Abstract
Rates of uterine leiomyomata (UL) are 2–3 times higher in African Americans than in European Americans. It is unclear whether inherited factors explain the ethnic disparity. To investigate the presence of risk alleles for UL that are highly differentiated in frequency between African Americans and European Americans, the authors conducted an admixture-based genome-wide scan of 2,453 UL cases confirmed by ultrasound or surgery in the Black Women's Health Study (1997–2009), a national prospective cohort study. Controls (n = 2,102) were women who did not report a UL diagnosis through 2009. Mean percentage of European ancestry was significantly lower among cases (20.00%) than among controls (21.63%; age-adjusted mean difference = −1.76%, 95% confidence interval: −2.40, −1.12; P < 0.0001), and the association was stronger in younger cases. Admixture analyses showed suggestive evidence of association at chromosomes 2, 4, and 10. The authors also genotyped a dense set of tag single nucleotide polymorphisms at different loci associated with UL in Japanese women but failed to replicate the associations. This suggests that genetic variation for UL differs in populations with and without African ancestry. The admixture findings further indicate that no single highly differentiated locus is responsible for the ethnic disparity in UL, raising the possibility that multiple variants jointly contribute to the higher incidence of UL in African Americans.
Keywords: African Americans, African continental ancestry group, European continental ancestry group, female, genetics, leiomyoma, prospective studies, uterine neoplasms
Uterine leiomyomata (UL), or fibroids, are benign neoplasms arising from smooth muscle cells of the myometrium and are clinically recognized in approximately 30% of reproductive-age women (1–3). Studies consistently document a 2- to 3-fold higher incidence of UL in African Americans than in European Americans (4, 5). African Americans tend to have younger ages at diagnosis, more tumors, and greater symptomatology than European Americans (6). Given that identified environmental risk factors do not explain the discrepancy in UL rates (4, 6–8), a role for inherited factors has been proposed (9, 10).
Evidence for the existence of UL susceptibility genes comes from familial aggregation studies (11–14), twin studies (15, 16), genetic linkage studies in families with UL-associated syndromes (17–19), and a genome-wide association study (GWAS) (20). Having several affected first-degree relatives increases UL risk (11, 21). Twin studies show a strong element of heritability in women undergoing hysterectomy for UL (15, 16). Larger-scale linkage studies based on sibling-pair analysis (22) have the ability to find common genetic variants of strong effect (>3- to 4-fold increased risk per copy) that contribute to UL risk (23). However, linkage studies—even those with large sample sizes—have low power to find genetic variants of weaker effect (<2.5-fold increased risk per copy) that may contribute to risk of many common diseases.
Admixture mapping involves scanning the genome in persons of mixed ancestry to identify regions where the proportion of a particular ancestry in cases is strikingly higher or lower than that seen elsewhere in their genomes. Such a region would indicate the presence of at least 1 genetic risk variant for the phenotype whose frequency differs between the ancestral populations. African-American populations offer the unique opportunity to perform admixture mapping because of the recent mixture between African and European populations (6 generations ago, on average (24)). Because the blocks of contiguous European or African ancestry have not had much time to break up due to recombination, admixture-generated linkage disequilibrium extends many millions of base pairs around phenotypically important genes. Therefore, one only needs to screen 1 marker every few million base pairs with admixture mapping to identify with high reliability regions of African and European ancestry, rather than the hundreds of thousands to a million markers required for GWAS.
We carried out an admixture-based genome-wide scan to search for UL risk alleles that are highly differentiated in frequency between African-American and European-American women. We analyzed DNA samples from 2,453 premenopausal women with incident UL diagnosed during 1997–2009 (cases) and 2,102 premenopausal women not diagnosed with UL during the same time period (controls) in the Black Women's Health Study, a prospective cohort study. We also sought to replicate results from a recent GWAS in Japanese women (20) that identified 3 single nucleotide polymorphisms (SNPs) on chromosomes 10q24.33, 22q13.1, and 11p15.5 associated with UL.
MATERIALS AND METHODS
Study population
The Black Women's Health Study is an ongoing prospective cohort study of 59,000 US women who self-identify as “black” (25). The study began in 1995 when women aged 21–69 years from across the United States completed a 14-page postal health questionnaire. Follow-up questionnaires have been mailed to participants every 2 years, and cohort retention exceeded 80% through 2009. During 2004–2007, we obtained saliva samples as a source of DNA from 26,814 participants using the mouthwash-swish method (26). Participants in the DNA substudy were slightly older than nonparticipants (49.7 years vs. 47.7 years) but were similar with respect to education (≤12 years: 18% vs. 19%), region (Northeast: 27% vs. 28%; South: 31% vs. 30%; Midwest: 24% vs. 23%; West: 18% vs. 19%), body mass index (weight (kg)/height (m)2; 28.1 vs. 27.8), and family history of UL (38.5% vs. 36.5%). The present analysis included 4,555 premenopausal women who were aged 23–50 years in 1997. The study protocol was approved by the Institutional Review Board of Boston University Medical Center (Boston, Massachusetts).
Assessment of UL
Ultrasonography is the standard method used to confirm UL diagnoses clinically (3), and it has high sensitivity (99%) and specificity (91%) relative to histologic evidence (27, 28). Every 2 years, beginning in 1999, women reported whether they had been diagnosed with “uterine fibroids,” the calendar year of first diagnosis, and whether their diagnosis was confirmed by ultrasound or surgery. Analyses were restricted to premenopausal women because UL are rare after menopause (3). We assessed the accuracy of self-reporting in a random sample of 248 incident cases and confirmed the diagnosis for 96% (122/127) by medical record (29). There were no systematic differences in demographic, lifestyle, or reproductive factors between cases who did and did not release their medical records, indicating that the validated group was representative of the larger case group (29).
Assessment of covariates
Baseline and biennial follow-up questionnaires collected data on reproductive, contraceptive, and medical history, height, current weight, smoking, alcohol, physical activity, geographic region, and various indicators of socioeconomic status. In 2007, women reported their recency of pelvic ultrasonography (<5, 5–9, or ≥10 years prior or never). Family history of UL (“Has your mother or any of your sisters ever been diagnosed with uterine fibroids?”) was ascertained in 2009.
