Abstract
Observational studies have found an inverse association between type 2 diabetes (T2D) and prostate cancer (PCa), and genome-wide association studies have found common variants near 3 loci associated with both diseases. The authors examined whether a genetic background that favors T2D is associated with risk of advanced PCa. Data from the National Cancer Institute's Breast and Prostate Cancer Cohort Consortium, a genome-wide association study of 2,782 advanced PCa cases and 4,458 controls, were used to evaluate whether individual single nucleotide polymorphisms or aggregations of these 36 T2D susceptibility loci are associated with PCa. Ten T2D markers near 9 loci (NOTCH2, ADCY5, JAZF1, CDKN2A/B, TCF7L2, KCNQ1, MTNR1B, FTO, and HNF1B) were nominally associated with PCa (P < 0.05); the association for single nucleotide polymorphism rs757210 at the HNF1B locus was significant when multiple comparisons were accounted for (adjusted P = 0.001). Genetic risk scores weighted by the T2D log odds ratio and multilocus kernel tests also indicated a significant relation between T2D variants and PCa risk. A mediation analysis of 9,065 PCa cases and 9,526 controls failed to produce evidence that diabetes mediates the association of the HNF1B locus with PCa risk. These data suggest a shared genetic component between T2D and PCa and add to the evidence for an interrelation between these diseases.
Keywords: carcinoma; diabetes mellitus, type 2; genetic predisposition to disease; genetics; genome-wide association study; humans; polymorphism, single nucleotide; prostatic neoplasms
Prostate cancer (PCa) and type 2 diabetes (T2D) are two of the most common chronic diseases afflicting the US aging male population (1, 2). Observational studies have consistently shown an apparent inverse association between T2D and risk of PCa, with meta-analysis risk ratios ranging from 0.84 to 0.91 (3, 4). The reduction in PCa risk has been reported to increase with years since T2D diagnosis, with men who have had T2D for more than 15 years being at a 22% reduced hazard of PCa (5). The association is poorly understood, with one hypothesis suggesting that the metabolic status of men with T2D could move gradually from hyperinsulinemia to endogenous insulin deficiency, which could mitigate the oncogenic action of insulin in the prostate (6, 7).
Recently, 3 shared genomic regions for T2D and PCa have been highlighted. The first region, located on chromosome 17, is in intron 2 of HNF1B, formerly known as TCF2. The major allele A of rs4430796 is positively associated with PCa risk (odds ratio (OR) = 1.22) and inversely associated with risk of T2D (OR = 0.91) (8–10). The second region is located on chromosome 7 near the JAZF1 locus, where the major allele G of rs10486567 is inversely associated with risk of PCa (aggressive PCa: OR = 0.89; nonaggressive PCa: OR = 0.74) (11), whereas the minor allele G of rs864745 is positively associated with T2D (OR = 1.10) (12). THADA is the third region, located on chromosome 2, with the minor allele A of rs1465618 being associated with PCa (OR = 1.08) (13) and the major allele T of rs7578597 associated with T2D (OR = 1.15) (12). However, the single nucleotide polymorphisms (SNPs) for T2D and PCa in the JAZF1 and THADA regions are weakly linked, with R2 values of 0.03 and 0.02, respectively. It is not clear that these associations are driven by the same haplotype (14, 15).
Stevens et al. (16) investigated the T2D-PCa relation further and concluded that diabetic status did not mediate the observed relation between the HNF1B and JAZF1 gene variants and PCa risk. In the Atherosclerosis Risk in Communities cohort, Meyer et al. (17) examined the relation of T2D-associated variants with risk of PCa and found that 4 of 13 T2D SNPs were nominally associated with PCa, which provides additional evidence that some of the T2D-PCa association could be driven by shared genetic factors. Another study by Pierce et al. (18) evaluated the ability of risk scores, consisting of 18 replicated T2D risk variants, to predict PCa risk and concluded that persons with increased genetic susceptibility to T2D have a reduced risk of PCa. However, in a recent study of 5 racial/ethnic groups in the Multiethnic Cohort and PAGE (Population Architecture using Genomics and Epidemiology), Waters et al. (19) found no association between T2D risk variants, either individually or in risk scores, and PCa risk.
With a large sample size and an expanded set of recently published T2D susceptibility loci, we aimed to investigate whether and to what extent individual T2D risk variants and aggregations of T2D replicated risk variants are associated with PCa risk. We used novel approaches to test both whether these risk variants are inversely associated with PCa risk in accordance with the inverse relation observed between T2D and PCa in observational studies and, more generally, whether these T2D loci are associated with PCa risk without regard to directionality of association. Additionally, using causal inference methods, our study attempted to more definitively investigate the potential for mediation of the effect of HNF1B on PCa risk through T2D phenotype.
MATERIALS AND METHODS
Genotyping data for PCa cases and controls came from the National Cancer Institute's Breast and Prostate Cancer Cohort Consortium (BPC3). The BPC3 is a consortium of prospective cohort studies, with contributors including the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (20), the American Cancer Society Cancer Prevention Study II Nutrition Cohort (21), the European Prospective Investigation into Cancer and Nutrition (22), the Health Professionals Follow-up Study, the Melbourne Collaborative Cohort Study (23), the Multiethnic Cohort Study (24), the Physicians' Health Study, and the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (25). In total, 9,065 PCa cases and 9,526 controls comprised the PCa nested case-control study. Diabetes phenotype was self-reported at study baseline, with data available for 96.7% of BPC3 participants. A genome-wide association scan was conducted on a subset of 2,782 European cases with advanced disease and 4,458 controls with European ancestry. Advanced PCa was defined as PCa cases that had either a high histologic grade (Gleason score ≥8) or extraprostatic extension (stage C/D). All controls were free of PCa at the time of selection and were sampled from the same cohort as the cases. Controls were age-matched to cases, and study indicator variables were used to adjust for sampling differences between studies. Informed consent was received from all study participants, and all study protocols were reviewed by the institutional review boards of the National Cancer Institute and each participating study center.
