Abstract
Actinobacillus pleuropneumoniae is the etiologic agent of porcine pleuropneumonia, which causes serious economic losses in the pig farming industry worldwide. Due to a lack of knowledge of its virulence factors and a lack of effective vaccines able to confer cross-serotype protection, it is difficult to place this disease under control. By analyzing its genome sequences, we found that type IV fimbrial subunit protein ApfA is highly conserved among different serotypes of A. pleuropneumoniae. Our study shows that ApfA is an adhesin since its expression was greatly upregulated (135-fold) upon contact with host cells, while its deletion mutant attenuated its capability of adhesion. The inactivation of apfA dramatically reduced the ability of A. pleuropneumoniae to colonize mouse lung, suggesting that apfA is a virulence factor. Purified recombinant ApfA elicited an elevated humoral immune response and conferred robust protection against challenges with A. pleuropneumoniae serovar 1 strain 4074 and serovar 7 strain WF83 in mice. Importantly, the anti-ApfA serum conferred significant protection against both serovar 1 and serovar 7 in mice. These studies indicate that ApfA promotes virulence through attachment to host cells, and its immunogenicity renders it a promising novel subunit vaccine candidate against infection with A. pleuropneumoniae.
INTRODUCTION
Actinobacillus pleuropneumoniae is a Gram-negative bacterium that causes porcine pleuropneumonia, a highly infectious fatal respiratory disease of pigs (1). The A. pleuropneumoniae organism is transmitted via respiratory droplets or through direct contact. It colonizes the epithelial cells of the lower respiratory tract of pigs (1, 2). The infected animals develop disease symptoms such as chronic necrotizing pneumonia, acute fibrinous pneumonia, and pleuritis with high mortality, which leads to large economic losses in the pig farming industry worldwide (3).
A total of 15 serotypes of A. pleuropneumoniae have been identified on the basis of their capsular and lipopolysaccharide (LPS) antigens (3). Although their virulence and regional prevalence vary, all the serotypes cause disease, and with the growing emergence of antibiotic resistance and the rising concern about food safety, vaccination to prevent A. pleuropneumoniae infections is of increasing relevance (2, 4). However, the diverse genetic makeup of A. pleuropneumoniae poses difficulties for developing a universally effective vaccine against this organism. The first commercial vaccines were killed bacterins, which are able to reduce the mortality caused by A. pleuropneumoniae infection but often fail to prevent severe morbidity (5). Bacterin-induced immunity is serotype specific; thus, protection can be achieved only for the serotypes which are contained in the vaccine (6). In contrast, natural or experimental infection with A. pleuropneumoniae generally elicits protection against homologous and heterologous serotype infections in pigs (7). The limited protection elicited by bacterins may be due to the absence of secreted or in vivo-induced immunogenic proteins or to alteration of the antigenic potency of certain bacterial antigens by the inactivation treatment (4). Therefore, developing vaccines by targeting the immunogenic proteins which provide cross-serovar protection may be an appropriate vaccine development direction for A. pleuropneumoniae.
Some key virulence factors of A. pleuropneumoniae, such as the ApxA exotoxins, the outer membrane proteins (OMP), and iron-acquisition factors, have previously been tested as subunit vaccines (4). Among these subunit vaccine candidates, the Apx exotoxins are virulence factors that play a predominant role in the pathogenesis of A. pleuropneumoniae. Four different Apx exotoxins (ApxI, ApxII, ApxIII, and ApxIV) have been found to be secreted by A. pleuropneumoniae. The ApxI, ApxII, and ApxIII toxins, which are secreted in different combinations by different serotypes, are strongly immunogenic and induce protective immunity (4). Since each of the Apx exotoxins confers only partial protection against porcine pleuropneumonia and the distribution of Apx exotoxins varies among the different serotypes, the current commercial vaccine (Porcilis APP, Intervet, Holland) is composed of three toxoids—ApxI, ApxII, and ApxIII—and one 42-kDa outer membrane protein. This subunit vaccine is effective at preventing acute disease, but it neither protects effectively against colonization nor confers cross-serotype protection (8).
Therefore, identifying novel and conserved antigens is crucial for vaccine development. Recent advances in genomic technologies and bioinformatics make it possible to perform global profiling of conserved outer membrane proteins (9). Analyzing the available 12 genomic sequences of different serotype A. pleuropneumoniae strains (10–12), we found that type IV fimbrial subunit protein ApfA was present in all these strains. Importantly, ApfA is highly conserved among these serotypes. Fimbriae are common mediators of bacterial adhesion to host mucosal epithelial cells. They make an important contribution to the pathogenesis of Gram-negative bacteria such as enteropathogenic and enterohemorrhagic Escherichia coli, Salmonella enterica serovar Typhi, Pseudomonas aeruginosa, Legionella pneumophila, Neisseria gonorrhoeae, Neisseria meningitidis, and Vibrio cholerae (13). Fimbriae are involved in adhesion and colonization, biofilm formation, twitching motility, protein export, and DNA uptake (13). We wondered whether ApfA also has such a function. Furthermore, fimbriae are usually highly immunogenic: fimbrial antigens have been used successfully as subunit vaccine candidates against bacterial pathogens, including Moraxella bovis, Dichelobacter nodosus, and enterotoxigenic Escherichia coli (ETEC) (14–16). Hence, the potential of ApfA as a vaccine candidate for preventing infection from different A. pleuropneumoniae serotypes is worthy of further investigation.
In this study, we investigated the ability of ApfA to adhere to host cells and its involvement in colonizing the lower respiratory tract. ApfA was further tested in active and passive immunizations to determine whether it could confer cross-serotype protection against both serovar 1 and serovar 7 infections, which are the most prevalent serotypes in China (11).
MATERIALS AND METHODS
Strains and media.
The bacterial strains used in this study are listed in Table 1. A. pleuropneumoniae 4074 was used for isolation of genomic DNA. A. pleuropneumoniae 4074 (serovar 1) and WF83 (serovar 7) strains were used for challenge experiments. All A. pleuropneumoniae strains were grown in tryptic soy broth (TSB) or on tryptic soy agar (TSA) (Difco Laboratories, Detroit, MI) supplemented with 10% (vol/vol) filtered cattle serum and 10 μg/ml of NAD when necessary. For screening A. pleuropneumoniae apfA mutant, 5 μg/ml chloramphenicol was added to the above media. For the selection of complementation strains, 25 μg/ml gentamicin was added to the same media.
Table 1.