Genotyping and quality control
DNA isolation and amplification
DNA was isolated from mouthwash-swish samples at the Boston University Molecular Core Genetics Laboratory using the QIAAMP DNA Mini Kit (Qiagen, Valencia, California). Whole genome amplification was performed with Qiagen RePLI-g Kits using the method of multiple displacement amplification. Amplified samples underwent purification and PicoGreen quantification at the Broad Institute Center for Genotyping and Analysis (Cambridge, Massachusetts) before being plated for genotyping.
DNA genotyping
All samples were genotyped at the Broad Institute Center for Genotyping and Analysis. UL cases were genotyped on the “Phase 3” admixture mapping panel, consisting of 1,509 ancestry informative markers (AIMs) in an Illumina GoldenGate custom assay in the BeadLab platform (Illumina, San Diego, California) (30). We filtered out AIMs that had a call rate less than 95% or that visually showed poor clustering in the genotyping calls. We also included only data for samples that had at least 95% genotyping completeness. The final admixture mapping analysis included 2,453 UL cases and 1,430 AIMs.
Controls (n = 2,173), defined as premenopausal women who had not been diagnosed with UL through 2009 and who reported no family history of UL, were genotyped for a panel of 30 AIMs. These AIMs were a subset of the SNPs in the admixture mapping panel, selected to have the greatest difference in frequency between European and African populations. This reduced set of AIMs was shown to produce estimates of European ancestry that were highly correlated with those based on the complete admixture panel (r = 0.89) (31). Genotyping of these 30 SNPs was carried out in a Sequenom iPLEX assay (Sequenom, San Francisco, California). One SNP and 71 samples failed the genotyping, leaving 29 SNPs and 2,102 controls in the final case-control analysis.
We successfully genotyped an additional 53 SNPs in all cases and controls using the Sequenom iPLEX assay: 14 SNPs were selected to increase the density of the admixture scan in the 2 regions with the strongest signal, and 37 SNPs were selected to fine-map 3 loci identified in a recent GWAS based on a Japanese population (20). To select SNPs for fine mapping, we first identified blocks around the index SNPs—centered on chromosome 10 index SNP rs7913069, chromosome 11 index SNP rs2280543, and chromosome 22 index SNP rs12484776—in the HapMap CHB (Han Chinese in Beijing) and JPT (Japanese in Tokyo) populations (http://hapmap.ncbi.nlm.nih.gov/). We then downloaded SNPs covering the entire linkage disequilibrium blocks from the HapMap YRI (Yoruba in Ibadan, Nigeria) database (http://hapmap.ncbi.nlm.nih.gov/), under the hypothesis that the true causal variant should be in linkage disequilibrium with the index SNP in populations of East Asian ancestry. We used the Tagger software implemented in Haploview, version 4.1 (http://www.broadinstitute.org/haploview/haploview), to select all tagging SNPs with a minor allele frequency of ≥5% and r2 ≥ 0.9. The 3 index SNPs were forced into the sets. We used logistic regression to assess the association between each SNP and UL, with further adjustment for local ancestry (percentage of European ancestry at the genomic location under the tested SNP, coded as a continuous variable) (32).
Admixture mapping in UL cases
We used both ANCESTRYMAP (24, 33, 34) and ADMIXMAP (35) to assess individual ancestry proportions and to scan the genome for regions of African ancestry that differed significantly from the genome average. ANCESTRYMAP compares the null hypothesis of no ancestry effect with a range of alternative hypotheses within a Bayesian framework. ANCESTRYMAP computes the log-genome score, which is the logarithm (base 10) of the posterior probability of the model for disease association divided by the posterior probability of the model for no disease association. Parameters of 100 for burn-in iterations and 200 for follow-on iterations were used for all Markov chain Monte Carlo runs (24). A log-genome score greater than 1.0 is considered suggestive evidence for association based on published criteria, while a score greater than 2.0 is considered genome-wide significant (24). ANCESTRYMAP also calculates a local LOD (logarithm (base 10) of odds) score, with values greater than 4 considered suggestive and values greater than 5 considered statistically significant based on published criteria (24).
ADMIXMAP is an independently developed Markov chain Monte Carlo algorithm for carrying out admixture scans, using a hybrid of Bayesian and frequentist methods (35). We ran ADMIXMAP using 1,000 burn-in iterations and 4,000 follow-on iterations. ADMIXMAP implements a score test that compares ancestry in each chromosomal position with genome-wide ancestry. Statistical significance is assessed using standard normal Z statistics, with a threshold of |Z| > 3.0 considered suggestive and a threshold of |Z| > 4.0 considered statistically significant (35). A negative Z score indicates that lower European ancestry at that particular locus compared with the genome-wide average is associated with higher UL risk, while a positive Z score indicates that higher European ancestry is associated with higher UL risk.
On average, African Americans have different proportions of European ancestry on chromosome X than on the rest of the genome, due to a history of more male European ancestors than female European ancestors. This can confound admixture mapping methods that compare locus-specific ancestry with the genome-wide average, particularly if no controls are available for fitting a model relating the distribution of ancestry on the autosomes to that on chromosome X (36, 37). To ensure that results were not confounded by different average European ancestry proportions on the X chromosome, we excluded chromosome X markers from all analyses.