A literature search was conducted to find robustly replicated disease susceptibility loci that are associated with T2D at genome-wide significance levels (P < 5 × 10−8). In total, 36 independent autosomal loci associated with T2D were identified, and published T2D risk alleles and odds ratios were extracted (9, 10, 12, 26–36).
Individual association tests were carried out for each T2D SNP with PCa risk in the BPC3 genome-wide association study (37). Quality control filters were used to remove samples with heterozygosity, underperforming samples or markers, markers with genotype frequencies that significantly departed from Hardy-Weinberg equilibrium, and subjects with significant evidence of non-European ancestry or sample structure. Of the 36 T2D SNPs, 19 were not directly genotyped on the Illumina HumanHap610 Quad Arrays (Illumina, San Diego, California) and were therefore imputed with MACH (http://www.sph.umich.edu/csg/abecasis/MaCH/) (38). MACH references the HapMap (http://hapmap.ncbi.nlm.nih.gov/) CEU population (Utah residents with Northern and Western European ancestry from the Centre d'Etude du Polymorphisme Humain (CEPH) collection) to infer expected genotype counts for each marker locus. MACH quality scores and R2 values were more than 0.85 and 0.75, respectively, for all imputed SNPs. Logistic regression models were used to test for T2D SNP associations with PCa risk. The number of T2D risk alleles was used as the exposure, and adjustment was made for cohort (indicator variables). A nominal association P value of 0.05 was used to assess whether T2D markers exhibited more significant associations with PCa than would be expected by chance. Additional binomial and permutation tests (39) (10,000 permutations) were carried out to test for a relation in risk allele directionality and significant departures of the PCa association statistics from the null distribution, respectively.
The T2D SNPs were combined to form a genetic risk score (GRS) using the --score command in PLINK (40). The GRS was calculated in two ways. The first method, referred to here as the count method, involved summing the number of T2D risk alleles at each locus (0, 1, or 2) and then summing across all T2D loci. This count method is an additive model that weights each locus equally and assumes no gene-gene interactions. The second method, referred to here as the weighted method, uses the log odds ratio of the published T2D loci to weight the sum of T2D risk alleles at each locus and then sums across all T2D loci. The weighted method is an additive model that weights each locus in accordance with the T2D literature and assumes no gene-gene interactions. The rationale for weighting is to create a score that is the best GRS for T2D and therefore can be used as an instrument for testing an association with PCa. For each GRS method, we included the GRS as a predictor in a logistic regression model with PCa case-control status as the outcome, and we adjusted for cohort with an indicator variable. Cohort-specific associations were also calculated.
Additionally, multilocus linear kernel tests were used to assess the joint relation between the 36 T2D variants and PCa risk. These linear models allow associations of multiple genetic loci to be tested simultaneously with one test statistic (41) and have been generalized for dichotomous outcomes (42). Unlike the GRS methods, these tests require no prespecification of risk allele directionality (i.e., that the risk allele is associated with increased risk of T2D and decreased risk of PCa).
The HNF1B locus was the only T2D locus significantly associated with PCa risk after adjustment for multiple comparisons, so it was carried forward for mediation analysis to evaluate whether T2D phenotype is a potential mediator of the relation between HNF1B and PCa. We used an expanded set of data on 9,065 PCa cases (including nonaggressive cases) and 9,526 controls from the BPC3 (43) with self-reported information on diabetes phenotype. Data on rs7501939 at HNF1B were generated as part of a previous project characterizing known PCa loci; this SNP is in high linkage disequilibrium with rs757210 (R2 = 0.81). This was the only T2D risk marker typed in the larger BPC3 data set. To assess mediation, we used the mediation framework proposed by Baron and Kenny (44), extended into the counterfactual framework by VanderWeele and Vansteelandt (45) as direct and indirect effects, and further generalized for use with dichotomous intermediate and outcome. This framework for mediation analysis is flexible to an interaction between exposure and an intermediate factor, has a causal interpretation, and can assess mediation on both the multiplicative and additive scales. Assessing mediation in this manner involved fitting both an outcome model and a mediator model. The outcome model was a logistic regression model that modeled PCa as the outcome, included parameters for the T2D variant of interest and diabetes phenotype, and adjusted for potential confounders of the exposure-outcome and intermediate-outcome relations, including cohort indicator, age at baseline, and body mass index (weight (kg)/height (m)2). The mediator model was a logistic regression model that modeled diabetes phenotype as the outcome, included a parameter for the T2D variant of interest, and controlled for potential confounders, including cohort indicator, age at baseline, and body mass index. In the mediator model, the case-control nature of the BPC3 needed to be accounted for to obtain consistent effect estimates. This was accomplished by fitting the model only in the PCa controls, who represent the study's base population, and assuming a rare outcome. Once both the outcome and mediator models were fitted, parameter estimates were used to calculate direct and indirect (mediated) effects by which to assess mediation (45).
