Abstract
At least one pilus island, PI-1 (70%), PI-2a (79%), or PI-2b (21%), was found among 898 Streptococcus agalactiae (group B streptococcus [GBS]) isolates recovered from humans, supporting the use of pilus proteins in vaccines. The stability and dominance of PI-1 and PI-2a in multiple serotypes and founder multilocus sequence types disseminated worldwide suggest it could be the PI combination present in ancestral GBS human pathogens.
TEXT
Streptococcus agalactiae (group B streptococcus [GBS]) is a commensal of the gastrointestinal and genital tracts. However, it is a leading cause of neonatal bacterial sepsis and meningitis and is increasingly associated with invasive disease in adults (1, 2).
Development of a vaccine is the most promising approach for the prevention of GBS infections given the potential adverse effects of intrapartum antimicrobial prophylaxis as well as the need for effective prevention of both adult and late perinatal disease (3). The identification of pilus-like structures on the surface of GBS, their potential importance in GBS colonization and pathogenicity, and the ubiquitous presence of their coding loci in the GBS genome rendered these structures important candidates in the development of a vaccine (4, 5). GBS pili are composed of three subunits: a backbone pilin protein and two ancillary proteins, a pilus-associated adhesin and a component that anchors the pili to the cell wall (6). These are encoded by two loci in different regions of the genome, designated pilus islands 1 and 2 (PI-1 and PI-2), the latter presenting two distinct variants, PI-2a and PI-2b (7).
While much attention has been given to the structural and immunological properties of pili as vaccine candidates, less information is available on the distribution of the different PIs among GBS clones. The aim of our work was to investigate the distribution of PI-1, PI-2a, and PI-2b among GBS.
We assembled a collection of 898 isolates associated with carriage in pregnant women (n = 276), noninvasive disease among nonpregnant adults (n = 98), and invasive disease (n = 524). Of the latter, 275 isolates were recovered from newborns (early-onset disease [EOD], n = 164 isolates; late-onset disease [LOD], n = 111 isolates) and 249 from adults. All isolates were characterized by serotyping and pulsed-field gel electrophoresis (PFGE), and nearly half the collection (n = 440/898, 49%), including representatives of all main PFGE-based genetic lineages, was characterized by multilocus sequence typing (MLST) (8–12). PFGE clusters were defined as isolates with ≥80% relatedness on a dendrogram created with the Dice coefficient and unweighted pair group method with arithmetic averages (UPGMA).
In the context of this study, all isolates were tested for the presence of alpha and alpha-like surface protein genes (alp) (13) and of PI-1, PI-2a, and PI-2b loci by PCR (14). Absence of PI-1 genes was confirmed by simultaneous amplification of the regions flanking the PI operon (14), and whenever one isolate tested negative for the presence of PI-2a or PI-2b, dot blot hybridization with specific probes was performed (data not shown).
PIs were differentially detected among GBS serotypes (Table 1). At least one PI was present in all isolates. Overall, PI-2a was detected in 79% (n = 707), PI-1 in 70% (n = 624), and PI-2b in 21% (n = 191) of the isolates. The most frequently found PIs were PI-1 and PI-2a (49%), followed by PI-2a alone (30%), PI-1 and PI-2b (21%), and PI-2b alone (0.6%). Simpson's index of diversity (SID) (15) was used to estimate the diversity found among the isolates studied (Table 2). The clonal diversity based on PFGE and MLST was high, indicating that our collection captures the overall diversity of the GBS population as expected from epidemiologically unrelated isolates. SID values remained unchanged among the subset of isolates characterized by MLST (having a smaller number of types by PFGE), indicating that these isolates represent the diversity of the entire collection (Table 1). As expected, the small number of distinct serotypes, alp genes, and PIs (only 4 PI combinations were observed) generated smaller SID values, leading to the identification of associations of PI with disease presentation and age groups that reached statistical significance.
Table 1.
Distribution of genes encoding pilus islands across serotypes
| Serotype | No. of isolates by PIa |
Total no. of isolates | |||
|---|---|---|---|---|---|
| PI-1 and PI-2a | PI-1 and PI-2b | PI-2a | PI-2b | ||
| Ia | 11 | 4 | 211 | 2 | 228 |
| Ib | 51 | 0 | 3 | 0 | 54 |
| II | 104 | 0 | 8 | 0 | 112 |
| III | 102 | 168 | 4 | 0 | 274 |
| IV | 11 | 5 | 0 | 1 | 17 |
| V | 119 | 3 | 21 | 1 | 144 |
| VI | 0 | 0 | 1 | 0 | 1 |
| VII | 1 | 0 | 2 | 0 | 3 |
| NTb | 39 | 6 | 19 | 1 | 65 |
| Total | 438 | 186 | 269 | 5 | 898 |
The PI combination including more than half of the isolates in serotypes including more than 10 isolates are indicated in bold.