Age-adjusted odds ratios and 95% confidence intervals for the association between global ancestry (in quartiles) and UL were estimated with logistic regression among cases and controls. We constructed models that additionally controlled for UL determinants, including age at menarche (years), parity (number of births), age at first birth (years), years since last birth (<5, 5–9, 10–14, 15, or ≥20 years), age at first oral contraceptive use (years), body mass index (<20, 20–24, 25–29, 30–34, or ≥35), smoking (current, past, or never), current alcohol consumption (<1, 1–6, or ≥7 drinks/week), education (≤12, 13–15, 16, or ≥17 years), marital status (married/partnered, divorced/separated/widowed, or single), occupation (white-collar, non-white-collar, unemployed, or missing), annual household income (≤$25,000, $25,001–$50,000, $50,001–$100,000, >$100,000, or missing), and region (South, Northeast, Midwest, or West). Because multivariable models gave slightly stronger results than age-adjusted models, we present the more conservative age-adjusted results. Given that early diagnosis may reflect a genetic predisposition to disease and that surgically confirmed cases often have more symptomatic disease (9), we stratified our data by age at diagnosis and surgery. Tests for trend were conducted by inserting the continuous variable for percentage of European ancestry into the regression model and evaluating the associated Wald test statistic (38). Two-sided t tests were used to assess the statistical significance of associations. SAS software, version 9.2, was used to conduct these analyses (32).
RESULTS
Mean age at the start of follow-up (1997) was 34.4 years for UL cases and 33.3 years for controls (Table 1). Mean age at diagnosis of UL cases was 38.3 years, with 31.3% of cases being diagnosed before age 35 years and 43.9% being confirmed by surgery. Approximately 93.5% of cases and 94.4% of controls had been born in the United States. Family history of UL was reported by 52.8% of all cases and 56.7% of cases aged <35 years at diagnosis; controls were selected not to have a family history of UL.
Table 1.
Baseline Characteristics of Women With Uterine Leiomyomata (Cases) and Controls, Black Women's Health Study, 1997–2009
| Cases |
||||||||
|---|---|---|---|---|---|---|---|---|
| Controls (n = 2,102) |
All Cases (n = 2,453) |
Surgical Cases (n = 1,076) |
Cases Aged <35 Years at Diagnosis (n = 768) |
|||||
| Mean | % | Mean | % | Mean | % | Mean | % | |
| Age, years | 33.3 | 34.4 | 34.9 | 28.1 | ||||
| Age at diagnosis (cases) or at end of follow-up, years | 42.9 | 38.3 | 38.1 | 30.9 | ||||
| Family history of UL as of 2009 | 0a | 52.8 | 55.0 | 56.7 | ||||
| Age at menarche, years | 12.4 | 12.2 | 12.2 | 12.0 | ||||
| Parity (no. of births)b | 1.9 | 1.9 | 2.0 | 1.6 | ||||
| Age at first birth, yearsb | 23.7 | 22.8 | 22.4 | 21.7 | ||||
| Years since last birth, yearsb | 8.1 | 9.9 | 9.7 | 5.1 | ||||
| Age at first OC use (ever users), years | 18.9 | 18.7 | 18.7 | 18.1 | ||||
| Education in 1995, years | 14.9 | 14.9 | 14.8 | 15.0 | ||||
| Body mass indexc | 27.6 | 27.5 | 27.6 | 27.1 | ||||
| Current alcohol intake (≥1 drink/day) | 3.5 | 3.0 | 2.9 | 2.1 | ||||
| Smoking history | ||||||||
| Current smoker | 9.6 | 10.8 | 12.4 | 7.3 | ||||
| Former smoker | 16.1 | 14.6 | 14.6 | 5.7 | ||||
| Geographic region of residence | ||||||||
| Northeast | 20.9 | 21.8 | 19.1 | 27.3 | ||||
| South | 34.7 | 36.3 | 36.4 | 37.6 | ||||
| Midwest | 24.5 | 24.0 | 25.9 | 22.0 | ||||
| West | 19.9 | 17.9 | 18.6 | 13.1 | ||||
| Born in United States | 94.4 | 93.5 | 93.0 | 92.1 | ||||
Abbreviations: OC, oral contraceptive; UL, uterine leiomyomata.
a Controls were selected not to have a family history of UL.
b Restricted to parous women only.
c Weight (kg)/height (m)2.
Global ancestry analyses
Mean percentage of European ancestry was significantly lower among cases than among controls (age-adjusted mean difference (β) = −1.76%, 95% confidence interval (CI): −2.40, −1.12) (Table 2). Larger differences in mean percentage of European ancestry were found among cases diagnosed at younger ages (<35 years: β = −2.76%, 95% CI: −3.71, −1.82), surgical cases aged <35 years (β = −2.31%, 95% CI: −3.62, −0.99), and cases with a family history of UL (β = −1.91%, 95% CI: −2.68, −1.14). When we restricted the control group to persons with a recent pelvic ultrasound, results were stronger (β = −2.26%, 95% CI: −3.29, −1.23). Results were similar when percentage of European ancestry was calculated after restriction to the 22 AIMs that we successfully genotyped in both cases and controls.
Table 2.