The PCa study was conducted between May and August of 2011. All statistical analyses were carried out in SAS 9.1 (SAS Institute Inc., Cary, North Carolina) and R 2.11.1 (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
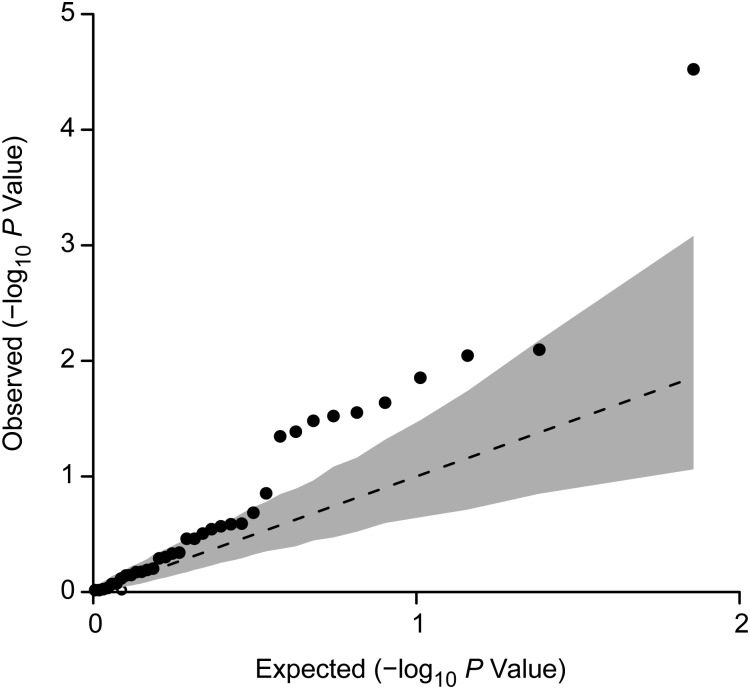
Results from the individual association tests showed that 10 of the 36 T2D markers had a P value less than 0.05 for association with PCa, significantly more than the 1.8 markers that would be expected by chance (P = 7.5 × 10−6) (Table 1). These markers include the HNF1B and JAZF1 loci, as well as NOTCH2, ADCY5, CDKN2A/B, TCF7L2, MTNR1B, FTO, and 2 independent loci at KCNQ1 (Table 1). After permutation adjustment for multiple comparisons, only HNF1B remained significant (adjusted P = 0.001). Small fluctuations in effect estimates of ≤3% were observed when adjustment for diabetes status was made in the models, with overall conclusions remaining the same (results not shown). We observed an inflation in the observed P values for these 36 SNPs (λGC = 2.0; Figure 1). When the observed λGC was compared with the distribution of permutation λGC values, the observed λGC was significantly elevated (P = 0.03), which indicated that the distribution of association P values was significantly lower than expected.
Table 1.
Individual Associations of 36 Independent Type 2 Diabetes Susceptibility Variants With Prostate Cancer Risk in the Breast and Prostate Cancer Cohort Consortiuma
| Chromosome | Reported Gene(s) | Single Nucleotide Polymorphism | Genotyped?b | Type 2 Diabetes Risk Allele | Frequency of Risk Allele | Odds Ratioc | 95% Confidence Interval | P Value | Adjusted P Value |
|---|---|---|---|---|---|---|---|---|---|
| 1 | NOTCH2 | rs10923931 | No | T | 0.11 | 0.86d | 0.76, 0.96 | 0.008* | 0.255 |
| 1 | PROX1 | rs340874 | Yes | C | 0.52 | 1.01 | 1.08, 0.94 | 0.845 | 1.000 |
| 2 | GCKR | rs780094 | Yes | C | 0.61 | 0.98d | 1.05, 0.91 | 0.498 | 1.000 |
| 2 | THADA | rs7578597 | Yes | T | 0.91 | 1.03 | 1.16, 0.91 | 0.644 | 1.000 |
| 2 | BCL11A | rs243021 | Yes | A | 0.47 | 1.02 | 0.95, 1.10 | 0.511 | 1.000 |
| 2 | IRS1 | rs2943641 | Yes | C | 0.64 | 0.95d | 1.02, 0.88 | 0.140 | 0.995 |
| 3 | PPARG | rs1801282 | No | C | 0.86 | 0.96d | 1.07, 0.87 | 0.465 | 1.000 |
| 3 | ADAMTS9 | rs4607103 | No | C | 0.76 | 0.99d | 1.08, 0.91 | 0.853 | 1.000 |
| 3 | ADCY5 | rs11708067 | No | A | 0.78 | 0.91d | 0.99, 0.84 | 0.028* | 0.630 |
| 3 | IGF2BP2 | rs4402960 | Yes | T | 0.32 | 1.03 | 0.95, 1.11 | 0.456 | 1.000 |
| 4 | WFS1 | rs10010131 | No | G | 0.60 | 1.00 | 1.07, 0.93 | 0.924 | 1.000 |
| 5 | ZBED3 | rs4457053 | No | G | 0.29 | 1.02 | 0.94, 1.10 | 0.672 | 1.000 |
| 6 | CDKAL1 | rs7754840 | Yes | C | 0.32 | 1.04 | 0.97, 1.13 | 0.270 | 1.000 |
| 7 | DGKB | rs2191349 | No | T | 0.52 | 1.00 | 1.07, 0.93 | 0.945 | 1.000 |
| 7 | JAZF1 | rs864745 | No | T | 0.50 | 1.08 | 1.16, 1.01 | 0.033* | 0.694 |
| 7 | GCK | rs4607517 | Yes | A | 0.15 | 1.06 | 0.96, 1.16 | 0.256 | 1.000 |
| 7 | KLF14 | rs972283 | No | G | 0.53 | 1.02 | 1.09, 0.95 | 0.627 | 1.000 |
| 8 | TP53INP1 | rs896854 | Yes | T | 0.51 | 1.02 | 1.09, 0.95 | 0.668 | 1.000 |
| 8 | SLC30A8 | rs13266634 | Yes | C | 0.68 | 1.00 | 1.08, 0.93 | 0.963 | 1.000 |
| 9 | CDKN2A/B | rs10811661 | No | T | 0.82 | 0.91d | 1.00, 0.83 | 0.045* | 0.809 |
| 9 | TLE4 | rs13292136 | No | C | 0.93 | 0.93d | 1.07, 0.81 | 0.312 | 1.000 |
| 10 | CDC123/CAMK1D | rs12779790 | No | G | 0.18 | 1.06 | 0.97, 1.16 | 0.206 | 1.000 |
| 10 | HHEX/IDE | rs1111875 | Yes | C | 0.58 | 1.01 | 1.09, 0.94 | 0.713 | 1.000 |
| 10 | TCF7L2 | rs7903146 | Yes | T | 0.28 | 0.90d | 0.83, 0.97 | 0.009* | 0.276 |
| 11 | KCNQ1 | rs231362 | No | G | 0.50 | 0.92d | 0.86, 0.98 | 0.014* | 0.393 |
| 11 | KCNQ1 | rs2237892 | Yes | C | 0.94 | 0.85d | 0.98, 0.74 | 0.030* | 0.659 |
| 11 | KCNJ11 | rs5215 | Yes | T | 0.61 | 0.99d | 1.06, 0.92 | 0.719 | 1.000 |