Characteristics of bacterial strains and primers used in this study
| Strain, plasmid, or primer | Relevant characteristics or sequence | Source or reference |
|---|---|---|
| Strains | ||
| 4074 | A. pleuropneumoniae reference strain of serovar 1, apfA positive | P. Blackall |
| WF83 | A. pleuropneumoniae reference strain of serovar 7, apfA positive | P. Blackall |
| 4074ΔapfA | A. pleuropneumoniae 4074 apfA mutation strain | This work |
| C4074ΔapfA | A. pleuropneumoniae apfA complementary strain | This work |
| Plasmids | ||
| pET-30a(+) | Expression vector, kanamycin resistant | Novagen |
| pJN105-sodC | Shuttle vector pJN105 with a sodC promoter, gentamicin resistant | 17 |
| pEMOC2 | Suicide vector, chloramphenicol resistant | 18 |
| Primers | ||
| apfA01 | 5′-CGTACGGTCGACGAATTGATGATCGTGATTGC-3′ | This work |
| apfA02 | 5′-CGCTAGGCGGCCGCTAAGTCCTCCCCTTTACATT-3′ | This work |
| apfA03 | 5′- CGTACGGAATTCATGCAAAAACTAAGTCTTATTC-3′ | This work |
| apfA04 | 5′- CGCTAGTCTAGATTAATTTGATGCGCAGAAATT-3′ | This work |
| apfA05 | 5′-CGTACGGAATTCTTTACTTTAATTGAATTGATGAT-3′ | This work |
| apfA06 | 5′-CGCTAGGTCGACTTAATTTGATGCGCAGAAATT-3′ | This work |
| apfA-S | 5′-CGAGATCTGCATATATAACACCG-3′ | This work |
| apfA-A | 5′-CAGAAATTTGCCGGAAATAAAC-3′ | This work |
| apfB-S | 5′-TTCGGGAAGACCAACAAACA-3′ | This work |
| apfB-A | 5′-TAAACAGGCCGTCCAACACT-3′ | This work |
| apfC-S | 5′-AACAAAGCGGGCGATTAGG-3′ | This work |
| apfC-A | 5′-GGCACGATAAAAAGGAGCATG-3′ | This work |
| ackA-S | 5′-GCGGAAGATAACTACGAGGACG-3′ | This work |
| ackA-A | 5′-AATGTTGAGTCGTCCGCAGTG-3′ | This work |
qRT-PCR.
Quantitative real-time PCR (qRT-PCR) assays were performed to compare the expression of type IV fimbriae cluster genes apfA, apfB, and apfC of A. pleuropneumoniae in contact or without contact with St. Jude porcine lung (SJPL) cells (kindly donated by Robert G. Webster, St. Jude Children's Research Hospital). This cell line has been used as a model of porcine lung epidermal cells (19, 20), although recent findings indicated that the SJPL cell line is not of porcine origin (21). The protocol used for A. pleuropneumoniae cultured alone or cocultured with SJPL cells was as previously described (22). Three biological samples were included. The total RNA was extracted using an SV total RNA isolation system (Promega, WI). The cDNAs were synthesized using the reverse transcriptase XL and random primer (TaKaRa, Dalian, China). Each cDNA sample was used as a template for a qRT-PCR, and the amplification mixture contained SYBR green (TaKaRa) and primers of apfA (apfA-S/-A), apfB (apfB-S/-A), apfC (apfC-S/-A), or the internal control ackA (ackA-S/-A) (23) (Table 1). Amplification and detection were performed using an ABI PRISM 7500 sequence detection system, and all reactions were done in triplicate. For each run, to normalize the amount of sample cDNA, the threshold cycle (CT) value of the endogenous control ackA gene was subtracted from the CT value of the target gene (ΔCT = CT target gene − CT ackA). To compare the target gene expression levels between the bacterial organisms that had contacted with cell lines and those had not contacted with cell lines, the following formula was used: ΔΔCT = ΔCT with cell contact − ΔCT without cell contact. The fold changes were expressed as 2−ΔΔCT. Data represent means ± standard deviations (SD) of the results of triplicate reactions for each gene transcript.
Construction of apfA mutant and complementary strains.
The strategy used for inactivation of apfA in A. pleuropneumoniae was described previously (24). A 360-bp DNA fragment of apfA (encoding amino acid residues 5 to 124) was amplified from the genomic DNA of A. pleuropneumoniae strain 4074 with a pair of primers of apfA01 and apfA02 (Table 1). The resultant PCR product was cloned into suicide plasmid pEMOC2 using SalI and NotI, creating an insertional plasmid, which was then electroporated into wild-type (wt) A. pleuropneumoniae 4074. The recombinants were selected on TSA plates containing 5 μg/ml chloramphenicol. The resultant 4074ΔapfA mutant strain was verified to have the plasmid inserted into the apfA locus by PCR and DNA sequencing analyses. To construct the complementary strain, the full-length apfA with its signal peptide sequence was amplified with primers apfA03 and apfA04 (Table 1). The PCR product was cloned between the EcoRI and the XbaI sites of the shuttle vector pJN105-sodC (17). The recombinant plasmid was confirmed by DNA sequencing and then electroporated into the 4074ΔapfA strain, and the transformants were selected on TSA plates containing gentamicin and chloramphenicol. The complementary strain was confirmed by PCR and reverse transcription-PCR analyses, and this complementary strain was termed C4074ΔapfA.
Cell adhesion and inhibition assays.
The SJPL cell line and porcine iliac artery endothelial cell line (PIEC cells; Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, China) were used to test the ability of adhesion of A. pleuropneumoniae. To quantify the adhesion of the different strains to both cell lines, 2.5 × 105 cells were seeded to wells of 24-well tissue culture plates (Corning, Inc., Corning, NY) and incubated overnight. The three bacterial strains, wt strain 4074, mutant strain 4074ΔapfA, and complementary strain C4074ΔapfA of A. pleuropneumoniae, were resuspended in the adequate cell culture medium to a bacterial density of 2.5 × 106 CFU/ml. A 1-ml volume of this suspension was added to each well at a multiplicity of infection (MOI) of 10:1, and the plates were incubated for 2 h. Nonadherent bacteria were removed by washing four times with sterile Dulbecco's phosphate-buffered saline (DPBS). Cells with adherent bacteria were released from the wells by adding 100 μl of 0.25% trypsin–EDTA (Gibco, Grand Island, NY) and suspended in 900 μl DPBS. Serial dilutions were performed by plating diluted cells on TSA plates to determine the number of bacteria that adhered to the host cells.
In an adhesion inhibition assay, before A. pleuropneumoniae 4074 was added, host cells were cultured with 100 μg/ml of purified recombinant ApfA (rApfA) or bovine serum albumin (BSA) for 2 h at 37°C in 5% CO2. After incubation with rApfA, the host cells were washed three times with sterile DPBS before the adhesion assay.
Confocal microscopic observation of adhesion.
To facilitate microscopic observation, SJPL cell monolayers were prepared by growing cells on sterile glass coverslips in 24-well tissue culture plates (Corning). Meanwhile, bacteria were labeled with carboxyfluorescein diacetate succinimidyl ester (CFDA SE) according to the manufacturer's instructions (CFDA SE cell proliferation assay and tracking kit; Beyotime Institute of Biotechnology, Hangzhou, China). CFDA SE passively diffuses into cells. Its acetate groups are cleaved by intracellular esterases to yield highly fluorescent carboxyfluorescein succinimidyl ester that binds covalently to proteins, forming green fluorescent conjugates. The labeled bacteria were then added to each well and incubated for 1 h. After that, slides were washed three times with DPBS to remove the nonadherent bacteria. The cell monolayers were fixed in 4% paraformaldehyde, and incubated for 10 min at room temperature. Afterward, the cells were permeabilized with 0.5% Triton X-100 for 5 min. Subsequently, cells were stained with the fluorescent dye 4′,6′diamidino-2-phenylindole (DAPI; Beyotime Institute of Biotechnology) for 4 min at room temperature. Finally, the cells were washed twice with DPBS and detected using a laser scanning confocal microscope (LSCM; Carl Zeiss, Germany).