Mean Percentage of European Ancestry Among Women With Uterine Leiomyomata (Cases) and Controls, Black Women's Health Study, 1997–2009
| Cases |
Controls |
Unadjusted Mean Difference (β) | Age-Adjusted Mean Difference (β)a | Age-Adjusted 95% CIa | P Value | |||
|---|---|---|---|---|---|---|---|---|
| No. | Mean % | No. | Mean % | |||||
| All cases and controls | 2,453 | 20.00 | 2,102 | 21.63 | −1.63 | −1.76 | −2.40, −1.12 | <0.0001 |
| All cases and controls (restricted)b | 2,438 | 19.92 | 2,083 | 21.20 | −1.29 | −1.40 | −1.89, −0.90 | <0.0001 |
| All cases and controls with recent ultrasound | 2,453 | 20.00 | 529 | 21.85 | −1.84 | −2.26 | −3.29, −1.23 | 0.0004 |
| Cases with a family history of UL and all controls | 1,187 | 19.79 | 2,102 | 21.63 | −1.85 | −1.91 | −2.68, −1.14 | <0.0001 |
| Cases aged <35 years at diagnosis and all controls | 770 | 18.40 | 2,102 | 21.63 | −3.23 | −2.76 | −3.71, −1.82 | <0.0001 |
| Cases aged 35–39 years at diagnosis and all controls | 648 | 20.05 | 2,102 | 21.63 | −1.58 | −1.54 | −2.51, −0.57 | 0.0018 |
| Cases aged ≥40 years at diagnosis and all controls | 1,035 | 21.16 | 2,102 | 21.63 | −0.48 | −1.01 | −1.95, −0.06 | 0.0362 |
| Surgical cases and all controls | 1,076 | 20.18 | 2,102 | 21.63 | −1.45 | −1.63 | −2.44, −0.82 | <0.0001 |
| Surgical cases aged <35 years and all controls | 324 | 18.94 | 2,102 | 21.63 | −2.70 | −2.31 | −3.62, −0.99 | 0.0006 |
| Parous women (1997) | 1,293 | 19.99 | 1,193 | 21.59 | −1.60 | −1.70 | −2.58, −0.83 | 0.0001 |
| Nulliparous women (1997) | 1,160 | 20.01 | 909 | 21.69 | −1.68 | −1.99 | −2.93, −1.04 | <0.0001 |
| Nulliparous surgical cases and all controls | 472 | 20.23 | 2,102 | 21.63 | −1.40 | −1.31 | −2.39, −0.22 | 0.0181 |
| BMIc <25 (1997) | 1,041 | 20.72 | 885 | 22.54 | −1.82 | −2.01 | −3.03, −0.99 | 0.0001 |
| BMI 25–29 (1997) | 740 | 20.07 | 619 | 21.43 | −1.36 | −1.57 | −2.72, −0.41 | 0.0080 |
| BMI ≥30 (1997) | 672 | 18.81 | 589 | 20.54 | −1.73 | −1.73 | −2.87, −0.60 | 0.0028 |
Abbreviations: BMI, body mass index; CI, confidence interval; UL, uterine leiomyomata.
a Adjusted for age in 1997 (years).
b The panel of ancestry informative markers was restricted to the 22 single nucleotide polymorphisms that cases and controls had in common.
c Weight (kg)/height (m)2.
In age-adjusted logistic regression analyses of the categorical admixture variable, we observed a statistically significant overall association between percentage of European ancestry and UL (Table 3). Odds of UL were 34% lower among persons in the highest quartile of percentage of European ancestry relative to the lowest quartile (odds ratio (OR) = 0.66, 95% CI: 0.56, 0.78). Additional control for UL risk factors made little difference in the odds ratio (OR = 0.64, 95% CI: 0.53, 0.76). The association was strongest among cases aged <35 years at diagnosis (OR = 0.56, 95% CI: 0.43, 0.73). Assuming a linear relation, each 10% increase in European ancestry was associated with 14% decreased odds of UL (OR = 0.86, 95% CI: 0.82, 0.91). Overall and age-specific results were similar for surgical cases (Table 3), as were results restricted to the 22 AIMs that cases and controls had in common (data not shown).
Table 3.
Oddsa of Uterine Leiomyomata According to Quartile of Percentage of European Ancestry, by Age at Diagnosis and Surgery Status, Black Women's Health Study, 1997–2009
| No. of Cases | No. of Controls | Quartile of Percentage of European Ancestry |
P-trend | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Q1 (<13%) |
Q2 (13%–18%) |
Q3 (19%–25%) |
Q4 (≥26%) |
||||||||
| OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | ||||
| All women | 2,453 | 2,102 | 1.00 | Referent | 0.77 | 0.65, 0.91 | 0.79 | 0.67, 0.93 | 0.66 | 0.56, 0.78 | <0.0001 |
| Cases aged <35 years | 770 | 2,102 | 1.00 | Referent | 0.81 | 0.64, 1.03 | 0.72 | 0.56, 0.92 | 0.56 | 0.43, 0.73 | <0.0001 |
| Cases aged ≥35 years | 1,683 | 2,102 | 1.00 | Referent | 0.76 | 0.63, 0.93 | 0.86 | 0.71, 1.04 | 0.73 | 0.60, 0.88 | 0.0014 |
| Surgical cases | 1,076 | 2,102 | 1.00 | Referent | 0.77 | 0.63, 0.96 | 0.81 | 0.66, 0.99 | 0.70 | 0.56, 0.86 | 0.0001 |
| Surgical cases aged <35 years | 324 | 2,102 | 1.00 | Referent | 0.82 | 0.58, 1.14 | 0.83 | 0.60, 1.16 | 0.66 | 0.46, 0.94 | 0.0006 |
| Surgical cases aged ≥35 years | 752 | 2,102 | 1.00 | Referent | 0.77 | 0.60, 0.98 | 0.82 | 0.64, 1.05 | 0.71 | 0.56, 0.90 | 0.0066 |
Abbreviations: CI, confidence interval; OR, odds ratio; Q, quartile.
a All odds ratios were adjusted for age in 1997 (years).
Admixture mapping analyses
Because ANCESTRYMAP requires a prior hypothesis for the risk model at each genomic locus, we assessed risk models ranging from a 0.4-fold risk to a 2.2-fold risk per European locus, placing a greater prior probability on models associated with decreased risk due to European ancestry (Appendix Table 1). Table 4 shows admixture scan results for selected UL phenotypes. In addition to the initial “Phase 3” panel data, these analyses also included genotyping data from 16 “density-increasing” SNPs on chromosomes 4 and 10, the two loci that gave weak hints of association in a first-round analysis. In ANCESTRYMAP, we found a statistically suggestive peak at chromosome 4p16 around rs9715724 (LOD = 4.10). In ADMIXMAP, we observed 2 statistically significant peaks: one at chromosome 4p16 around SNP rs9715724 (Z = −4.20) and the other at chromosome 10q26 around rs7100028 (Z = −4.15). We also found suggestive evidence for a peak at chromosome 2q33 around rs7573626 (Z = 3.42). Subgroup analyses within ANCESTRYMAP did not reveal any suggestive peaks. However, in ADMIXMAP, we observed peaks on chromosome 2 for cases aged <35 years (rs2271767: Z = 3.69), surgical cases (rs920249: Z = 4.46), nulliparous surgical cases (rs6710083: Z = 4.10), and surgical cases aged <35 years (rs920249: Z = 5.10), all localized within an approximately 10-megabase region on 2q32-33, with the latter 3 peaks reaching statistical significance; results for chromosomes 4 and 10 were generally weaker (rs numbers are available upon request).