| 11 | CENTD2 | rs1552224 | Yes | A | 0.83 | 1.00 | 1.10, 0.91 | 0.963 | 1.000 |
| 11 | MTNR1B | rs10830963 | No | G | 0.28 | 1.10 | 1.01, 1.19 | 0.023* | 0.561 |
| 12 | HMGA2 | rs1531343 | No | C | 0.10 | 0.98d | 0.88, 1.10 | 0.764 | 1.000 |
| 12 | TSPAN8/LGR5 | rs7961581 | No | C | 0.26 | 1.05 | 0.97, 1.13 | 0.259 | 1.000 |
| 12 | HNF1A/TCF1 | rs7957197 | No | T | 0.80 | 0.96d | 1.05, 0.88 | 0.346 | 1.000 |
| 15 | ZFAND6 | rs11634397 | No | G | 0.66 | 1.04 | 1.12, 0.96 | 0.346 | 1.000 |
| 15 | PRC1 | rs8042680 | Yes | A | 0.32 | 1.04 | 0.97, 1.12 | 0.286 | 1.000 |
| 16 | FTO | rs9939609 | No | A | 0.40 | 0.93d | 0.86, 1.00 | 0.041* | 0.775 |
| 17 | HNF1B/TCF2 | rs757210 | Yes | T | 0.35 | 0.85d | 0.79, 0.92 | 3e−05* | 0.001e |
Abbreviations: CI, confidence interval; OR, odds ratio; T2D, type 2 diabetes; RA, risk allele; SNP, single nucleotide polymorphism.
* P < 0.05.
a Association tests were carried out in the Breast and Prostate Cancer Cohort Consortium using a log-additive genetic model with adjustment made for cohort indicators.
b Indicates whether or not variants were genotyped. Variants that were not directly genotyped were imputed.
c Odds ratio for the increase in prostate cancer risk associated with a 1-unit increase in the number of type 2 diabetes risk alleles carried at each locus.
d Association for prostate cancer was in the inverse direction.
e Significant after permutation correction for multiple testing.
Figure 1.
Quantile-quantile plot comparing the uniformly distributed −log10 P values for the 36 type 2 diabetes (T2D) susceptibility markers with −log10 P values observed in the Breast and Prostate Cancer Cohort Consortium data set when the authors tested for an association with prostate cancer (PCa) risk by means of a Wald test. The dotted line shows the expected −log10 P value distribution. The black points represent observed P values for the association of each T2D locus with PCa risk. The gray region is the 95% confidence interval for 10,000 permutations. The inflation index (λGC) of 1.95 is significantly elevated (P = 0.02), which indicates an overall inflation in association P values but gives no information about the directionality of association between the T2D variants and PCa risk.
We used exact binomial tests to assess whether significantly more T2D risk alleles were inversely associated with PCa risk than would be expected by chance. By chance alone, 1.8 of the 36 markers would be expected to be significant, of which, under the null, 0.9 would be expected to be significantly associated with increased risk of PCa and 0.9 would be expected to be significantly associated with decreased risk of PCa. In our data, we observed 2 T2D loci that were significantly associated with increased PCa risk, which did not differ statistically from the 0.9 loci expected by chance (P = 0.23). However, the 8 T2D loci we observed to be significantly associated with reduced risk of PCa were significantly more than the 0.9 that would be expected by chance (P = 2.45 × 10−6), which indicates that more T2D risk alleles than expected are associated with reduced risk of PCa.
Associations for GRS using both the unweighted count and the weighted log odds method are shown in Table 2. The risk score for the unweighted count did not show evidence for an association of these genetic variants with PCa risk. However, a significant association was observed for the weighted log odds method when HNF1B was both included in (P = 0.002) and excluded from (P = 0.015) the GRS. No changes in results were observed when we adjusted for diabetes status in the models (results not shown). Study-specific analyses showed that the log odds-weighted GRS was statistically significant only in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial, although the test for heterogeneity indicated no significant departures from homogeneity (P = 0.60).
Table 2.