In vivo colonization assay.
All the animal experiment protocols described in the study were approved by the Laboratory Animal Monitoring Committee of Huazhong Agricultural University and performed accordingly. After challenge, animals were monitored for 10 days, and lethal disease was recorded.
A total of six 6-week-old female BALB/c mice were randomly assigned to two groups of three each. The mice in each group were inoculated intraperitoneally (i.p.) with 2 × 106 CFU of wt 4074 or 4074ΔapfA. At 24 h postinfection, the intact lungs of three mice in each group were removed into 2 ml of DPBS and homogenized. Then, all samples were diluted and plated onto TSA plates to determine the numbers of the live bacteria by counting the bacterial CFU.
Cloning, expression, and purification of rApfA.
The apfA gene without the signal peptide sequence was amplified using primers apfA05 and apfA06 (Table 1). The PCR product, digested with EcoRI-SalI (TaKaRa), was cloned into the vector pET-30a (+) to obtain pET-30a-rApfA.
E. coli strain BL21 cells harboring the recombinant plasmid pET-30a-rApfA were cultivated to the mid-log phase at 37°C, and then a final concentration of 0.8 mM isopropyl-β-d-thiogalactoside (IPTG) was added to the medium. After 3 h of induction, the cells were harvested by centrifugation and resuspended in sterile DPBS. Cells were sonicated on ice, and the recombinant protein was purified from inclusion bodies by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography, as described in the QIAexpress manual (Qiagen, Valencia, CA). The purity and quantity of the rApfA were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and the Bradford method, respectively. The purified protein was stored at −20°C.
ELISA.
An enzyme-linked immunosorbent assay (ELISA) was performed to test IgG titers as described previously (25). Microtiter plates were coated with 250 ng/100 μl of purified rApfA at 4°C overnight, and washed three times with washing buffer (PBS [pH 7.2] containing 0.05% Tween 20). The plates were then blocked with 150 μl of 1% BSA for 1 h at 37°C, and treated with washing buffer. Serially diluted mouse sera (starting at 1:100) were added and incubated for 30 min at 37°C. Detection of bound IgG was achieved by incubating plates with IgG-horseradish peroxidase (IgG-HRP) (Southern Biotech, Birmingham, AL) diluted 1:5,000 in washing buffer for 30 min at 37°C, followed by adding 100 μl of the activated substrate solution (sodium citrate buffer containing 1 mg/ml 3,3,5,5-tetramethylbenzidine and 0.03% H2O2). After incubation in dark for 10 min, the reaction was stopped by adding 50 μl of 0.25% hydrofluoric acid to each well. The plates were read with an ELISA reader at 630 nm. Antibody titers were determined as the reciprocal of the dilution of serum yielding 50% of the maximum optical density (OD) value above the background.
ELISA was also applied to test whether ApfA can raise strong antibody titers in A. pleuropneumoniae-infected piglets. Twenty convalescent-phase serum samples from piglets infected with A. pleuropneumoniae (pig farms in Hubei Province, China) were tested positive by an ApxIIA-ELISA kit (Wuhan Keqian Biological Products Co., Ltd., China). These samples were then subjected to rApfA-based ELISA. Ten serum samples from healthy piglets were used as the negative control.
Mouse immunization and challenge.
A total of 60 BALB/c mice (6-week-old females) were randomly assigned to three groups of 20 each. A 50-μg volume of purified rApfA protein was emulsified with an equal volume of Marcol 52 (Esso)-based adjuvant and was then administered to the first group of mice by i.p. injection. Subsequent booster immunization was given at day 14 post-primary immunization. The second group of mice immunized with commercial inactivated A. pleuropneumoniae bacterin (Swine Infections Pleuropneumonia Trivalent Vaccine [serovar 1, 2, and 7]; Wuhan Keqian Biological Products Co., Ltd.) was used as a positive control. The third group of mice was dosed with DPBS emulsified in the same oil adjuvant and was used as a negative control. At day 14 post-booster immunization, blood samples were drawn by tail vein bleeding and 10 mice in each group were i.p. challenged with 1.5 × 108 CFU of A. pleuropneumoniae WF83 (serovar 7) or 8 × 107 CFU of A. pleuropneumoniae 4074 (serovar 1).
Passive protection assays.
Passive immunization was performed as previously described (25). Three groups of 20 6-week-old female BALB/c mice were intravenously (i.v.) injected with 100 μl of anti-rApfA hyperimmune serum, anti-bacterin hyperimmune serum, and control serum from adjuvant immunized mice. At 24 h after immunization, 10 mice in each group were i.p. challenged with 1.5 × 108 CFU of A. pleuropneumoniae WF83 (serovar 7) or 8 × 107 CFU of A. pleuropneumoniae 4074 (serovar 1).
Statistical analysis.
Unless otherwise specified, data were presented as the mean ± SD. Comparisons between data sets were performed using the t test. The survival rate was analyzed with the log rank (Mantel-Cox) test. For all tests, statistical significance was defined at P < 0.05. The Prism software program (Graph-Pad Software, Inc.) was used for all statistical analyses.
RESULTS
The apfA gene is conserved among A. pleuropneumoniae serovars.
Ten genomes of A. pleuropneumoniae reference strains of serovars 1, 2, 3, 4, 6, 9, 10, 11, 12, and 13 were sequenced recently by our laboratory (11, 12), and serovars 5b and 7 are available in the NCBI database (10). We found that the apfA gene exists in all these A. pleuropneumoniae serotypes. These sequences were aligned via BLASTP, and the 12 A. pleuropneumoniae strains were found to share a high amino acid sequence identity (>98%) for ApfA protein (Table 2). Only in serovar 13 strain N273, ApfA has a shortened peptide (coverage = 44%, identity = 100%, compared with the serovar 1 sequence). This result suggests that AfpA is highly conserved among the different A. pleuropneumoniae serotypes.
Table 2.
Type IV fimbrial protein ApfA is conserved in A. pleuropneumoniae reference strains of different serovars
| Serovar | % identity of ApfA in serovar: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5b | 6 | 7 | 9 | 10 | 11 | 12 | 13 | |
| 1 | 100 | |||||||||||
| 2 | 100 | 100 | ||||||||||
| 3 | 100 | 100 | 100 | |||||||||
| 4 | 100 | 100 | 100 | 100 | ||||||||
| 5b | 99 | 99 | 99 | 99 | 100 | |||||||
| 6 | 100 | 100 | 100 | 100 | 99 | 100 | ||||||
| 7 | 100 | 100 | 100 | 100 | 99 | 100 | 100 | |||||
| 9 | 100 | 100 | 100 | 100 | 99 | 100 | 100 | 100 | ||||
| 10 | 100 | 100 | 100 | 100 | 99 | 100 | 100 | 100 | 100 | |||
| 11 | 100 | 100 | 100 | 100 | 99 | 100 | 100 | 100 | 100 | 100 | ||
| 12 | 99 | 99 | 99 | 99 | 99 | 99 | 99 | 99 | 99 | 99 | 100 | |
| 13a | 100 | 100 | 100 | 100 | 98 | 100 | 100 | 100 | 100 | 100 | 98 | 100 |
Coverage = 44%.