Table 4.
Results From a Genetic Admixture Mapping Scan Carried Out Among 2,453 Women With Uterine Leiomyomata (Cases), Black Women's Health Study, 1997–2009
| No. of Cases | No. of Markers | Genome- wide Scorea | Maximum LOD or Z Score |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Chromosome 2 |
Chromosome 4 |
Chromosome 10 |
|||||||
| ANCESTRY-MAPa | ADMIX- MAPb | ANCESTRY-MAPa | ADMIX- MAPb | ANCESTRY-MAPa | ADMIX- MAPb | ||||
| All cases | 2,453 | 1,382 | 1.39 | −0.04 | 3.42c | 4.10c | −4.20d | 2.45 | −4.15d |
| Cases with a family history of UL | 1,187 | 1,378 | 0.42 | 0.21 | 2.96 | 2.91 | −3.65c | 0.55 | −2.26 |
| Cases aged <35 years | 770 | 1,375 | −0.04 | 1.21 | 3.69c | 1.60 | −2.62 | 1.71 | −2.74 |
| Surgical cases | 1,076 | 1,381 | 0.37 | 2.93 | 4.46d | 1.03 | −2.72 | 1.21 | −3.07c |
| Surgical cases aged <35 years | 324 | 1,367 | 0.82 | 3.61 | 5.10d | 1.62 | −2.72 | 0.55 | −1.73 |
| Nulliparous cases | 1,100 | 1,378 | −0.14 | 1.02 | 3.56c | 1.82 | −3.57c | 0.11 | −1.89 |
| Parous cases | 1,352 | 1,382 | 0.85 | −0.41 | 2.61 | 1.80 | −2.87 | 3.51 | −4.46d |
| Nulliparous surgical cases | 459 | 1,374 | 0.16 | 2.35 | 4.10d | 2.26 | −3.67c | 0.08 | −1.70 |
| Cases with BMIe <25 | 741 | 1,381 | 0.19 | 0.63 | 2.76 | 1.67 | −3.82c | 2.29 | 3.32c |
| Cases with BMI 25–29 | 787 | 1,377 | −0.42 | 0.64 | 2.87 | 1.33 | −3.22c | 0.76 | −3.20c |
| Cases with BMI ≥30 | 924 | 1,379 | 1.03 | 0.30 | −2.42 | 0.66 | −2.00 | 1.25 | −2.52 |
Abbreviations: BMI, body mass index; LOD, logarithm (base 10) of odds; UL, uterine leiomyomata.
a Analyses were performed in ANCESTRYMAP (suggestive result: LOD score >4; significant result: LOD score >5).
b Analyses were performed in ADMIXMAP (suggestive result: |Z score| >3; significant result: |Z score| >4). A negative Z score indicates an inverse association between European ancestry and UL risk.
c Statistically suggestive peak.
d Statistically significant peak.
e Weight (kg)/height (m)2.
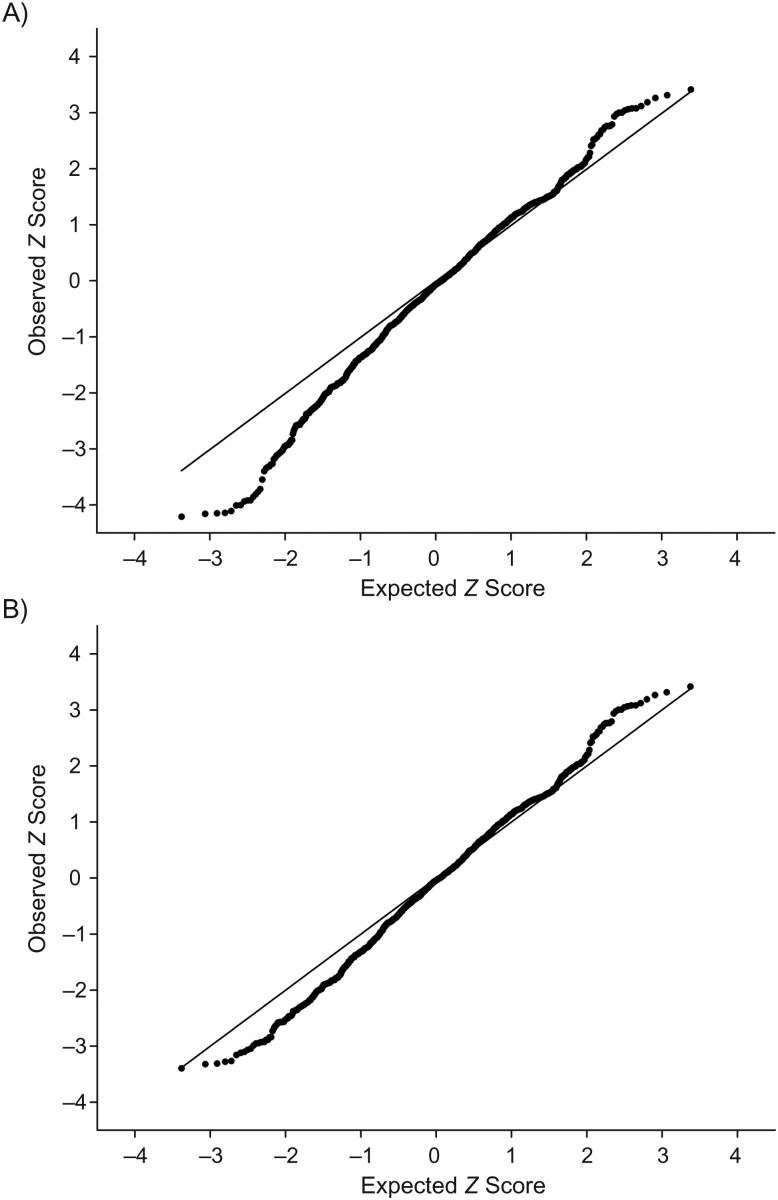
Figure 1 shows the quantile-quantile plots of the ADMIXMAP Z score. Although we observed inflation of the test statistic at the bottom of the curve (Figure 1A), there was no appreciable inflation of the Z score after we removed the AIMs in chromosomes 4 and 10 (Figure 1B).