Individual Cohort and Combined Results for Unweighted and Log Odds Ratio-Weighted Type 2 Diabetes Genetic Risk Score in the Breast and Prostate Cancer Cohort Consortiuma
| Cohort | No. in Cohort | No. of Cases | Totalb | Meanc |
GRS |
GRS (-HNF1B)d |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR | 95% CI | P Value | OR | 95% CI | P Value | ||||
| Unweighted count | |||||||||||
| ATBC | 1,490 | 245 | 72 | 36.48 | 36.44 | 1.00 | 0.97, 1.04 | 0.894 | 1.00 | 0.97, 1.04 | 0.841 |
| CPSII | 1,258 | 636 | 72 | 37.48 | 37.55 | 1.00 | 0.97, 1.03 | 0.740 | 1.00 | 0.97, 1.03 | 0.839 |
| EPIC | 857 | 431 | 72 | 37.47 | 37.66 | 0.99 | 0.95, 1.02 | 0.460 | 1.00 | 0.96, 1.04 | 0.984 |
| HPFS | 418 | 214 | 72 | 37.70 | 37.47 | 1.02 | 0.97, 1.07 | 0.539 | 1.02 | 0.97, 1.08 | 0.419 |
| MEC | 503 | 244 | 72 | 37.80 | 37.89 | 0.99 | 0.95, 1.04 | 0.779 | 1.00 | 0.96, 1.05 | 0.936 |
| PHS | 553 | 298 | 72 | 37.59 | 37.81 | 0.99 | 0.95, 1.03 | 0.521 | 1.00 | 0.95, 1.04 | 0.800 |
| PLCO | 2,161 | 714 | 72 | 37.36 | 37.64 | 0.98 | 0.96, 1.00 | 0.111 | 0.98 | 0.96, 1.01 | 0.191 |
| Combinede | 7,240 | 2,782 | 72 | 37.42 | 37.31 | 0.99 | 0.98, 1.00 | 0.168 | 1.00 | 0.98, 1.01 | 0.534 |
| Weighted log OR | |||||||||||
| ATBC | 1,490 | 245 | 8.16 | 4.33 | 4.34 | 0.93 | 0.68, 1.29 | 0.675 | 0.94 | 0.68, 1.30 | 0.718 |
| CPSII | 1,258 | 636 | 8.16 | 4.45 | 4.47 | 0.89 | 0.69, 1.14 | 0.358 | 0.90 | 0.70, 1.16 | 0.416 |
| EPIC | 857 | 431 | 8.16 | 4.45 | 4.47 | 0.90 | 0.67, 1.20 | 0.460 | 1.01 | 0.75, 1.36 | 0.961 |
| HPFS | 418 | 214 | 8.16 | 4.49 | 4.46 | 1.11 | 0.73, 1.68 | 0.635 | 1.17 | 0.76, 1.80 | 0.481 |
| MEC | 503 | 244 | 8.16 | 4.49 | 4.54 | 0.78 | 0.53, 1.15 | 0.215 | 0.83 | 0.56, 1.23 | 0.352 |
| PHS | 553 | 298 | 8.16 | 4.45 | 4.52 | 0.76 | 0.53, 1.07 | 0.118 | 0.80 | 0.56, 1.15 | 0.232 |
| PLCO | 2,161 | 714 | 8.16 | 4.43 | 4.49 | 0.74 | 0.61, 0.91 | 0.004 | 0.76 | 0.62, 0.93 | 0.008 |
| Combinede | 7,240 | 2,782 | 8.16 | 4.44 | 4.45 | 0.84 | 0.75, 0.94 | 0.002 | 0.87 | 0.78, 0.97 | 0.015 |
Abbreviations: ATBC, Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study; CI, confidence interval; CPSII, American Cancer Society Cancer Prevention Study II Nutrition Cohort; EPIC, European Prospective Investigation into Cancer and Nutrition; GRS, genetic risk score; HPFS, Health Professionals Follow-up Study; MEC, Multiethnic Cohort Study; OR, odds ratio; PCa, prostate cancer; PHS, Physicians' Health Study; PLCO, Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial; T2D, type 2 diabetes.
a Logistic regression models were used to regress GRS on risk of PCa.
b Total indicates the maximum bound for the respective GRS, with a value close to this total indicating high genetic predisposition for T2D.
c Mean GRS was calculated for PCa cases and PCa controls.
d HNF1B was excluded from the GRS and included as a separate covariate.
e For combined estimates, cohort indicators were added to adjust for cohort effects.
The multilocus kernel test that jointly tested for a PCa association with all 36 T2D loci without specifying weight or directionality of risk alleles was statistically significant (P = 0.0001). When HNF1B was removed from the list of included markers and the remaining 35 markers were fitted, the P value was attenuated but remained significant (P = 0.01), which indicated that a substantial portion of the association was a result of the HNF1B locus but that other T2D loci were associated with PCa as well.
We conducted mediation analyses for the HNF1B locus to investigate whether the locus had effects that act directly on PCa risk or whether the effects of the locus were mediated through diabetes phenotype (Table 3). The outcome model produced significant evidence for an association between HNF1B and PCa risk (OR = 0.83, 95% confidence interval (CI): 0.79, 0.86; P = 6.37 × 10−19) and an association between diabetes phenotype and PCa risk (OR = 0.76, 95% CI: 0.66, 0.87; P = 8.13 × 10−5). The mediator model indicated that the minor T allele of rs7501939 was not statistically significantly associated with an increased risk of diabetes among the 9,526 PCa controls (OR = 1.10, 95% CI: 0.97, 1.25; P = 0.14), although the per-allele odds ratio for association with T2D was consistent with previous reports (8–10). When these results were combined together, the estimated direct effect of HNF1B on PCa risk was statistically significant (OR = 0.83, 95% CI: 0.79, 0.86; P = 1.02 × 10−18), but the mediated (indirect) effect through diabetes phenotype was nonsignificant (OR = 1.00, 95% CI: 1.00, 1.00; P = 0.71). These results are in agreement with the standard mediation analysis, which produced an insignificant 0.5% change in the parameter estimate for the effect of HNF1B when diabetes status was included as a covariate.
Table 3.