NT, nontypeable.
Table 2.
Number of types and diversity of the isolates when characterized by each typing method
| Typing method | All isolates (n = 898) |
Isolates characterized by MLST (n = 440) |
||
|---|---|---|---|---|
| No. of types found | SID (CI95) | No. of types found | SID (CI95) | |
| Serotype | 9 | 0.793 (0.781–0.805) | 9 | 0.777 (0.755–0.798) |
| alp locus | 6 | 0.697 (0.682–0.712) | 6 | 0.687 (0.664–0.711) |
| PI combination | 4 | 0.630 (0.614–0.646) | 4 | 0.653 (0.638–0.668) |
| PFGE (80%)a | 88 | 0.906 (0.895–0.918) | 67 | 0.915 (0.900–0.929) |
| MLST | 54 | 0.894 (0.878–0.910) | ||
PFGE clusters were defined as groups of isolates with ≥80% relatedness on the dendrogram.
While our results are in agreement with previous work in terms of PI distribution in serotypes and age groups (5), additional correlations were found, probably reflecting the fact that our collection includes a high number of isolates recovered from multiple sources. When the GBS populations recovered from carriage and neonatal infections were compared, PI-1 and PI-2a were associated with maternal colonization and PI-1 and PI-2b with neonatal disease (Fisher's exact test, all P < 0.05). In contrast to the association of PI-1 and PI-2b with neonatal invasive infection, PI-1 and PI-2a were associated with invasive disease in adults (P < 0.001). On the other hand, a closer examination of neonatal disease onset revealed that EOD was associated with PI-1 and PI-2a and PI-2a alone, supporting a maternal source of disease, while PI-1 and PI-2b were overrepresented in LOD (all P < 0.05). However, we consider that these associations between PI distribution and disease presentation or age groups are likely to reflect the different representation of the GBS clones in these groups, already evaluated by PFGE and MLST, that are more discriminatory (9, 11, 12). Since other clonal properties, in addition to the presence of PIs, could also influence the results, interpretation of these data should be performed with caution. Adjusted Wallace coefficients (AW) (16) were calculated to evaluate the relationship between PI and typing methods. The AW between PFGE and PI was high (AWPFGE→PI = 0.903, 95% confidence interval [CI95] = 0.859 to 0.947), indicating that the overwhelming majority of pairs of isolates grouped into the same PFGE cluster also share the same PI combination. An even higher correspondence of the sequence type (ST) with PI (AWST→PI = 0.921, CI95 = 0.877 to 0.965) corroborates a clonal distribution of PIs in spite of prior evidence for the transfer of large DNA fragments in GBS (14, 17).
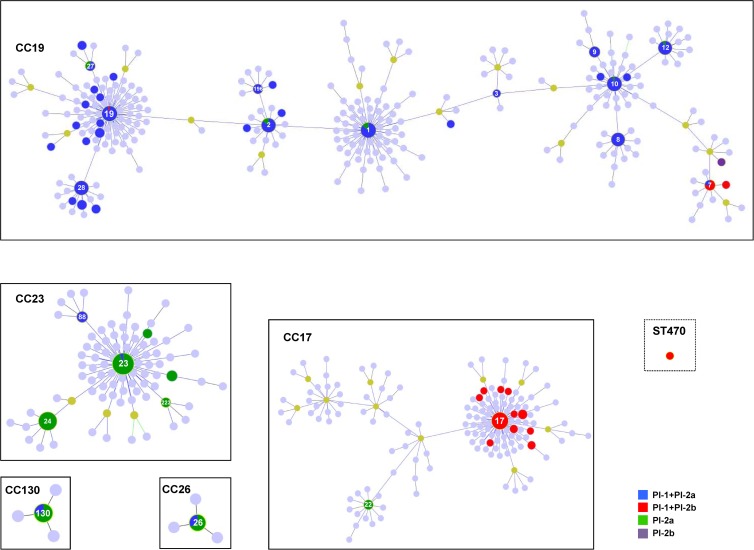
Such association between STs and PIs is clearly demonstrated by goeBURST (18) implemented in PHYLOViZ (19). The algorithm divided the 54 STs identified in our data set into 5 clonal complexes (CCs) and one singleton (Fig. 1). When the distribution of the PIs in the collection is displayed, a nearly exclusive association of particular PI combinations and CCs is observed (Fig. 1). The concomitant presence of PI-1 and PI-2a was the most frequent combination among GBS isolates and nearly ubiquitous in CC19, encompassing nearly 50% of all known GBS STs (287/591 by August 2012). This large CC now groups several genetic lineages that were initially identified as having separate origins, such as ST1 and ST10 (now identified as subfounders) and may be better viewed as a collection of more distantly related lineages (20). Among the 200 isolates in the collection included in CC19, 184 (92%) shared the PI-1 and PI-2a combination, including the founder ST19 and most subfounders. Its significant prevalence across all serotypes (Ia to VII), the diversity of STs in which it was found, and association with the majority of putative founder genotypes are consistent with considering PI-1 and PI-2a as the PI combination present in early GBS associating with carriage and infection in humans. Sequencing of pilus-coding genes revealed a higher variability of PI-2a genes than PI-1 and PI-2b genes (5), consistent with adaptation or drift of an ancestral genotype to the different lifestyles and multiple genetic backgrounds where it is now found.