Figure 1.
Quantile-quantile plots of the test statistic Z score obtained using ADMIXMAP software (35), Black Women's Health Study, 1997–2009. A) Results including all autosome ancestry informative markers (AIMs); B) results excluding AIMs within the two peaks on chromosomes 4 and 10.
Lack of replication of Japanese GWAS findings
We genotyped 35 SNPs on chromosomes 10, 11, and 22 in an attempt to tag the genetic variation around SNPs associated with UL in a Japanese population (20). When we individually examined the 3 best tags for the associations (Table 5), the odds ratios for SNPs rs2280543 (chromosome 11) and rs12484776 (chromosome 22) were weaker but pointed in the same direction as those from the Japanese GWAS, and the 95% confidence intervals overlapped. In contrast, the 95% confidence intervals for SNP rs7913069 (chromosome 10) did not overlap between the studies. None of the other 32 SNPs reached statistical significance (data available upon request). Results were consistent across strata of age, body mass index, parity status, and family history of UL. Results were also similar when the control group was restricted to persons with a recent pelvic ultrasound.
Table 5.
Odds of Uterine Leiomyomata for Single Nucleotide Polymorphisms Previously Identified in a Genome-Wide Association Study of a Japanese Population, Black Women's Health Study, 1997–2009
| Single Nucleotide Polymorphism | Chromosome | Risk Allele | Japanese Populationa |
Black Women's Health Study Population |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Cases | No. of Controls | RAF in Controls, % | ORb | 95% CI | No. of Cases | No. of Controls | RAF in Controls, % | ORb,c | 95% CIc | |||
| rs7913069 | 10 | A | 5,037 | 4,672 | 7 | 1.47 | 1.23, 1.75 | 2,017 | 2,005 | 14.6 | 0.97 | 0.86, 1.10 |
| rs2280543 | 11 | G | 5,045 | 4,672 | 88 | 1.39 | 1.17, 1.64 | 2,018 | 2,007 | 93.4 | 1.13 | 0.94, 1.35 |
| rs12484776 | 22 | G | 5,042 | 4,670 | 36 | 1.23 | 1.11, 1.37 | 2,017 | 2,005 | 9.7 | 1.10 | 0.95, 1.28 |
Abbreviations: CI, confidence interval; OR, odds ratio; RAF, risk allele frequency.
a Data were obtained from Cha et al. (20).
b Odds ratio for a 1-unit increase in the risk allele.
c Model additionally controlled for local ancestry.
DISCUSSION
To our knowledge, this was the first genome-wide association scan for UL in African Americans and the first admixture mapping study of UL in any population. We found strong evidence for an inverse association between percentage of European ancestry and UL risk, particularly among cases aged <35 years at diagnosis. However, we found only suggestive evidence for an association with European ancestry at specific loci (chromosomes 2, 4, and 10), finding stronger results among younger and surgical cases for chromosome 2 only. Finally, we failed to replicate results from a recent GWAS in Japanese women (20).
Our main finding was the detection of an inverse association between European ancestry and UL risk. A potential concern is that ancestry estimates were computed with a larger number of SNPs in cases than in controls. However, the results were consistent when we restricted the AIMs analyzed to the 22 SNPs that cases and controls had in common. We found only suggestive evidence of significant associations with specific genomic regions. This contrasts with prostate cancer (39) and nondiabetic end-stage renal disease (40), which both have an epidemiology similar to UL in that they occur at higher frequency in African Americans than in European Americans. For both diseases, admixture mapping in smaller samples than we analyzed here found a locus that by itself explained a substantial part of the higher risk of disease in African Americans. Our failure to detect a single locus underpinning the higher risk for UL might indicate that there are multiple loci in the genome with relatively small effects that contribute to the increased risk in African Americans.
While we found evidence for a stronger effect of European ancestry (averaged across the genome) on UL risk among younger cases, admixture analyses among younger cases revealed stronger peaks only on chromosome 2, not chromosomes 4 and 10. If the individual loci on chromosomes 4 and 10 genuinely modulate risk for UL, it is possible that these loci are not associated with age at diagnosis (i.e., the association of European ancestry with age at diagnosis is driven by other loci). Such a pattern has been observed for prostate cancer risk in African Americans, where some but not all loci at 8q24 are associated with age at diagnosis (39, 41). In this instance, we would expect there to be a loss in power due to reduced sample size in the younger-onset cases, decreasing our ability to detect a true association. An alternative possibility is that these loci are not causally related to UL risk, as they were only marginally significant in our admixture scans.
Our failure to replicate results from the Japanese GWAS does not necessarily indicate that these genomic regions are not causally associated with UL. For two of the SNPs, the associations pointed in the same direction as the Japanese study and the confidence intervals overlapped (20). There are several potential reasons why we failed to replicate results from the Japanese study. First, the Japanese GWAS, like any study that finds the first association at a locus, was subject to a “winner's curse” of overestimating the effect size. Second, Japanese are known to have a different pattern of genetic variation than African Americans; in particular, the reported disease-associated SNP may be in strong linkage disequilibrium with the truly associated SNP in Japanese but not in African Americans. Thus, we may not have succeeded in genotyping a SNP in linkage disequilibrium with the true causal variant in African Americans. Finally, the causal variant may not exist in African Americans.
Retention of the baseline cohort was approximately 80% through 2009, reducing potential for differential loss to follow-up. Because we did not screen women for the presence of UL, we may have misclassified a large number of true cases (5). However, our validation study of UL indicated high accuracy in reporting (>96%), and results were similar when we restricted the control group to women with a recent pelvic ultrasound, who were most likely to be free of UL. Our findings were also stronger among younger women, among whom misclassification of UL is reduced (5). The results were consistent across subcategories of UL risk factors, and control for these factors had little impact on the results. The large sample size and high incidence of UL in this population conferred excellent statistical power to detect relatively small differences in risk. Most cases had symptoms at diagnosis, and 87% of diagnoses were made because of UL-related symptoms or a palpable tumor upon pelvic examination (29). Therefore, our results probably apply to women with symptomatic UL, which represents the disease burden in reproductive-age women.