Mediation Analysis for the Association Between HNF1B (rs7501939) and Prostate Cancer With Diabetes Phenotype as a Potential Intermediate in the Breast and Prostate Cancer Cohort Consortiuma
| Odds Ratio | 95% Confidence Interval | P Value | |
|---|---|---|---|
| HNF1B-T2D association | 1.10 | 0.97, 1.25 | 0.14 |
| T2D-prostate cancer association | 0.76 | 0.66, 0.87 | 8.13 × 10−05 |
| Natural indirect effect | 1.00 | 1.00, 1.00 | 0.71 |
| Natural direct effect | 0.83 | 0.79, 0.86 | 1.02 × 10−18 |
| Total effect | 0.83 | 0.79, 0.86 | 6.37 × 10−19 |
Abbreviation: T2D, type 2 diabetes.
a All analyses were conducted in the Breast and Prostate Cancer Cohort Consortium and were adjusted for cohort indicator, age at baseline (years), and body mass index (weight (kg)/height (m)2).
DISCUSSION
Our study suggests that genetic variants associated with T2D are also associated with PCa risk. Ten of 36 T2D susceptibility markers were nominally associated with PCa risk at NOTCH2, ADCY5, JAZF1, CDKN2A/B, TCF7L2, KCNQ1, MTNR1B, FTO, and HNF1B, although only the HNF1B locus remained significantly associated with PCa risk after adjustment for multiple testing. However, log odds ratio-weighted GRS and kernel machine models also were associated with PCa risk both with and without inclusion of the HNF1B locus, which suggests that other genetic variants associated with T2D risk also contribute to PCa risk. Finally, mediation analysis provided insufficient evidence that the association of the HNF1B locus with PCa risk is mediated through diabetes phenotype.
Our study adds to the evidence that a genetic background favorable to the development of T2D is associated with PCa risk. The HNF1B locus was most strongly associated with PCa risk in this analysis and accounted for some but not all of the association between the T2D variants and PCa risk in the GRS and the kernel regression. The noted inflation in our association P values for other T2D SNPs is consistent with what others have observed (17, 18) and indicates that more germline variants are held in common between T2D and PCa than would be expected by chance.
Our study's large sample size and recently published T2D susceptibility loci permitted us to detect potentially novel genetic relations between T2D and PCa that have not been reported previously. Seven loci (NOTCH2, ADCY5, CDKN2A/B, TCF7L2, KCNQ1, MTNR1B, and FTO) not previously associated with PCa at genome-wide significance levels were seen as nominally associated in our study, one of which (FTO) was also reported by Pierce et al. (18). Four of these loci (CDKN2A/B, TCF7L2, KCNQ1, and MTNR1B) are associated with altered beta cell dysfunction or impaired insulin release and could result in less insulin production, thus blunting insulin effects in increasing PCa risk (46). Additionally, our second most highly associated locus, the NOTCH2 locus (P = 0.008; permutation P = 0.26), is of interest. NOTCH2 is a member of the NOTCH family of receptors, which modulate cellular differentiation, proliferation, and apoptosis (47). The locus has been reported to be associated with both T2D and breast cancer (48, 49). Evidence from gene expression data indicates that NOTCH2 is expressed in developing prostate stroma and that NOTCH signaling affects stromal survival only in the presence of testosterone (50). Therefore, the regulatory ability of NOTCH2 and its sensitivity to the presence of testosterone might be important in prostate carcinogenesis, although additional studies are needed to investigate this further.
Our use of GRS and kernel machine models allowed us to investigate the cumulative effect of T2D susceptibility variants on PCa risk. Although another study was successful in showing an association between unweighted T2D GRS and PCa (18), our study did not find a relation between unweighted T2D risk scores and PCa. A potential explanation for our lack of association is that with the most recent T2D loci added to our risk score, including T2D variants found through meta-analyses with lower-than-average effect sizes, the number of SNPs doubled, and the range of effect estimates for each variant might have widened. Our study did find a significant association between the log odds-weighted T2D risk scores and PCa. This association was significant when the HNF1B locus was both included in and excluded from the GRS. Although one of the larger cohorts, the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial, seems to have been responsible for most of this association, a test of heterogeneity indicated that there was no significant evidence for heterogeneity. The fact that the log odds ratio-weighted GRS was significant and the unweighted risk score was insignificant indicates that some T2D variants could have a stronger influence on PCa risk than others. The GRS approach makes the assumption that all T2D loci included in the GRS have T2D risk alleles that function in the same direction when PCa risk is considered. This might not be the case, with some T2D-associated loci possibly having the same rather than the (expected) opposite direction of effect on PCa. Multilocus kernel tests allowed us to assess the cumulative effect of these 36 T2D variants on PCa risk without requiring an assumption about risk allele directionality. Results from the multilocus kernel tests indicated that the 36 T2D variants were significantly associated with PCa risk when HNF1B was both included in and excluded from the models, which suggests that common pathways could be involved in both T2D and PCa.
A potential limitation of this study is that information on diabetes phenotype was self-reported (43). However, previous studies have shown that self-reporting of diabetes has up to 97% agreement with medical records (51, 52). Another limitation is that we could not differentiate between cases of type 1 diabetes and T2D, although the median age (62 years; interquartile range, 55–70) and ethnicity of our study population were such that the majority of diabetes cases were likely to be T2D (53). Furthermore, BPC3 data on T2D status were available only at baseline, and although this could have resulted in underestimation of the true prevalence of diabetes in our study population, it did guard against potential reverse causality.
Our study showed a highly significant inverse relation between T2D and PCa. The estimate was adjusted for body mass index, age at baseline, and cohort indicator and is unlikely to be due to chance or uncontrolled bias. To our knowledge, this is the largest case-control study in which this inverse association has been examined, and our estimate (OR = 0.76) is comparable to, albeit slightly stronger than, the point estimates reported in meta-analyses and other studies, including prior reports from 2 cohorts in the BPC3 (i.e., relative risks ranged from 0.84 to 0.91) (3–5, 54).
