen JN. Antimicrobial Agents and Chemotherapy. 2009;53:3505–3507. doi: 10.1128/AAC.01728-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beyenal H, Tanyolac A, Lewandowski Z. Water Science and Technology. 1998;38:171–178. [Google Scholar]
- 54.Beyenal H, Lewandowski Z. Water Research. 2000;34:528–538. [Google Scholar]
- 55.Renslow RS, Majors PD, McLean JS, Fredrickson JK, Ahmed B, Beyenal H. Biotechnol. Bioeng. 2010;106:928–937. doi: 10.1002/bit.22755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jain A, Gazzola G, Panzera A, Zanoni M, Marsii E. Electrochimica Acta. 2011;56:10776–10785. [Google Scholar]
- 57.Picioreanu C, Head IM, Katuri KP, van Loosdrecht MCM, Scott K. Water Research. 2007;41:2921–2940. doi: 10.1016/j.watres.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 58.Picioreanu C, Katuri KP, Head IM, van Loosdrecht MCM, Scott K. Water Science and Technology. 2008;57:965–971. doi: 10.2166/wst.2008.095. [DOI] [PubMed] [Google Scholar]
- 59.Marcus AK, Torres CI, Rittmann BE. Biotechnol. Bioeng. 2007;98:1171–1182. doi: 10.1002/bit.21533. [DOI] [PubMed] [Google Scholar]
- 60.Picioreanu C, van Loosdrecht MCM, Curtis TP, Scott K. Bioelectrochemistry. 2010;78:8–24. doi: 10.1016/j.bioelechem.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 61.Marcus AK, Torres CI, Rittmann BE. Electrochimica Acta. 2010;55:6964–6972. [Google Scholar]
- 62.Marcus AK, Torres CI, Rittmann BE. Bioresource Technology. 2011;102:253–262. doi: 10.1016/j.biortech.2010.03.100. [DOI] [PubMed] [Google Scholar]
- 63.Strycharz SM, Malanoski AP, Snider RM, Yi H, Lovley DR, Tender LM. Energy & Environmental Science. 2011;4:896–913. [Google Scholar]
- 64.Merkey BV, Chopp DL. Bulletin of Mathematical Biology. 2012;74:834–857. doi: 10.1007/s11538-011-9690-0. [DOI] [PubMed] [Google Scholar]
- 65.Miller TL, Wolin MJ. Applied Microbiology. 1974;27:985–987. doi: 10.1128/am.27.5.985-987.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Babauta JT, Nguyen HD, Harrington TD, Renslow R, Beyenal H. Biotechnol. Bioeng. 2012 doi: 10.1002/bit.24538. In Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao B, Majors PD, Ahmed B, Renslow RS, Silvia CP, Shi L, Kjelleberg S, Fredrickson JK, Beyenal H. Environ. Microbiol. 2012;14:2901–2910. doi: 10.1111/j.1462-2920.2012.02850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewandowski Z, Beyenal H. Fundamentals of Biofilm Research. CRC Press; Boca Raton: 2007. [Google Scholar]
- 69.IUPAC . Compendium of Chemical Terminology (the “Gold Book”) 2nd edn. Blackwell Scientific Publications; Oxford: 1997. [Google Scholar]
- 70.Fan LS, Leyva-Ramos R, Wisecarver KD, Zehner BJ. Biotechnol Bioeng. 1990;35:279–286. doi: 10.1002/bit.260350309. [DOI] [PubMed] [Google Scholar]
- 71.Yi HN, Nevin KP, Kim BC, Franks AE, Klimes A, Tender LM, Lovley DR. Biosens. Bioelectron. 2009;24:3498–3503. doi: 10.1016/j.bios.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 72.Nevin KP, Richter H, Covalla SF, Johnson JP, Woodard TL, Orloff AL, Jia H, Zhang M, Lovley DR. Environ. Microbiol. 2008;10:2505–2514. doi: 10.1111/j.1462-2920.2008.01675.x. [DOI] [PubMed] [Google Scholar]