The expression of apfA is enhanced by contact with host cells.
Fimbriae are common mediators of bacterial adhesion to host mucosal epithelial cells, and the expression of fimbriae in A. pleuropneumoniae is rapidly lost upon subculture (26). Here, we examined whether the expression of apfA is involved in the interaction with the host following SJPL cell contact. A. pleuropneumoniae strain 4074 was incubated with SJPL cells for 2 h, and the expression of apfABC (the type IV fimbriae gene cluster) was determined via qRT-PCR. The results show that the transcription of apfA, apfB, and apfC was increased 135-fold (P < 0.01), 25-fold (P < 0.01), and 11-fold (P < 0.01), respectively (Fig. 1A), compared to control strain 4074 without contact with SJPL cells. The significantly enhanced expression of the apfABC cluster suggests that type IV fimbriae are deeply involved in the interaction between the bacteria and the host.
Fig 1.
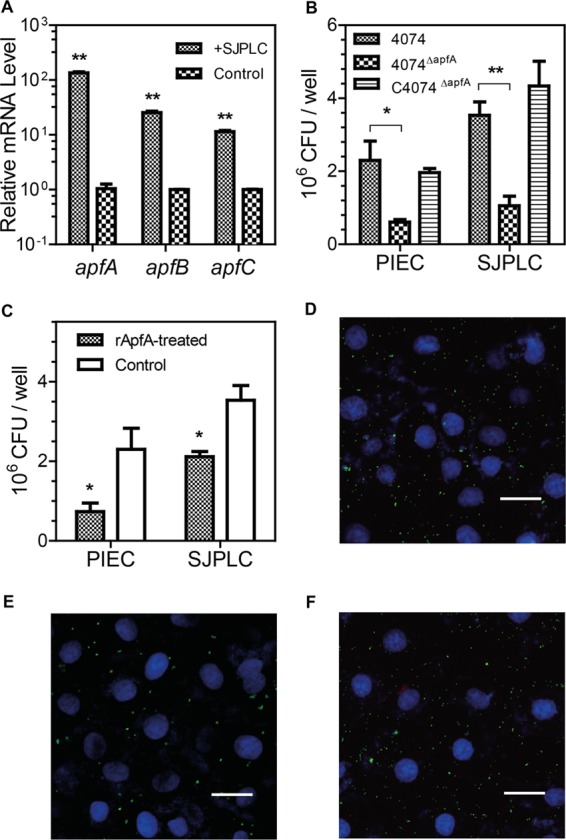
The expression of type IV fimbriae is induced upon contact with host cells, and ApfA mediates the adhesion of A. pleuropneumoniae to host cells. (A) The relative mRNA levels of the apfA, apfB, and apfC genes were upregulated after A. pleuropneumoniae 4074 was cocultured with SJPL cells (SJPLC), compared to those of bacteria without cell coculture. The mRNA level was determined by qRT-PCR. (B) Adhesion capabilities of wt strain 4074, mutant strain 4074ΔapfA, and complementary strain C4074ΔapfA to SJPL and PIEC cells. (C) Blockage of A. pleuropneumoniae adhesion to SJPL and PIEC cells by rApfA. SJPL and PIEC cells were pretreated with rApfA (rApfA-treated) or BSA (Control) before being subjected to contact with bacteria. The data shown are means ± SD of data determined with triplicate samples of one experiment representative of three independent experiments. The asterisks show significant differences (*, P < 0.05; **, P < 0.01). Confocal microscopy showed the adhesion of wt strain 4074 (D), mutant strain 4074ΔapfA (E), and complementary strain C4074ΔapfA (F) to SJPL cells. A. pleuropneumoniae cells were labeled with CFDA (green fluorescence). SJPL cell nuclei were stained with DAPI (blue fluorescence) (scale bars = 50 μm).
Type IV fimbriae are critical to mediating the adhesion of A. pleuropneumoniae to host cells.
Although previous reports on A. pleuropneumoniae suggested that fimbriae are involved in adhesion (2, 26, 27), no direct evidence had yet supported this hypothesis. To determine whether A. pleuropneumoniae ApfA has a role in adhesion, the apfA mutant strain (4074ΔapfA) was examined for adhesion using the complementary strain (C4074ΔapfA) and wt 4074 as controls.
The adhesion capabilities of wt strain 4074, mutant strain 4074ΔapfA, and complementary strain C4074ΔapfA to SJPL and PIEC cells were compared. The C4074ΔapfA strain exhibited a capability of adhering to SJPL and PIEC cells similar to that of wt strain 4074, since these two strains had similar numbers of CFU attached to cells per well (P > 0.05) (Fig. 1B). In contrast, the 4074ΔapfA mutant lost 70% (P < 0.01) of wt strain 4074's adhesion to SJPL and 73% (P < 0.05) of its adhesion to PIEC cells. To further determine the role of ApfA in the A. pleuropneumoniae adhesion process, a laser scanning confocal microscopic observation was conducted. As expected, mutant strain 4074ΔapfA (Fig. 1E) had significantly fewer bacteria adhering to SJPL cells than wt 4074 (Fig. 1D) or C4074ΔapfA (Fig. 1F).
To further investigate whether ApfA has a direct effect on the interaction of A. pleuropneumoniae with host cells, we conducted an adhesion inhibition assay (Fig. 1C). When SJPL and PIEC cells were pretreated with purified rApfA proteins, strain 4074 showed reduced adhesion to these eukaryotic cells, suggesting that rApfA proteins inhibit the adhesion capability of strain 4074. Taken together, these results indicate that type IV fimbriae are essential for mediating the adhesion of A. pleuropneumoniae to host cells.
Mutant strain 4074ΔapfA showed reduced colonization in lung tissue.
The colonization capabilities of wt strain 4074 and mutant strain 4074ΔapfA were compared using a mouse model (Fig. 2). Mice were inoculated with wt 4074 or mutant 4074ΔapfA. Live bacteria were recovered from lung tissues at 24 h after infection. The number of CFU recovered from mice infected with wt 4074 (1.45 ± 0.11 × 106 CFU/g lung) was significantly higher than that recovered from mice infected with mutant 4074ΔapfA (3.73 ± 0.36 × 105 CFU/g lung) (P < 0.01), which suggests that a lack of type IV fimbriae dampens the colonization capability of A. pleuropneumoniae in lung tissue.
Fig 2.