We further assessed the potential for T2D phenotype to mediate the effect of HNF1B with PCa risk. Results indicated a highly significant direct association between HNF1B and PCa risk, but there was no significant evidence for an indirect association. Although other investigators have observed a significant relation between HNF1B and T2D risk (8, 9), we did not, which indicates that our sample set might have lacked sufficient statistical power to detect this effect. The lack of a mediation role for diabetes phenotype in the HNF1B-PCa association has been reported elsewhere in a smaller subset of the BPC3 data (16), although larger studies are needed to more definitively rule out the potential for mediation.
The majority of our analysis, excluding the mediation analysis, was conducted on data from a genome-wide association study of advanced PCa. Although there is concern that results from our study might not be generalizable to other subtypes of PCa, the overwhelming number of similarities between our analysis and others indicates that T2D risk variants have a similar effect on advanced PCa risk and on total PCa risk. This is in agreement with association studies comparing PCa germline variants that show very few examples of different effects by disease aggressiveness.
In conclusion, our data provide additional evidence for a relation between T2D and PCa. Current investigations of a shared genetic background that could underlie this observed association are still in their infancy but suggest that a genetic predisposition to T2D might also be associated with PCa risk. Future studies should further investigate the potential genetic factors that link these two common chronic diseases.
ACKNOWLEDGMENTS
Author affiliations: Program in Molecular and Genetic Epidemiology, Department of Epidemiology, Harvard School of Public Health, Boston, Massachusetts (Mitchell J. Machiela, Sara Lindström, David J. Hunter, Peter Kraft); Cancer Epidemiology Unit, Nuffield Department of Clinical Medicine, University of Oxford, Oxford, United Kingdom (Naomi E. Allen, Ruth Travis); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California (Christopher A. Haiman, Brian E. Henderson, Daniel O. Stram); Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, Maryland (Demetrius Albanes, Sonja I. Berndt, Stephen Chanock); Navarre Public Health Institute, Pamplona, Spain (Aurelio Barricarte); National Institute for Public Health and the Environment, Bilthoven, the Netherlands (H. Bas Bueno-de-Mesquita); Department of Gastroenterology and Hepatology, University Medical Centre Utrecht, Utrecht, the Netherlands (H. Bas Bueno-de-Mesquita); Division of Aging, Brigham and Women's Hospital, Boston, Massachusetts (J. Michael Gaziano); Massachusetts Veterans Epidemiology Research and Information Center, VA Boston Healthcare System, Boston, Massachusetts (J. Michael Gaziano); Epidemiology Research Program, American Cancer Society, Atlanta, Georgia (Susan M. Gapstur, Eric J. Jacobs, Victoria L. Stevens); Department of Nutrition, Harvard School of Public Health, Boston, Massachusetts (Edward Giovannucci, Meir J. Stampfer, Walter C. Willett); Department of Epidemiology, Harvard School of Public Health, Boston, Massachusetts (Edward Giovannucci, Meir J. Stampfer, Walter C. Willett); University of Hawaii Cancer Center, Honolulu, Hawaii (Laurence N. Kolonel, Loic Le Marchand); Fondazione IRCCS [Istituto Di Ricovero e Cura a Carattere Scientifico], Istituto Nazionale dei Tumori, Milan, Italy (Vittorio Krogh); Channing Laboratory, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts (Jing Ma, Meir J. Stampfer); and Institute of Cancer Epidemiology, Danish Cancer Society, Copenhagen, Denmark (Anne Tjønneland).
This work was supported by the National Cancer Institute (cooperative agreements U01-CA98233-07 with David J. Hunter, U01-CA98710-06 with Susan M. Gapstur, U01-CA98216-06 with Elio Riboli and Rudolf Kaaks, and U01-CA98758-07 with Brian E. Henderson and the Intramural Research Program of the National Institutes of Health/National Cancer Institute, Division of Cancer Epidemiology and Genetics), as well as National Institutes of Health grants T32-CA09001 and T32-GM074897.
Conflict of interest: none declared.
REFERENCES
- 1.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Mokdad AH, Ford ES, Bowman BA, et al. Diabetes trends in the U.S.: 1990–1998. Diabetes Care. 2000;23(9):1278–1283. doi: 10.2337/diacare.23.9.1278. [DOI] [PubMed] [Google Scholar]
- 3.Kasper JS, Giovannucci E. A meta-analysis of diabetes mellitus and the risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15(11):2056–2062. doi: 10.1158/1055-9965.EPI-06-0410. [DOI] [PubMed] [Google Scholar]
- 4.Bonovas S, Filioussi K, Tsantes A. Diabetes mellitus and risk of prostate cancer: a meta-analysis. Diabetologia. 2004;47(6):1071–1078. doi: 10.1007/s00125-004-1415-6. [DOI] [PubMed] [Google Scholar]
- 5.Kasper JS, Liu Y, Giovannucci E. Diabetes mellitus and risk of prostate cancer in the Health Professionals Follow-Up Study. Int J Cancer. 2009;124(6):1398–1403. doi: 10.1002/ijc.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaaks R, Stattin P. Obesity, endogenous hormone metabolism, and prostate cancer risk: a conundrum of “highs” and “lows.”. Cancer Prev Res (Phila) 2010;3(3):259–262. doi: 10.1158/1940-6207.CAPR-10-0014. [DOI] [PubMed] [Google Scholar]
- 7.Giovannucci E, Michaud D. The role of obesity and related metabolic disturbances in cancers of the colon, prostate, and pancreas. Gastroenterology. 2007;132(6):2208–2225. doi: 10.1053/j.gastro.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 8.Winckler W, Weedon MN, Graham RR, et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56(3):685–693. doi: 10.2337/db06-0202. [DOI] [PubMed] [Google Scholar]