- 73.Babauta JT, Nguyen HD, Beyenal H. Environ Sci Technol. 2011;45:6654–6660. doi: 10.1021/es200865u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McLean JS, Majors PD, Reardon CL, Bilskis CL, Reed SB, Romine MF, Fredrickson JK. Journal of Microbiological Methods. 2008;74:47–56. doi: 10.1016/j.mimet.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 75.Nevin KP, Lovley DR. Geomicrobiol. J. 2002;19:141–159. [Google Scholar]
- 76.Malvankar NS, Tuominen MT, Lovley DR. Energy & Environmental Science. 2012;5:6247–6249. [Google Scholar]
- 77.Strycharz-Glaven SM, Snider RM, Guiseppi-Elie A, Tender LM. Energy & Environmental Science. 2011;4:4366–4379. [Google Scholar]
- 78.Strycharz-Glaven SM, Tender LM. Energy & Environmental Science. 2012;5:6250–6255. [Google Scholar]
- 79.Franks AE, Nevin KP, Jia HF, Izallalen M, Woodard TL, Lovley DR. Energy & Environmental Science. 2009;2:113–119. [Google Scholar]
- 80.Torres CI, Marcus AK, Rittmann BE. Biotechnol. Bioeng. 2008;100:872–881. doi: 10.1002/bit.21821. [DOI] [PubMed] [Google Scholar]
- 81.Brutinel ED, Gralnick JA. Applied Microbiology and Biotechnology. 2012;93:41–48. doi: 10.1007/s00253-011-3653-0. [DOI] [PubMed] [Google Scholar]
- 82.El-Naggar MY, Wanger G, Leung KM, Yuzvinsky TD, Southam G, Yang J, Lau WM, Nealson KH, Gorby YA. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18127–18131. doi: 10.1073/pnas.1004880107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Childers SE, Ciufo S, Lovley DR. Nature. 2002;416:767–769. doi: 10.1038/416767a. [DOI] [PubMed] [Google Scholar]
- 84.Lovley DR, Holmes DE, Nevin KP. In: Advances in Microbial Physiology, Vol. 49. Poole RK, editor. Vol. 49. 2004. pp. 219–286. [DOI] [PubMed] [Google Scholar]
- 85.Agmon N. Chemical Physics Letters. 1995;244:456–462. [Google Scholar]
- 86.Wraight CA. Biochimica Et Biophysica Acta-Bioenergetics. 2006;1757:886–912. doi: 10.1016/j.bbabio.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 87.Müller F. Free Radical Biology and Medicine. 1987;3:215–230. doi: 10.1016/0891-5849(87)90009-8. [DOI] [PubMed] [Google Scholar]
- 88.Gibson QH, Massey V, Atherton NM. Biochem J. 1962;85:369–383. doi: 10.1042/bj0850369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Swinehart JH. Journal of the American Chemical Society. 1966;88:1056–1058. doi: 10.1021/ja00957a037. [DOI] [PubMed] [Google Scholar]
- 90.Bouhenni RA, Vora GJ, Biffinger JC, Shirodkar S, Brockman K, Ray R, Wu P, Johnson BJ, Biddle EM, Marshall MJ, Fitzgerald LA, Little BJ, Beliaev JKAS, Ringeisen BR, Saffarini DA. Electroanalysis. 2010;22:856–864. [Google Scholar]
- 91.Ross DE, Brantley SL, Tien M. Applied and Environmental Microbiology. 2009;75:5218–5226. doi: 10.1128/AEM.00544-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johs A, Shi L, Droubay T, Ankner JF, Liang L. Biophysical Journal. 2010;98:3035–3043. doi: 10.1016/j.bpj.2010.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lower BH, Yongsunthon R, Shi L, Wildling L, Gruber HJ, Wigginton NS, Reardon CL, Pinchuk GE, Droubay TC, Boily J-F, Lower SK. Applied and Environmental Microbiology. 2009;75:2931–2935. doi: 10.1128/AEM.02108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.