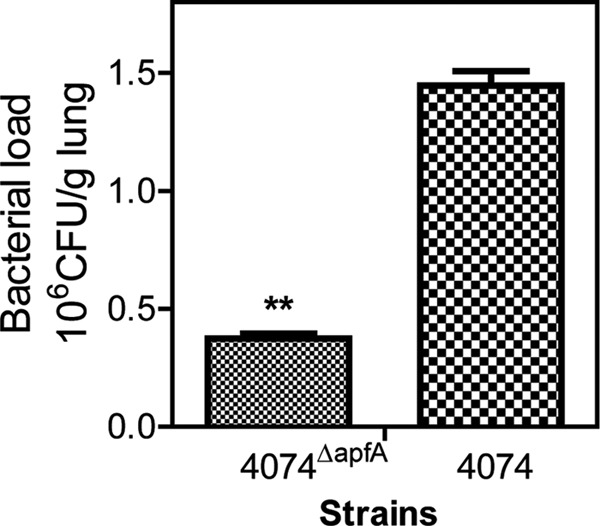
ApfA mediates the colonization of lung by A. pleuropneumoniae. The bacterial burdens in lungs of mice infected with wt strain 4074 or mutant strain 4074ΔapfA were evaluated at 24 h postinfection. The data shown are representative of the results of one of three independent experiments. The asterisks show significant differences (**, P < 0.01).
Piglets which have convalesced from A. pleuropneumoniae infection exhibit positive anti-ApfA sera.
The length without a signal peptide sequence of gene apfA is 408 bp, which encodes a protein of 136 amino acids with a predicted molecular mass of 15 kDa. This 408-bp apfA gene was fused to a His tag by cloning it to plasmid pET-30a (+). Then pET-30a-rApfA was transformed to E. coli (BL21) for rApfA synthesis. The rApfA proteins were purified via Ni-NTA affinity chromatography as previously described (25) (Fig. 3A).
Fig 3.
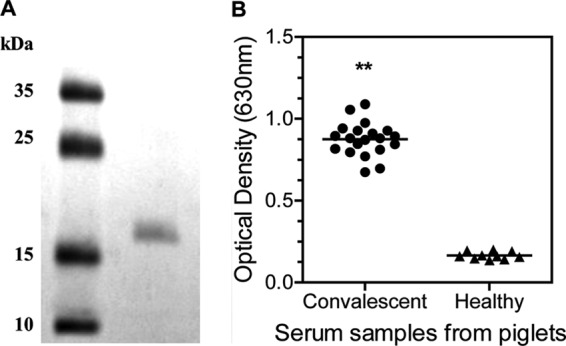
Naturally infected piglets show high anti-ApfA immune responses. (A) SDS-PAGE analysis of rApfA purified from E. coli cells. Left lane, protein marker; right lane, protein rApfA (2 μg). (B) Piglets infected with A. pleuropneumoniae show an elevated anti-ApfA immune response. The convalescent-phase sera are serum samples from pigs infected with A. pleuropneumoniae, and the negative sera are samples from healthy pigs. The anti-rApfA IgG level was determined using ELISA. The asterisks show significant differences (**, P < 0.01).
Since fimbriae are highly immunogenic and ApfA played an important role in the A. pleuropneumoniae-host cell interaction (Fig. 1), we further investigated whether ApfA serves as an immune target in natural infection in pigs. Pig convalescent-phase serum samples that tested positive and control serum samples that tested negative in an ApxIIA-ELISA kit (Wuhan Keqian Biological Products Co., Ltd.) were examined in a rApfA-based ELISA. All of the convalescent-phase sera showed significantly higher absorbance than the negative sera (P < 0.01) (Fig. 3B), which suggests that infecting the piglets with A. pleuropneumonia stimulated strong anti-ApfA immune responses, indicating that ApfA is a potent immunogen.
Immunization with rApfA confers protection against lethal infection with serovar 1 or 7.
We assessed whether rApfA could raise a protective immune response against A. pleuropneumoniae infection. Fourteen days after a booster immunization, serum samples were collected and the IgG antibodies raised by rApfA were determined using rApfA-based ELISA. Immunization with rApfA induced significantly higher levels of anti-rApfA titers than dosing with DPBS in mice (Fig. 4A, P < 0.01).
Fig 4.
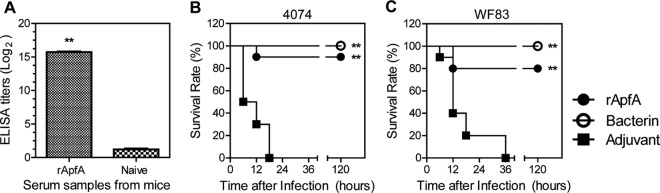
Immunization with rApfA protein protects mice against lethal challenges with strain 4074 (serovar 1) and strain WF83 (serovar 7). (A) A high level of specific anti-rApfA IgG antibody was induced in mice by immunization with rApfA. (B and C) Survival of mice immunized with rApfA, bacterin (positive control), and adjuvant (negative control) following i.p. infection with (B) strain 4074 (serovar 1) and (C) strain WF83 (serovar 7). The data shown are representative of the results of one of three independent experiments; the survival data were analyzed using the log rank (Mantel-Cox) test. The asterisks show significant differences (**, P < 0.01).
Next, a challenge study was performed to determine whether ApfA induces protective immunity in mice. In this experiment, commercial vaccine bacterin was included as a positive control. Mice were immunized with rApfA, bacterin, or DPBS. At 2 weeks post-booster immunization, they were challenged with a lethal dose (1.5 × 108 CFU of strain WF83 or 8 × 107 CFU of strain 4074) of A. pleuropneumoniae (Fig. 4B and C). The survival percentages of mice were recorded for 5 days after the challenge. All mice in the DPBS-dosed group developed anorexia and depression and died within 36 h of the challenge, while bacterin-immunized mice all survived. The rApfA vaccine showed a 90% protective efficacy against wt strain 4074 (serovar 1) in mice, which is significantly higher than that of DPBS (P < 0.01). For serovar 7 strain WF83, rApfA conferred 80% protection, which is also significantly higher than that of the DPBS control (P < 0.01). These results indicate that rApfA confers protection against infection with either serovar 1 or serovar 7, similar to commercial bacterin.
Passive immunization with anti-rApfA serum protects mice against lethal infection with serovar 1 or serovar 7.
To determine whether the protection was due to rApfA-stimulated humoral immunity, naïve mice were passively immunized with anti-rApfA serum via i.v. injection. The results showed that the anti-rApfA serum provided protection against both strain WF83 (serovar 7, 60%, P < 0.05) and strain 4074 (serovar 1, 40%, P < 0.01), while the antibacterin serum provided mice 80% protection against both strains (Fig. 5). In contrast, all mice administered a control serum from adjuvant-immunized mice died within 24 h postchallenge. These results indicate that the anti-rApfA serum could confer cross-serotype protection.
Fig 5.