Fig 1.
goeBURST diagram of PI distribution among the GBS isolates characterized by MLST (n = 440). Light-blue and light-green circles indicate STs that are not present in the collection analyzed but that could be found in the Pubmlst database; the latter were identified as subfounders. CCs are identified by the number of the putative founder ST identified by goeBURST. Within each CC, the STs present within our collection are represented in dark blue, red, darker green, and purple, with the size of the circles being proportional to the number of isolates (in a logarithmic scale). The figure was prepared using the PHYLOViZ software (http://www.phyloviz.net/).
The presence of PI-2a alone was restricted mostly to a single clonal complex (CC23) in which serotype Ia is dominant in human isolates (20). Within this CC, while ST23 is disseminated worldwide, ST24 is particularly successful in the Iberian Peninsula and Mediterranean region, significantly contributing to GBS disease in all age groups (9, 11, 12). Out of 113 isolates belonging to CC23, only 6 (5.3%) did not carry exclusively PI-2a (four isolates also presented with a serotype different from Ia). Conversely, in other CCs, very few isolates (n = 20/785, 2.5%) did not carry PI-1. Moreover, analysis of PI distribution across serotypes revealed that the absence of PI-1 is significantly associated only with serotype Ia (P < 0.05). Taken together, these data suggest that PI-1 is a mostly stable component of the GBS genome and that the loss of PI-1 is a rare event that probably accompanied the recent emergence of CC23 in humans.
Similarly, the acquisition of PI-2b seems to be infrequent in human isolates, consistent with the presence of this operon being mostly restricted to serotype III (P < 0.05) and a particular clonal complex (CC17). This frequently designated “hypervirulent” lineage has been extensively studied in past decades due to its significant overrepresentation in late neonatal infections. Since these serotype III isolates present a unique combination of virulence factors (20), and PI-2b is seldom found among other CCs, it is possible that PI-2b is also a particular acquisition of this lineage and partly responsible for its particular virulence and tropism. The characterization of cattle isolates revealed that the related ST67 lineage lacks PI-1 (20), suggesting independent evolution in different hosts of these two related lineages.
In agreement with the high AW relating STs and PIs, the data presented here argue for essentially stable PI content of individual GAS clones. The epidemiological characterization of GBS populations has revealed a significant number of genetically distinct lineages, most capable of both colonizing and causing invasive disease in different age groups, although with different propensity (8–12). The stability and dominance of PI-1 and PI-2a distribution in these worldwide disseminated lineages suggest that this PI combination reflects an ancestral genotype that is potentially well adapted to multiple genetic backgrounds. The identification of different PIs among more recently recognized and particularly successful lineages at causing specific disease manifestations could reflect ongoing GBS adaptation to the human host and supports continued surveillance of PI distribution. While our study has focused only on the presence of the PI operons within the GBS genome and neither on pili expression at the bacterial surface nor on the variability of these loci that is known to occur, the ubiquitous presence of PIs in GBS associated with humans underpins considering them as vaccine candidates. Our results indicate that a vaccine including components from PI-1 and PI-2a could provide potential coverage against 99.4% of isolates, supporting their use in a future vaccine as previously suggested (5).
ACKNOWLEDGMENTS
E.R.M. was supported by a grant from Fundação para a Ciência e a Tecnologia (SFRH/BPD/80038/2011). J.M.-C. has received research grants administered through his university and received honoraria for consulting and serving on the speakers' bureaus of Pfizer, Bial, GlaxoSmithKline, and Novartis. M.R. has received honoraria for consulting and serving on a speakers' bureau of Pfizer. The other authors declare no conflicts of interests. No company or financing body had any interference in the decision to publish.