- 9.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39(8):977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 10.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8(9):657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 11.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 12.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Wellcome Trust Case Control Consortium. Nat Genet. 2008;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eeles RA, Kote-Jarai Z, Al Olama AA, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41(10):1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson AD, Handsaker RE, Pulit SL, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prokunina-Olsson L, Fu YP, Tang W, et al. Refining the prostate cancer genetic association within the JAZF1 gene on chromosome 7p15.2. Cancer Epidemiol Biomarkers Prev. 2010;19(5):1349–1355. doi: 10.1158/1055-9965.EPI-09-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens VL, Ahn J, Sun J, et al. HNF1B and JAZF1 genes, diabetes, and prostate cancer risk. Prostate. 2010;70(6):601–607. doi: 10.1002/pros.21094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer TE, Boerwinkle E, Morrison AC, et al. Diabetes genes and prostate cancer in the Atherosclerosis Risk in Communities study. Cancer Epidemiol Biomarkers Prev. 2010;19(2):558–565. doi: 10.1158/1055-9965.EPI-09-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierce BL, Ahsan H. Genetic susceptibility to type 2 diabetes is associated with reduced prostate cancer risk. Hum Hered. 2010;69(3):193–201. doi: 10.1159/000289594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waters KM, Wilkens LR, Monroe KR, et al. No association of type 2 diabetes risk variants and prostate cancer risk: the Multiethnic Cohort and PAGE. Cancer Epidemiol Biomarkers Prev. 2011;20(9):1979–1981. doi: 10.1158/1055-9965.EPI-11-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ATBC Cancer Prevention Study Group. The Alpha-Tocopherol, Beta-Carotene Lung Cancer Prevention Study: design, methods, participant characteristics, and compliance. Ann Epidemiol. 1994;4(1):1–10. doi: 10.1016/1047-2797(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 21.Calle EE, Rodriguez C, Jacobs EJ, et al. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer. 2002;94(9):2490–2501. doi: 10.1002/cncr.101970. [DOI] [PubMed] [Google Scholar]
- 22.Riboli E, Hunt KJ, Slimani N, et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5(6B):1113–1124. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 23.Severi G, Morris HA, MacInnis RJ, et al. Circulating insulin-like growth factor-I and binding protein-3 and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15(6):1137–1141. doi: 10.1158/1055-9965.EPI-05-0823. [DOI] [PubMed] [Google Scholar]
- 24.Kolonel LN, Henderson BE, Hankin JH, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151(4):346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prorok PC, Andriole GL, Bresalier RS, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials. 2000;21(6):273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 26.Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26(1):76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 27.Gloyn AL, Weedon MN, Owen KR, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52(2):568–572. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 28.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 29.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 30.Sandhu MS, Weedon MN, Fawcett KA, et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet. 2007;39(8):951–953. doi: 10.1038/ng2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 32.Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40(9):1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 33.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41(10):1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 34.Prokopenko I, Langenberg C, Florez JC, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41(1):77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumacher FR, Berndt SI, Siddiq A, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20(19):3867–3875. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Willer CJ, Ding J, et al. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westfall PH, Zaykin KV, Young SS. Multiple tests for genetic effects in association studies. In: Looney SW, editor. Methods in Molecular Biology. Totowa, NJ: Humana Press, Inc; 2002. pp. 143–168. [DOI] [PubMed] [Google Scholar]
- 40.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwee LC, Liu D, Lin X, et al. A powerful and flexible multilocus association test for quantitative traits. Am J Hum Genet. 2008;82(2):386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu MC, Kraft P, Epstein MP, et al. Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet. 2010;86(6):929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindstrom S, Schumacher F, Siddiq A, et al. Characterizing associations and SNP-environment interactions for GWAS-identified prostate cancer risk markers—results from BPC3. PLoS One. 2011;6(2):e17142. doi: 10.1371/journal.pone.0017142. doi:10.1371/journal.pone.0017142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51(6):1173–1182. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 45.Vanderweele TJ, Vansteelandt S. Odds ratios for mediation analysis for a dichotomous outcome. Am J Epidemiol. 2010;172(12):1339–1348. doi: 10.1093/aje/kwq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grarup N, Sparsø T, Hansen T. Physiologic characterization of type 2 diabetes-related loci. Curr Diab Rep. 2010;10(6):485–497. doi: 10.1007/s11892-010-0154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 48.Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41(5):579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu YP, Edvardsen H, Kaushiva A, et al. NOTCH2 in breast cancer: association of SNP rs11249433 with gene expression in ER-positive breast tumors without TP53 mutations. Mol Cancer. 2010;9:113. doi: 10.1186/1476-4598-9-113. doi:10.1186/1476-4598-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orr B, Grace OC, Vanpoucke G, et al. A role for notch signaling in stromal survival and differentiation during prostate development. Endocrinology. 2009;150(1):463–472. doi: 10.1210/en.2008-0383. [DOI] [PubMed] [Google Scholar]
- 51.Midthjell K, Holmen J, Bjørndal A, et al. Is questionnaire information valid in the study of a chronic disease such as diabetes? The Nord-Trøndelag diabetes study. J Epidemiol Community Health. 1992;46(5):537–542. doi: 10.1136/jech.46.5.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okura Y, Urban LH, Mahoney DW, et al. Agreement between self-report questionnaires and medical record data was substantial for diabetes, hypertension, myocardial infarction and stroke but not for heart failure. J Clin Epidemiol. 2004;57(10):1096–1103. doi: 10.1016/j.jclinepi.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 53.Centers for Disease Control and Prevention. National Diabetes Fact Sheet: General Information and National Estimates on Diabetes in the United States, 2007. Atlanta, GA: Centers for Disease Control and Prevention; 2008. [Google Scholar]
- 54.Waters KM, Henderson BE, Stram DO, et al. Association of diabetes with prostate cancer risk in the Multiethnic Cohort. Am J Epidemiol. 2009;169(8):937–945. doi: 10.1093/aje/kwp003. [DOI] [PMC free article] [PubMed] [Google Scholar]