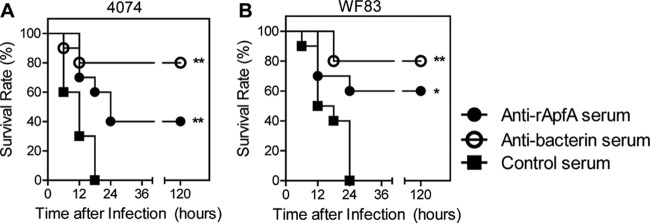
Serum from mice immunized with rApfA afforded passive protection against lethal challenges with strain 4074 (serovar 1) and strain WF83 (serovar 7). Serum against rApfA or bacterin significantly increased the survival rates after a challenge with (A) strain 4074 (serovar 1) or (B) strain WF83 (serovar 7), while control mice immunized with serum from adjuvant-immunized mice all succumbed to infection. The data shown shown are representative of the results of one of three independent experiments; the survival data were analyzed using the log rank (Mantel-Cox) test. The asterisks show significant differences (*, P < 0.05; **, P < 0.01).
DISCUSSION
The diversity among the 15 serotypes of A. pleuropneumoniae has made it difficult to develop vaccines that are effective against multiple serovars (4). The objective of this study was to find a universal virulence factor that can be used as an immune target so that cross-serotype protection can be achieved. Our results show that ApfA is an important adhesin to host cells and is also a promising candidate for a subunit vaccine that confers cross-serovar protection against A. pleuropneumoniae challenges.
The inactivation of apfA significantly reduced the adhesion capability of A. pleuropneumoniae to host cells, while the apfA complementary strain had this capability restored. This demonstrates that the type IV fimbriae of A. pleuropneumoniae are involved in the process of adhesion to host cells (Fig. 1B). We noticed that a few 4074ΔapfA cells still attached to SJPL cells. This observation implies that additional virulence factors may exist and be involved in the adhesion of A. pleuropneumoniae to alveolar epithelial cells. In fact, as an important initial step during infection, previous reports showed that factors such as oligosaccharide of LPS, 60-kDa outer membrane protein (OMP), 55-kDa OMP (28), and the Flp pilus (29) are all involved in the adhesion process. Since some adhesins are serotype specific or geographically distributed, diverse mechanisms may exist in different A. pleuropneumoniae serotypes. For instance, our previous study showed that the existence of Flp pili contributed to the adhesion capability of some A. pleuropneumoniae strains (29). However, serovar 9 and 11 strains which lack Flp pili could still adhere to host cells and display hypervirulence, indicating the possible existence of other adhesins. In contrast to the genetic diversity, ApfA was found to be highly conserved in different serotype strains. This indicates that ApfA is likely a universal adhesion factor of A. pleuropneumoniae.
Some adhesins have specific receptors on the surfaces of host cells to facilitate the bacterium-host interaction. For instance, a previous study found that transmembrane glycoprotein CD46, which is present on all human cells except erythrocytes and is involved in complement activation, is considered to be a pilus receptor for N. gonorrhoeae (30). In A. pleuropneumoniae, LPS showed specific patterns of binding to glycosphingolipids in respiratory epithelial cells (31). In this study, we found that when host cells were preincubated with rApfA, the adhesion of wt strain 4074 was significantly reduced (Fig. 1C). This implies that ApfA has a specific host receptor or binding site on epithelial cells, although further evidence is needed to verify this hypothesis.
Generally, the expression of bacterial surface appendages is regulated precisely. Otherwise, formidable consequences would result. This has been illustrated by the overexpression of ETEC colonization factor antigen I (CFA/I) fimbriae (32) and flagella (33), which resulted in a dramatic attenuation of wt Salmonella. The expression of flagella, for example, has the potential to turn on to provide a great advantage in the early stage of infection (34) and then turn off to minimize host recognition once infection is established (35). Similarly, we found that the expression of type IV fimbriae is accurately regulated. Gene apfA was upregulated when A. pleuropneumoniae bacilli came in contact with epithelial cells (Fig. 1A), suggesting that A. pleuropneumoniae also has a delicate regulation mechanism governing the expression of apfA. This result is in accordance with a previous study, which showed that the type IV fimbrial promoter is induced upon contact with host cells via the luxAB reporter system (22). Besides, two recent studies that looked at A. pleuropneumoniae gene expression during natural and experimental infections in pigs also indicated the upregulation of genes coding for type IV fimbriae (36, 37).
Colonization, the ability of a pathogen to adhere to host cells and multiply within the host, is often a prerequisite for the development of disease (1). To further verify the contribution ApfA makes to A. pleuropneumoniae colonization of the lower respiratory tract, we examined lung samples from mice infected with wt 4074 and mutant 4074ΔapfA at 24 h postinfection (Fig. 2). The results showed that mutant strain 4074ΔapfA is less capable of colonizing lung than wt 4074, suggesting that AfpA is a critical colonization factor. In addition, serum samples from convalescent and healthy pigs were tested with a rApfA-based ELISA. The level of the specific anti-rApfA IgG antibody was significantly higher in convalescent-phase sera than in the sera of healthy pigs, which confirms the high immunogenicity of ApfA. Thus, ApfA has the potential to be developed as a subunit vaccine against A. pleuropneumoniae. Considering that antigenic variation of the pilin subunit can be an important obstacle to the development of an effective pilus-based vaccine for cross-protection, as in Neisseria gonorrhoeae (38), the conserved ApfA of A. pleuropneumoniae has the potential to be developed as a cross-serovar protective vaccine.
Our results demonstrate that the vaccination of mice with rApfA provides 80% to 90% protection against lethal challenges with two distinct A. pleuropneumoniae serotype strains. This shows that ApfA is a valuable candidate for a subunit vaccine and deserves further evaluation in pigs. Currently, the single-antigen vaccines that have been tested in pigs are not desirable. For instance, although immunization with recombinant ApxIIA or TbpB proteins induced a strong humoral immune response in pigs, and a significant increase in survival was achieved in pigs challenged with the A. pleuropneumoniae serovar 7 strain, these immunized pigs were not protected against an A. pleuropneumoniae serovar 1 challenge (39). In another study, rApxIVA, which exists in all A. pleuropneumoniae strains and is expressed only in vivo, was used for vaccination. Vaccinated pigs challenged with either serovar 1 or serovar 2 showed severe respiratory symptoms and lung lesions, similar to those of the animals in the control group, despite the highly homologous sequence in different serotypes and high antibody titers against rApxIVA (40).
Since single antigens often provide only partial protection against A. pleuropneumoniae infection, multiantigen subunit vaccines may have enhanced protective efficacy. For instance, pigs receiving a mixture of ApxIIA and TbpB proteins had a tendency to recover faster than those that were vaccinated with only one protein (39). Thus, we anticipate that rApfA will improve protection upon its addition to a multiantigen subunit vaccine. A recent publication showed that the immunization of pigs with a combined hexa-antigen vaccine consisting of rApfA, rApxIA, rApxIIA, rApxIIIA, rApxIVA, and rTbpB proteins provided protective immunity against heterogeneous A. pleuropneumoniae serovar 9 infection at a significantly higher level than that acquired upon vaccination with the penta-antigen subunit vaccine without rApfA (41). Thus, the addition of rApfA may improve the protective efficacy of the currently used vaccines. However, adding rApfA coupled with rApxIV to a tetra-antigen vaccine did not improve its protective efficacy against a high-dose intranasal challenge with A. pleuropneumoniae serovar 1 or 2 in mice (42). So far, no protective efficacy in mice is available for ApxIV; it is difficult to estimate the contribution of rApfA to an elicited immune response because rApxIV is involved at the same time. Nevertheless, in agreement with a recent study showing that a DNA vaccine encoding ApfA conferred partial protection against serovar 2 infection in mice (43), our passive immunization shows that the anti-rApfA serum alone can provide significant protection against a lethal challenge with A. pleuropneumoniae serovar 1 or 7 in mice.