In summary, we found evidence of an inverse association between percentage of European ancestry and UL risk when comparing cases with controls, an association which strengthened with younger age at diagnosis. Suggestive genome-wide associations between UL and European ancestry were found at chromosomes 2, 4, and 10. Analysis of additional samples is needed to confirm whether these individual loci are causally associated with UL. Further work, particularly genome-wide association analysis, is needed to understand the genetic basis for the higher risk of UL in African Americans.
ACKNOWLEDGMENTS
Author affiliations: Slone Epidemiology Center at Boston University, Boston, Massachusetts (Lauren A. Wise, Edward A. Ruiz-Narvaez, Julie R. Palmer, Yvette C. Cozier, Rose G. Radin, Lynn Rosenberg); Department of Genetics, Harvard Medical School, Boston, Massachusetts (Arti Tandon, David Reich); and Broad Institute of Harvard and MIT, Cambridge, Massachusetts (Arti Tandon, Nick Patterson, David Reich).
This work was supported by grant R01HD057966 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development and by grants R01CA058420 and R01CA098663 from the National Cancer Institute, Division of Cancer Control and Population Science (http://www.cancercontrol.cancer.gov).
The authors gratefully acknowledge the staff of the Black Women's Health Study for their ongoing contributions. They also thank Drs. Elizabeth A. Stewart and Donna Day Baird for their guidance on the project.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health.
Conflict of interest: none declared.
Appendix Table 1.
Risk Models Evaluated in the Main Genetic Admixture Scan of 2,453 Women With Uterine Leiomyomata, Black Women's Health Study, 1997–2009a
| Risk Due to European Ancestry | Genome-wide Score With Risk | Weight of Model in Our Prior | % Weight |
|---|---|---|---|
| 0.4b | −16.78 | 6 | 0.77 |
| 0.45 | −10.97 | 8 | 1.03 |
| 0.5 | −6.56 | 18 | 2.32 |
| 0.55 | −3.27 | 45 | 5.80 |
| 0.6 | −0.91 | 70 | 9.02 |
| 0.65 | 0.68 | 90 | 11.60 |
| 0.7 | 1.59 | 100 | 12.89 |
| 0.75 | 1.92 | 86 | 11.08 |
| 0.8 | 1.81 | 68 | 8.76 |
| 0.85 | 1.46 | 52 | 6.70 |
| 0.9 | 0.92 | 40 | 5.15 |
| 1.1 | 0.25 | 6 | 0.77 |
| 1.2 | 0.06 | 19 | 2.45 |
| 1.3 | −1.50 | 40 | 5.15 |
| 1.4 | −4.27 | 52 | 6.70 |
| 1.5 | −7.91 | 32 | 4.12 |
| 1.6 | −11.87 | 18 | 2.32 |
| 1.7 | −15.96 | 11 | 1.42 |
| 1.8 | −19.83 | 8 | 1.03 |
| 1.9 | −20.00 | 4 | 0.52 |
| 2.0 | −20.00 | 1 | 0.13 |
| 2.1 | −20.00 | 1 | 0.13 |
| 2.2c | −20.00 | 1 | 0.13 |
aThe genome log-factor score was 1.361, and the maximum genome-wide score was 4.039.
b 0.4-fold decreased risk per European allele.
c 2.2-fold increased risk per European allele.
REFERENCES
- 1.Buttram VC, Reiter RC. Uterine leiomyomata: etiology, symptomatology, and management. Fertil Steril. 1981;36(4):433–445. doi: 10.1016/s0015-0282(16)45789-4. [DOI] [PubMed] [Google Scholar]
- 2.Coronado GD, Marshall LM, Schwartz SM. Complications in pregnancy, labor, and delivery with uterine leiomyomas: a population-based study. Obstet Gynecol. 2000;95(5):764–769. doi: 10.1016/s0029-7844(99)00605-5. [DOI] [PubMed] [Google Scholar]
- 3.Stewart EA. Uterine fibroids. Lancet. 2001;357(9252):293–298. doi: 10.1016/S0140-6736(00)03622-9. [DOI] [PubMed] [Google Scholar]
- 4.Marshall LM, Spiegelman D, Barbieri RL, et al. Variation in the incidence of uterine leiomyoma among premenopausal women by age and race. Obstet Gynecol. 1997;90(6):967–973. doi: 10.1016/s0029-7844(97)00534-6. [DOI] [PubMed] [Google Scholar]
- 5.Baird DD, Dunson DB, Hill MC, et al. High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. Am J Obstet Gynecol. 2003;188(1):100–107. doi: 10.1067/mob.2003.99. [DOI] [PubMed] [Google Scholar]
- 6.Kjerulff KH, Guzinski GM, Langenberg PW, et al. Hysterectomy and race. Obstet Gynecol. 1993;82(5):757–764. [PubMed] [Google Scholar]
- 7.Kjerulff KH, Langenberg P, Guzinski GM. The socioeconomic correlates of hysterectomies in the United States. Am J Public Health. 1993;83(1):106–108. doi: 10.2105/ajph.83.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kjerulff KH, Langenberg P, Seidman JD, et al. Uterine leiomyomas: racial differences in severity, symptoms, and age at diagnosis. J Reprod Med. 1996;41(7):483–490. [PubMed] [Google Scholar]
- 9.Schwartz SM, Marshall LM, Baird DD. Epidemiologic contributions to understanding the etiology of uterine leiomyomata. Environ Health Perspect. 2000;108(suppl 5):821–827. doi: 10.1289/ehp.00108s5821. [DOI] [PubMed] [Google Scholar]
- 10.Stewart EA, Morton CC. The genetics of uterine leiomyomata: what clinicians need to know. Obstet Gynecol. 2006;107(4):917–921. doi: 10.1097/01.AOG.0000206161.84965.0b. [DOI] [PubMed] [Google Scholar]
- 11.Vikhlyaeva EM, Khodzhaeva ZS, Fantschenko ND. Familial predisposition to uterine leiomyomas. Int J Gynaecol Obstet. 1995;51(2):127–131. doi: 10.1016/0020-7292(95)02533-i. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz S, Voigt L, Tickman E, et al. Familial aggregation of uterine leiomyomata [abstract] Am J Epidemiol. 2000;151(suppl):S10. [Google Scholar]
- 13.Kurbanova MKH, Koroleva AG, Sergeev AS. Genetic analysis of the predisposition to uterine myoma: prevalence and morbidity. Genetika. 1989;25(6):1122–1124. [PubMed] [Google Scholar]