Footnotes
Published ahead of print 26 December 2012
REFERENCES
- 1. Schuchat A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Skoff TH, Farley MM, Petit S, Craig AS, Schaffner W, Gershman K, Harrison LH, Lynfield R, Mohle-Boetani J, Zansky S, Albanese BA, Stefonek K, Zell ER, Jackson D, Thompson T, Schrag SJ. 2009. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990–2007. Clin. Infect. Dis. 49:85–92 [DOI] [PubMed] [Google Scholar]
- 3. Edwards MS. 2008. Group B streptococcal conjugate vaccine: a timely concept for which the time has come. Hum. Vaccin. 4:444–448 [DOI] [PubMed] [Google Scholar]
- 4. Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL. 2005. Genome analysis reveals pili in group B Streptococcus. Science 309:105. [DOI] [PubMed] [Google Scholar]
- 5. Margarit I, Rinaudo CD, Galeotti CL, Maione D, Ghezzo C, Buttazzoni E, Rosini R, Runci Y, Mora M, Buccato S, Pagani M, Tresoldi E, Berardi A, Creti R, Baker CJ, Telford JL, Grandi G. 2009. Preventing bacterial infections with pilus-based vaccines: the group B streptococcus paradigm. J. Infect. Dis. 199:108–115 [DOI] [PubMed] [Google Scholar]
- 6. Dramsi S, Caliot E, Bonne I, Guadagnini S, Prevost MC, Kojadinovic M, Lalioui L, Poyart C, Trieu-Cuot P. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 60:1401–1413 [DOI] [PubMed] [Google Scholar]
- 7. Rosini R, Rinaudo CD, Soriani M, Lauer P, Mora M, Maione D, Taddei A, Santi I, Ghezzo C, Brettoni C, Buccato S, Margarit I, Grandi G, Telford JL. 2006. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 61:126–141 [DOI] [PubMed] [Google Scholar]
- 8. Figueira-Coelho J, Ramirez M, Salgado MJ, Melo-Cristino J. 2004. Streptococcus agalactiae in a large Portuguese teaching hospital: antimicrobial susceptibility, serotype distribution, and clonal analysis of macrolide-resistant isolates. Microb. Drug Resist. 10:31–36 [DOI] [PubMed] [Google Scholar]
- 9. Martins ER, Andreu A, Correia P, Juncosa T, Bosch J, Ramirez M, Melo-Cristino J. 2011. Group B streptococci causing neonatal infections in Barcelona are a stable clonal population: 18-year surveillance. J. Clin. Microbiol. 49:2911–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martins ER, Florindo C, Martins F, Aldir I, Borrego MJ, Brum L, Ramirez M, Melo-Cristino J. 2007. Streptococcus agalactiae serotype Ib as an agent of meningitis in two adult nonpregnant women. J. Clin. Microbiol. 45:3850–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martins ER, Melo-Cristino J, Ramirez M. 2012. Dominance of serotype Ia among group B streptococci causing invasive infections in nonpregnant adults in Portugal. J. Clin. Microbiol. 50:1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martins ER, Pessanha MA, Ramirez M, Melo-Cristino J, the Portuguese Group for the Study of Streptococcal Infections. 2007. Analysis of group B streptococcal isolates from infants and pregnant women in Portugal revealing two lineages with enhanced invasiveness. J. Clin. Microbiol. 45:3224–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Creti R, Fabretti F, Orefici G, von Hunolstein C. 2004. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J. Clin. Microbiol. 42:1326–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martins ER, Melo-Cristino J, Ramirez M. 2010. Evidence for rare capsular switching in Streptococcus agalactiae. J. Bacteriol. 192:1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carrico JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, Almeida JS, Ramirez M. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Severiano A, Pinto FR, Ramirez M, Carrico JA. 2011. Adjusted Wallace coefficient as a measure of congruence between typing methods. J. Clin. Microbiol. 49:3997–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brochet M, Rusniok C, CouvÉ E, Dramsi S, Poyart C, Trieu-Cuot P, Kunst F, Glaser P. 2008. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 105:15961–15966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Francisco AP, Bugalho M, Ramirez M, Carriço JA. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carriço JA. 2012. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sorensen UB, Poulsen K, Ghezzo C, Margarit I, Kilian M. 2010. Emergence and global dissemination of host-specific Streptococcus agalactiae clones. mBio 1(3):e00178–10 doi:10.1128/mBio.00178-10 24 [DOI] [PMC free article] [PubMed] [Google Scholar]