Despite high antibody titers against rApfA, the vaccination of mice with rApfA alone provided effective but not full protection against a lethal challenge with A. pleuropneumoniae. Unlike the antigenic variation in pili in other organisms (38), apfA was conserved in all the sequenced serotypes. Therefore, the observed partial protection of immunized animals against pleuropneumonia was not due to variation in pilin antigens. A more plausible explanation is that, although ApfA-specific antibodies markedly inhibit the pilus-mediated binding of A. pleuropneumoniae to alveolar epithelial cells, some additional components contribute to bacterial colonization. The efficiency of providing 80% to 90% protection by immunization with rApfA is considerable, indicating the importance of ApfA in virulence despite the existence of other adhesion mechanisms. In light of its performance in mice, our laboratory will test the protective efficacy of ApfA in pigs to determine whether it can serve as a useful vaccine for livestock.
In conclusion, we have shown that the type IV fimbria is not only important in the adhesion of A. pleuropneumoniae to host cells but is also an important virulence factor in colonization. Moreover, our results demonstrate that immunization with rApfA induced a mouse-specific serum antibody response that reduced the mortality of mice after different A. pleuropneumoniae serotype challenges. Thus, rApfA is a promising candidate for a vaccine to be included in the next generation of subunit vaccines that provide cross-serovar protection against porcine pleuropneumonia.
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program of China (973 Program, 2012CB518802), Hi-Tech R & D Program of China (863 Program, 2012AA101304), and National Natural Science Foundation of China (31070113).
Footnotes
Published ahead of print 26 December 2012
REFERENCES
- 1. Bossé JT, Janson H, Sheehan BJ, Beddek AJ, Rycroft AN, Kroll JS, Langford PR. 2002. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microb. Infect. 4:225–235 [DOI] [PubMed] [Google Scholar]
- 2. Dom P, Haesebrouck F, Ducatelle R, Charlier G. 1994. In vivo association of Actinobacillus pleuropneumoniae serotype 2 with the respiratory epithelium of pigs. Infect. Immun. 62:1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Inzana TJ, Champion A. 2007. Use of an inhibition enzyme-linked immunosorbent assay for quantification of capsular polysaccharide or proteins in vaccines. Clin. Vaccine Immunol. 14:323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramjeet M, Deslandes V, Goure J, Jacques M. 2008. Actinobacillus pleuropneumoniae vaccines: from bacterins to new insights into vaccination strategies. Anim. Health Res. Rev. 9:25–45 [DOI] [PubMed] [Google Scholar]
- 5. Higgins R, Larivière S, Mittal KR, Martineau GP, Rousseau P, Cameron J. 1985. Evaluation of a killed vaccine against porcine pleuropneumonia due to Haemophilus pleuropneumoniae. Can. Vet. J. 26:86–89 [PMC free article] [PubMed] [Google Scholar]
- 6. Nielsen R. 1985. Haemophilus pleuropneumoniae (Actinobacillus pleuropneumoniae) serotypes 8, 3 and 6. Serological response and cross immunity in pigs. Nord. Vet. Med. 37:217–227 [PubMed] [Google Scholar]
- 7. Haesebrouk F, Van de Kerkhof A, Dom P, Chiers K, Ducatelle R. 1996. Cross protection between Actinobacillus pleuropneumoniae biotypes-serotypes in pigs. Vet. Microbiol. 52:277–284 [DOI] [PubMed] [Google Scholar]
- 8. Tumamao JQ, Bowles RE, van den Bosch H, Klaasen HL, Fenwick BW, Blackall PJ. 2004. An evaluation of the role of antibodies to Actinobacillus pleuropneumoniae serovar 1 and 15 in the protection provided by sub-unit and live streptomycin dependent pleuropneumonia vaccines. Aust. Vet. J. 82:773–780 [DOI] [PubMed] [Google Scholar]
- 9. Chen Z, Peng B, Wang S, Peng X. 2004. Rapid screening of highly efficient vaccine candidates by immunoproteomics. Proteomics 4:3203–3213 [DOI] [PubMed] [Google Scholar]
- 10. Foote SJ, Bossé JT, Bouevitch AB, Langford PR, Young NM, Nash JH. 2008. The complete genome sequence of Actinobacillus pleuropneumoniae L20 (serovar 5b). J. Bacteriol. 190:1495–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu Z, Zhou Y, Li L, Zhou R, Xiao S, Wan Y, Zhang S, Wang K, Li W, Li L, Jin H, Kang M, Dalai B, Li T, Liu L, Cheng Y, Zhang L, Xu T, Zheng H, Pu S, Wang B, Gu W, Zhang XL, Zhu GF, Wang S, Zhao GP, Chen H. 2008. Genome biology of Actinobacillus pleuropneumoniae JL03, an isolate of serovar 3 prevalent in China. PLoS One 3:e1450 doi:10.1371/journal.pone.0001450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu Z, Chen X, Li L, Li T, Wang S, Chen H, Zhou R. 2010. Comparative genomic characterization of Actinobacillus pleuropneumoniae. J. Bacteriol. 192:5625–5636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2:363–378 [DOI] [PubMed] [Google Scholar]
- 14. Lepper AW, Atwell JL, Lehrbach PR, Schwartzkoff CL, Egerton JR, Tennent JM. 1995. The protective efficacy of cloned Moraxella bovis pili in monovalent and multivalent vaccine formulations against experimentally induced infectious bovine keratoconjunctivitis (IBK). Vet. Microbiol. 45:129–138 [DOI] [PubMed] [Google Scholar]
- 15. Walduck AK, Opdebeeck JP. 1996. Effect of adjuvants on antibody responses of sheep immunised with recombinant pili from Dichelobacter nodosus. Aust. Vet. J. 74:451–455 [DOI] [PubMed] [Google Scholar]
- 16. Yang X, Thornburg T, Holderness K, Suo Z, Cao L, Lim T, Avci R, Pascual DW. 2011. Serum antibodies protect against intraperitoneal challenge with enterotoxigenic Escherichia coli. J. Biomed. Biotechnol. 2011:632396 doi:10.1155/2011/632396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li L, Zhou R, Li T, Kang M, Wan Y, Xu Z, Chen H. 2008. Enhanced biofilm formation and reduced virulence of Actinobacillus pleuropneumoniae luxS mutant. Microb. Pathog. 45:192–200 [DOI] [PubMed] [Google Scholar]
- 18. Baltes N, Hennig-Pauka I, Gerlach GF. 2002. Both transferrin binding proteins are virulence factors in Actinobacillus pleuropneumoniae serotype 7 infection. FEMS Microbiol. Lett. 209:283–287 [DOI] [PubMed] [Google Scholar]