- 14.Kurbanova MKH, Koroleva AG, Sergeev AS. Genetic-epidemiologic analysis of uterine myoma: assessment of repeated risk. Genetika. 1989;25(10):1896–1898. [PubMed] [Google Scholar]
- 15.Treloar SA, Martin NG, Dennerstein L, et al. Pathways to hysterectomy: insights from longitudinal twin research. Am J Obstet Gynecol. 1992;167(1):82–88. doi: 10.1016/s0002-9378(11)91632-9. [DOI] [PubMed] [Google Scholar]
- 16.Luoto R, Kaprio J, Rutanen EM, et al. Heritability and risk factors of uterine fibroids—the Finnish Twin Cohort study. Maturitas. 2000;37(1):15–26. doi: 10.1016/s0378-5122(00)00160-2. [DOI] [PubMed] [Google Scholar]
- 17.Reed WB, Walker R, Horowitz R. Cutaneous leiomyomata with uterine leiomyomata. Acta Derm Venereol. 1973;53(5):409–416. [PubMed] [Google Scholar]
- 18.Garcia Muret MP, Pujol RM, Alomar A, et al. Familial leiomyomatosis cutis et uteri (Reed's syndrome) Arch Derm Res. 1988;280(suppl):S29–S32. [PubMed] [Google Scholar]
- 19.Launonen V, Vierimaa O, Kiuru M, et al. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci USA. 2001;98(6):3387–3392. doi: 10.1073/pnas.051633798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cha PC, Takahashi A, Hosono N, et al. A genome-wide association study identifies three loci associated with susceptibility to uterine fibroids. Nat Genet. 2011;43(5):447–450. doi: 10.1038/ng.805. [DOI] [PubMed] [Google Scholar]
- 21.Van Voorhis BJ, Romitti PA, Jones MP. Family history as a risk factor for development of uterine leiomyomas: results of a pilot study. J Reprod Med. 2002;47(8):663–669. [PubMed] [Google Scholar]
- 22.Gross K, Morton C, Stewart EA. Finding genes for uterine fibroids [abstract] Obstet Gynecol. 2000;95(4 suppl):S60. [Google Scholar]
- 23.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273(5281):1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 24.Patterson N, Hattangadi N, Lane B, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74(5):979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg L, Adams-Campbell LL, Palmer JR. The Black Women's Health Study: a follow-up study for causes and preventions of illness. J Am Med Womens Assoc. 1995;50(2):56–58. [PubMed] [Google Scholar]
- 26.Cozier YC, Palmer JR, Rosenberg L. Comparison of methods for collection of DNA samples by mail in the Black Women's Health Study. Ann Epidemiol. 2004;14(2):117–122. doi: 10.1016/S1047-2797(03)00132-7. [DOI] [PubMed] [Google Scholar]
- 27.Dueholm M, Lundorf E, Hansen ES, et al. Accuracy of magnetic resonance imaging and transvaginal ultrasonography in the diagnosis, mapping, and measurement of uterine myomas. Am J Obstet Gynecol. 2002;186(3):409–415. doi: 10.1067/mob.2002.121725. [DOI] [PubMed] [Google Scholar]
- 28.Loutradis D, Antsaklis A, Creatsas G, et al. The validity of gynecological ultrasonography. Gynecol Obstet Invest. 1990;29(1):47–50. doi: 10.1159/000293299. [DOI] [PubMed] [Google Scholar]
- 29.Wise LA, Palmer JR, Stewart EA, et al. Age-specific incidence rates for self-reported uterine leiomyomata in the Black Women's Health Study. Obstet Gynecol. 2005;105(3):563–568. doi: 10.1097/01.AOG.0000154161.03418.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan JB, Oliphant A, Shen R, et al. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Narvaez EA, Rosenberg L, Wise LA, et al. Validation of a small set of ancestral informative markers for control of population admixture in African Americans. Am J Epidemiol. 2011;173(5):587–592. doi: 10.1093/aje/kwq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.SAS Institute Inc. SAS/STAT 9.2 User's Guide. Cary, NC: SAS Institute Inc; 2008. [Google Scholar]
- 33.Hoggart CJ, Parra EJ, Shriver MD, et al. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72(6):1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKeigue PM, Carpenter JR, Parra EJ, et al. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann Hum Genet. 2000;64(2):171–186. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]
- 35.Hoggart CJ, Shriver MD, Kittles RA, et al. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74(5):965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shlush LI, Bercovici S, Wasser WG, et al. Admixture mapping of end stage kidney disease genetic susceptibility using estimated mutual information ancestry informative markers. BMC Med Genomics. 2010;3:47. doi: 10.1186/1755-8794-3-47. (doi:10.1186/1755–8794-1183-1147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lind JM, Hutcheson-Dilks HB, Williams SM, et al. Elevated male European and female African contributions to the genomes of African American individuals. Hum Genet. 2007;120(5):713–722. doi: 10.1007/s00439-006-0261-7. [DOI] [PubMed] [Google Scholar]
- 38.Breslow NE, Day NE. Statistical Methods in Cancer Research. Vol 2. The Design and Analysis of Cohort Studies. Lyon, France: IARC Press; 1987. pp. 1–406. [PubMed] [Google Scholar]
- 39.Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103(38):14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kao WH, Klag MJ, Meoni LA, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40(10):1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39(5):638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]