- 19. Seo SH, Goloubeva O, Webby R, Webster RG. 2001. Characterization of a porcine lung epithelial cell line suitable for influenza virus studies. J. Virol. 75:9517–9525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Auger E, Deslandes V, Ramjeet M, Contreras I, Nash JH, Harel J, Gottschalk M, Olivier M, Jacques M. 2009. Host-pathogen interactions of Actinobacillus pleuropneumoniae with porcine lung and tracheal epithelial cells. Infect. Immun. 77:1426–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Silversides DW, Music N, Jacques M, Gagnon CA, Webby R. 2010. Investigation of the species origin of the St. Jude porcine lung epithelial cell line (SJPL) made available to researchers. J. Virol. 84:5454–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boekema BK, van Putten JP, Stockhofe-Zurwieden N, Smith HE. 2004. Host cell contact-induced transcription of the type IV fimbria gene cluster of Actinobacillus pleuropneumoniae. Infect. Immun. 72:691–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deslandes V, Nash JH, Harel J, Coulton JW, Jacques M. 2007. Transcriptional profiling of Actinobacillus pleuropneumoniae under iron-restricted conditions. BMC Genomics 8:72 doi:10.1186/1471-2164-8-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones CH, Bolken TC, Jones KF, Zeller GO, Hruby DE. 2001. Conserved DegP protease in gram-positive bacteria is essential for thermal and oxidative tolerance and full virulence in Streptococcus pyogenes. Infect. Immun. 69:5538–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li J, Xia J, Tan C, Zhou Y, Wang Y, Zheng C, Chen H, Bei W. 2011. Evaluation of the immunogenicity and the protective efficacy of a novel identified immunogenicprotein, SsPepO, of Streptococcus suis serotype 2. Vaccine 29:6514–6519 [DOI] [PubMed] [Google Scholar]
- 26. Utrera V, Pijoan C. 1991. Fimbriae in Actinobacillus pleuropneumoniae strains isolated from pig respiratory tracts. Vet. Rec. 128:357–358 [DOI] [PubMed] [Google Scholar]
- 27. Van Overbeke I, Chiers K, Charlier G, Vandenberghe I, Van Beeumen J, Ducatelle R, Haesebrouck F. 2002. Characterization of the in vitro adhesion of Actinobacillus pleuropneumoniae to swine alveolar epithelial cells. Vet. Microbiol. 88:59–74 [DOI] [PubMed] [Google Scholar]
- 28. Chiers K, De Waele T, Pasmans F, Ducatelle R, Haesebrouck F. 2010. Virulence factors of Actinobacillus pleuropneumoniae involved in colonization, persistence and induction of lesions in its porcine host. Vet. Res. 41:65 doi:10.1051/vetres/2010037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li T, Xu Z, Zhang T, Li L, Chen H, Zhou R. 2012. The genetic analysis of the flp locus of Actinobacillus pleuropneumoniae. Arch. Microbiol. 194:167–176 [DOI] [PubMed] [Google Scholar]
- 30. Källström H, Liszewski MK, Atkinson JP, Jonsson AB. 1997. Membrane cofactor protein (MCP or CD46) is a cellular pilus receptor for pathogenic Neisseria. Mol. Microbiol. 25:639–647 [DOI] [PubMed] [Google Scholar]
- 31. Abul-Milh M, Paradis SE, Dubreuil JD, Jacques M. 1999. Binding of Actinobacillus lipopolysaccharides to glycosphingolipids evaluated by thin-layer chromatography. Infect. Immun. 67:4983–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang X, Suo Z, Thornburg T, Holderness K, Cao L, Lim T, Walters N, Kellerman L, Loetterle L, Avci R, Pascual DW. 2012. Expression of Escherichia coli virulence usher protein attenuates wild-type Salmonella. Virulence 3:29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang X, Thornburg T, Suo Z, Jun S, Robison A, Li J, Lim T, Cao L, Hoyt T, Avci R, Pascual DW. 2012. Flagella overexpression attenuates Salmonella pathogenesis. PLoS One 7:e46828 doi:10.1371/journal.pone.0046828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robertson JM, McKenzie NH, Duncan M, Allen-Vercoe E, Woodward MJ, Flint HJ, Grant G. 2003. Lack of flagella disadvantages Salmonella enterica serovar Enteritidis during the early stages of infection in the rat. J. Med. Microbiol. 52:91–99 [DOI] [PubMed] [Google Scholar]
- 35. Sano G, Takada Y, Goto S, Maruyama K, Shindo Y, Oka K, Matsui H, Matsuo K. 2007. Flagella facilitate escape of Salmonella from oncotic macrophages. J. Bacteriol. 189:8224–8232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deslandes V, Denicourt M, Girard C, Harel J, Nash JH, Jacques M. 2010. Transcriptional profiling of Actinobacillus pleuropneumoniae during the acute phase of a natural infection in pigs. BMC Genomics 11:98 doi:10.1186/1471-2164-11-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klitgaard K, Friis C, Jensen TK, Angen Ø, Boye M. 2012. Transcriptional portrait of Actinobacillus pleuropneumoniae during acute disease–potential strategies for survival and persistence in the host. PLoS One 7:e35549 doi:10.1371/journal.pone.0035549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boslego JW, Tramont EC, Chung RC, McChesney DG, Ciak J, Sadoff JC, Piziak MV, Brown JD, Brinton CC, JR, Wood SW, Bryan JR. 1991. Efficacy trial of a parenteral gonococcal pilus vaccine in men. Vaccine 9:154–162 [DOI] [PubMed] [Google Scholar]
- 39. Rossi-Campos A, Anderson C, Gerlach GF, Klashinsky S, Potter AA, Willson PJ. 1992. Immunization of pigs against Actinobacillus pleuropneumoniae with two recombinant protein preparations. Vaccine 10:512–518 [DOI] [PubMed] [Google Scholar]
- 40. Wang C, Wang Y, Shao M, Si W, Liu H, Chang Y, Peng W, Kong X, Liu S. 2009. Positive role for rApxIVN in the immune protection of pigs against infection by Actinobacillus pleuropneumoniae. Vaccine 27:5816–5821 [DOI] [PubMed] [Google Scholar]
- 41. Sadilkova L, Nepereny J, Vrzal V, Sebo P, Osicka R. 2012. Type IV fimbrial subunit protein ApfA contributes to protection against porcine pleuropneumonia. Vet. Res. 43:2 doi:10.1186/1297-9716-43-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shao M, Wang Y, Wang C, Guo Y, Peng Y, Liu J, Li G, Liu H, Liu S. 2010. Evaluation of multicomponent recombinant vaccines against Actinobacillus pleuropneumoniae in mice. Acta Vet. Scand. 52:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu YC, Li MC, Chen YM, Chu CY, Lin SF, Yang WJ. 2011. DNA vaccine encoding type IV pilin of Actinobacillus pleuropneumoniae induces strong immune response but confers limited protective efficacy against serotype 2 challenge. Vaccine 29:7740–7746 [DOI] [PubMed] [Google Scholar]


